

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0062.3
(108 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
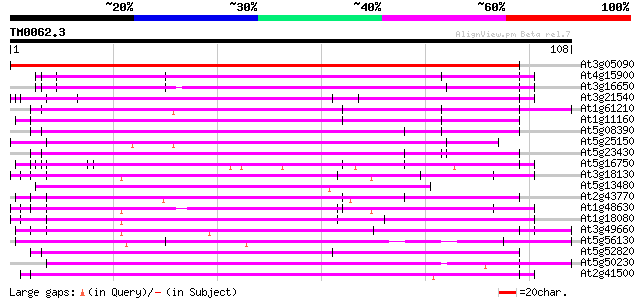
Score E
Sequences producing significant alignments: (bits) Value
At3g05090 unknown protein 183 1e-47
At4g15900 PRL1 protein 64 1e-11
At3g16650 PP1/PP2A phosphatases pleiotropic regulator PRL2 64 2e-11
At3g21540 WD repeat like protein 63 3e-11
At1g61210 60 1e-10
At1g11160 hypothetical protein 60 1e-10
At5g08390 katanin p80 subunit - like protein 60 2e-10
At5g25150 transcription initiation factor IID-associated factor-... 60 2e-10
At5g23430 unknown protein 60 2e-10
At5g16750 WD40-repeat protein 57 2e-09
At3g18130 protein kinase C-receptor/G-protein, putative 55 8e-09
At5g13480 putative protein 54 1e-08
At2g43770 putative splicing factor 54 1e-08
At1g48630 unknown protein 54 1e-08
At1g18080 putative guanine nucleotide-binding protein beta sub... 54 2e-08
At3g49660 putative WD-40 repeat - protein 53 2e-08
At5g56130 unknown protein (At5g56130) 53 3e-08
At5g52820 Notchless protein homolog 53 3e-08
At5g50230 putative protein 52 5e-08
At2g41500 putative U4/U6 small nuclear ribonucleoprotein 52 5e-08
>At3g05090 unknown protein
Length = 753
Score = 183 bits (465), Expect = 1e-47
Identities = 84/98 (85%), Positives = 93/98 (94%)
Query: 1 MNEGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIR 60
MN+ GT+LVSGGTEKV+RVWDPR+GSK+MKL+GHTDN+R LLLDSTGRFCLSGSSDSMIR
Sbjct: 221 MNDTGTMLVSGGTEKVLRVWDPRTGSKSMKLRGHTDNVRVLLLDSTGRFCLSGSSDSMIR 280
Query: 61 LWDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
LWDLGQQRCLH+YAVHTDSVWALA +FSHVYSGGRD
Sbjct: 281 LWDLGQQRCLHTYAVHTDSVWALACNPSFSHVYSGGRD 318
>At4g15900 PRL1 protein
Length = 486
Score = 63.9 bits (154), Expect = 1e-11
Identities = 26/89 (29%), Positives = 46/89 (51%)
Query: 10 SGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQRC 69
+G ++ +++WD +G + L GH + +R L + + + S D ++ WDL Q +
Sbjct: 193 TGSADRTIKIWDVATGVLKLTLTGHIEQVRGLAVSNRHTYMFSAGDDKQVKCWDLEQNKV 252
Query: 70 LHSYAVHTDSVWALASTSTFSHVYSGGRD 98
+ SY H V+ LA T + +GGRD
Sbjct: 253 IRSYHGHLSGVYCLALHPTLDVLLTGGRD 281
Score = 49.7 bits (117), Expect = 2e-07
Identities = 24/77 (31%), Positives = 42/77 (54%)
Query: 7 LLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQ 66
+L++GG + V RVWD R+ + L GH + + ++ T ++GS D+ I+ WDL
Sbjct: 274 VLLTGGRDSVCRVWDIRTKMQIFALSGHDNTVCSVFTRPTDPQVVTGSHDTTIKFWDLRY 333
Query: 67 QRCLHSYAVHTDSVWAL 83
+ + + H SV A+
Sbjct: 334 GKTMSTLTHHKKSVRAM 350
Score = 48.9 bits (115), Expect = 4e-07
Identities = 25/96 (26%), Positives = 45/96 (46%)
Query: 6 TLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLG 65
T + S G +K V+ WD GH + L L T L+G DS+ R+WD+
Sbjct: 231 TYMFSAGDDKQVKCWDLEQNKVIRSYHGHLSGVYCLALHPTLDVLLTGGRDSVCRVWDIR 290
Query: 66 QQRCLHSYAVHTDSVWALASTSTFSHVYSGGRDFSV 101
+ + + + H ++V ++ + T V +G D ++
Sbjct: 291 TKMQIFALSGHDNTVCSVFTRPTDPQVVTGSHDTTI 326
Score = 38.9 bits (89), Expect = 4e-04
Identities = 18/71 (25%), Positives = 38/71 (53%)
Query: 31 LKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQRCLHSYAVHTDSVWALASTSTFS 90
++GH +R++ D + + +GS+D I++WD+ + H + V LA ++ +
Sbjct: 172 IQGHLGWVRSVAFDPSNEWFCTGSADRTIKIWDVATGVLKLTLTGHIEQVRGLAVSNRHT 231
Query: 91 HVYSGGRDFSV 101
+++S G D V
Sbjct: 232 YMFSAGDDKQV 242
Score = 29.6 bits (65), Expect = 0.26
Identities = 14/50 (28%), Positives = 24/50 (48%)
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDS 57
+V+G + ++ WD R G L H ++RA+ L S S+D+
Sbjct: 317 VVTGSHDTTIKFWDLRYGKTMSTLTHHKKSVRAMTLHPKENAFASASADN 366
>At3g16650 PP1/PP2A phosphatases pleiotropic regulator PRL2
Length = 479
Score = 63.5 bits (153), Expect = 2e-11
Identities = 27/89 (30%), Positives = 45/89 (50%)
Query: 10 SGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQRC 69
+G ++ +++WD +G + L GH +R L + + + S D ++ WDL Q +
Sbjct: 187 TGSADRTIKIWDVATGVLKLTLTGHIGQVRGLAVSNRHTYMFSAGDDKQVKCWDLEQNKV 246
Query: 70 LHSYAVHTDSVWALASTSTFSHVYSGGRD 98
+ SY H V+ LA T V +GGRD
Sbjct: 247 IRSYHGHLHGVYCLALHPTLDVVLTGGRD 275
Score = 43.9 bits (102), Expect = 1e-05
Identities = 25/96 (26%), Positives = 42/96 (43%), Gaps = 1/96 (1%)
Query: 6 TLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLG 65
T + S G +K V+ WD GH + L L T L+G DS+ R+WD+
Sbjct: 225 TYMFSAGDDKQVKCWDLEQNKVIRSYHGHLHGVYCLALHPTLDVVLTGGRDSVCRVWDIR 284
Query: 66 QQRCLHSYAVHTDSVWALASTSTFSHVYSGGRDFSV 101
+ + H V+++ + T V +G D ++
Sbjct: 285 TKMQIF-VLPHDSDVFSVLARPTDPQVITGSHDSTI 319
Score = 43.9 bits (102), Expect = 1e-05
Identities = 24/78 (30%), Positives = 43/78 (54%), Gaps = 1/78 (1%)
Query: 7 LLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQ 66
++++GG + V RVWD R+ + L H ++ ++L T ++GS DS I+ WDL
Sbjct: 268 VVLTGGRDSVCRVWDIRTKMQIFVLP-HDSDVFSVLARPTDPQVITGSHDSTIKFWDLRY 326
Query: 67 QRCLHSYAVHTDSVWALA 84
+ + + H +V A+A
Sbjct: 327 GKSMATITNHKKTVRAMA 344
Score = 38.5 bits (88), Expect = 6e-04
Identities = 19/71 (26%), Positives = 37/71 (51%)
Query: 31 LKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQRCLHSYAVHTDSVWALASTSTFS 90
L+GH +R++ D + + +GS+D I++WD+ + H V LA ++ +
Sbjct: 166 LQGHLGWVRSVAFDPSNEWFCTGSADRTIKIWDVATGVLKLTLTGHIGQVRGLAVSNRHT 225
Query: 91 HVYSGGRDFSV 101
+++S G D V
Sbjct: 226 YMFSAGDDKQV 236
Score = 31.2 bits (69), Expect = 0.089
Identities = 16/77 (20%), Positives = 36/77 (45%)
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQ 67
+++G + ++ WD R G + H +RA+ L +S S+D++ + +
Sbjct: 310 VITGSHDSTIKFWDLRYGKSMATITNHKKTVRAMALHPKENDFVSASADNIKKFSLPKGE 369
Query: 68 RCLHSYAVHTDSVWALA 84
C + ++ D + A+A
Sbjct: 370 FCHNMLSLQRDIINAVA 386
>At3g21540 WD repeat like protein
Length = 949
Score = 62.8 bits (151), Expect = 3e-11
Identities = 29/97 (29%), Positives = 50/97 (50%)
Query: 2 NEGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRL 61
N+ G++L SG + + +WD S +L+GH D + L+ G+ +S S D +R+
Sbjct: 115 NKVGSMLASGSKDNDIILWDVVGESGLFRLRGHRDQVTDLVFLDGGKKLVSSSKDKFLRV 174
Query: 62 WDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
WDL Q C+ + H VW++ + +V +G D
Sbjct: 175 WDLETQHCMQIVSGHHSEVWSVDTDPEERYVVTGSAD 211
Score = 44.3 bits (103), Expect = 1e-05
Identities = 25/97 (25%), Positives = 40/97 (40%)
Query: 2 NEGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRL 61
+ +L+ G + +R+WD G+ + H + AL + G SGS D+ I L
Sbjct: 73 SSASSLVAVGYADGSIRIWDTEKGTCEVNFNSHKGAVTALRYNKVGSMLASGSKDNDIIL 132
Query: 62 WDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
WD+ + L H D V L + S +D
Sbjct: 133 WDVVGESGLFRLRGHRDQVTDLVFLDGGKKLVSSSKD 169
Score = 42.4 bits (98), Expect = 4e-05
Identities = 21/101 (20%), Positives = 45/101 (43%)
Query: 1 MNEGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIR 60
++ G L+V+G +K +++W G + H D++ + + S D +++
Sbjct: 583 ISSDGELIVTGSQDKNLKIWGLDFGDCHKSIFAHGDSVMGVKFVRNTHYLFSIGKDRLVK 642
Query: 61 LWDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRDFSV 101
WD + L + H +W LA ++ + +G D S+
Sbjct: 643 YWDADKFEHLLTLEGHHAEIWCLAISNRGDFLVTGSHDRSM 683
Score = 41.6 bits (96), Expect = 7e-05
Identities = 19/60 (31%), Positives = 32/60 (52%)
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQ 67
L S G +++V+ WD + L+GH I L + + G F ++GS D +R WD ++
Sbjct: 632 LFSIGKDRLVKYWDADKFEHLLTLEGHHAEIWCLAISNRGDFLVTGSHDRSMRRWDRSEE 691
Score = 38.9 bits (89), Expect = 4e-04
Identities = 22/85 (25%), Positives = 39/85 (45%)
Query: 14 EKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQRCLHSY 73
+ V+V+ S + L GH + + + S G ++GS D +++W L C S
Sbjct: 554 DSTVKVFYMDSLKFYLSLYGHKLPVMCIDISSDGELIVTGSQDKNLKIWGLDFGDCHKSI 613
Query: 74 AVHTDSVWALASTSTFSHVYSGGRD 98
H DSV + +++S G+D
Sbjct: 614 FAHGDSVMGVKFVRNTHYLFSIGKD 638
Score = 38.5 bits (88), Expect = 6e-04
Identities = 18/60 (30%), Positives = 32/60 (53%)
Query: 3 EGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLW 62
+GG LVS +K +RVWD + + GH + ++ D R+ ++GS+D +R +
Sbjct: 158 DGGKKLVSSSKDKFLRVWDLETQHCMQIVSGHHSEVWSVDTDPEERYVVTGSADQELRFY 217
Score = 29.3 bits (64), Expect = 0.34
Identities = 16/83 (19%), Positives = 39/83 (46%), Gaps = 3/83 (3%)
Query: 1 MNEGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIR 60
++E TLL+S +V ++W+P +GS + L+ ++ + G+ ++
Sbjct: 414 LSEDNTLLMSTSHSEV-KIWNPSTGSCLRTIDSGYG--LCSLIVPQNKYGIVGTKSGVLE 470
Query: 61 LWDLGQQRCLHSYAVHTDSVWAL 83
+ D+G + H ++W++
Sbjct: 471 IIDIGSATKVEEVKAHGGTIWSI 493
>At1g61210
Length = 282
Score = 60.5 bits (145), Expect = 1e-10
Identities = 27/92 (29%), Positives = 47/92 (50%)
Query: 7 LLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQ 66
L+++G + V+++WD GH N A+ G F SGSSD+ +++WD+ +
Sbjct: 102 LVLAGASSGVIKLWDVEEAKMVRAFTGHRSNCSAVEFHPFGEFLASGSSDANLKIWDIRK 161
Query: 67 QRCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
+ C+ +Y H+ + + T V SGG D
Sbjct: 162 KGCIQTYKGHSRGISTIRFTPDGRWVVSGGLD 193
Score = 53.5 bits (127), Expect = 2e-08
Identities = 23/79 (29%), Positives = 41/79 (51%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G L SG ++ +++WD R KGH+ I + GR+ +SG D+++++WDL
Sbjct: 142 GEFLASGSSDANLKIWDIRKKGCIQTYKGHSRGISTIRFTPDGRWVVSGGLDNVVKVWDL 201
Query: 65 GQQRCLHSYAVHTDSVWAL 83
+ LH + H + +L
Sbjct: 202 TAGKLLHEFKFHEGPIRSL 220
Score = 44.3 bits (103), Expect = 1e-05
Identities = 22/60 (36%), Positives = 32/60 (52%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G +VSGG + VV+VWD +G + K H IR+L +GS+D ++ WDL
Sbjct: 184 GRWVVSGGLDNVVKVWDLTAGKLLHEFKFHEGPIRSLDFHPLEFLLATGSADRTVKFWDL 243
Score = 42.0 bits (97), Expect = 5e-05
Identities = 26/112 (23%), Positives = 51/112 (45%), Gaps = 10/112 (8%)
Query: 7 LLVSGGTEKVVRVWDPRSGSKTMK----------LKGHTDNIRALLLDSTGRFCLSGSSD 56
L ++GG + V +W + MK L GHT + ++ DS L+G+S
Sbjct: 50 LFITGGDDYKVNLWAIGKPTSLMKNDAIAFYWQSLCGHTSAVDSVAFDSAEVLVLAGASS 109
Query: 57 SMIRLWDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRDFSVSAVFLKK 108
+I+LWD+ + + + ++ H + A+ + SG D ++ ++K
Sbjct: 110 GVIKLWDVEEAKMVRAFTGHRSNCSAVEFHPFGEFLASGSSDANLKIWDIRK 161
>At1g11160 hypothetical protein
Length = 974
Score = 60.5 bits (145), Expect = 1e-10
Identities = 29/97 (29%), Positives = 47/97 (47%)
Query: 2 NEGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRL 61
N L+++G + V+++WD GH N A+ G F SGSSD+ +R+
Sbjct: 16 NSEEVLVLAGASSGVIKLWDLEESKMVRAFTGHRSNCSAVEFHPFGEFLASGSSDTNLRV 75
Query: 62 WDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
WD ++ C+ +Y HT + + + V SGG D
Sbjct: 76 WDTRKKGCIQTYKGHTRGISTIEFSPDGRWVVSGGLD 112
Score = 57.4 bits (137), Expect = 1e-09
Identities = 26/79 (32%), Positives = 41/79 (50%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G L SG ++ +RVWD R KGHT I + GR+ +SG D+++++WDL
Sbjct: 61 GEFLASGSSDTNLRVWDTRKKGCIQTYKGHTRGISTIEFSPDGRWVVSGGLDNVVKVWDL 120
Query: 65 GQQRCLHSYAVHTDSVWAL 83
+ LH + H + +L
Sbjct: 121 TAGKLLHEFKCHEGPIRSL 139
Score = 44.3 bits (103), Expect = 1e-05
Identities = 22/60 (36%), Positives = 32/60 (52%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G +VSGG + VV+VWD +G + K H IR+L +GS+D ++ WDL
Sbjct: 103 GRWVVSGGLDNVVKVWDLTAGKLLHEFKCHEGPIRSLDFHPLEFLLATGSADRTVKFWDL 162
Score = 37.4 bits (85), Expect = 0.001
Identities = 19/70 (27%), Positives = 35/70 (49%)
Query: 29 MKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQRCLHSYAVHTDSVWALASTST 88
M L GHT + ++ +S L+G+S +I+LWDL + + + ++ H + A+
Sbjct: 1 MSLCGHTSPVDSVAFNSEEVLVLAGASSGVIKLWDLEESKMVRAFTGHRSNCSAVEFHPF 60
Query: 89 FSHVYSGGRD 98
+ SG D
Sbjct: 61 GEFLASGSSD 70
Score = 24.6 bits (52), Expect = 8.3
Identities = 11/52 (21%), Positives = 21/52 (40%)
Query: 7 LLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSM 58
LL +G ++ V+ WD + + +RA+ G+ G D +
Sbjct: 147 LLATGSADRTVKFWDLETFELIGTTRPEATGVRAIAFHPDGQTLFCGLDDGL 198
>At5g08390 katanin p80 subunit - like protein
Length = 823
Score = 60.1 bits (144), Expect = 2e-10
Identities = 28/92 (30%), Positives = 45/92 (48%)
Query: 7 LLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQ 66
L+ +G +++WD L GH N ++ G F SGS D+ +++WD+ +
Sbjct: 166 LVAAGAASGTIKLWDLEEAKVVRTLTGHRSNCVSVNFHPFGEFFASGSLDTNLKIWDIRK 225
Query: 67 QRCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
+ C+H+Y HT V L T + SGG D
Sbjct: 226 KGCIHTYKGHTRGVNVLRFTPDGRWIVSGGED 257
Score = 53.9 bits (128), Expect = 1e-08
Identities = 23/79 (29%), Positives = 39/79 (49%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G SG + +++WD R KGHT + L GR+ +SG D+++++WDL
Sbjct: 206 GEFFASGSLDTNLKIWDIRKKGCIHTYKGHTRGVNVLRFTPDGRWIVSGGEDNVVKVWDL 265
Query: 65 GQQRCLHSYAVHTDSVWAL 83
+ LH + H + +L
Sbjct: 266 TAGKLLHEFKSHEGKIQSL 284
Score = 46.2 bits (108), Expect = 3e-06
Identities = 25/79 (31%), Positives = 37/79 (46%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G +VSGG + VV+VWD +G + K H I++L +GS+D ++ WDL
Sbjct: 248 GRWIVSGGEDNVVKVWDLTAGKLLHEFKSHEGKIQSLDFHPHEFLLATGSADKTVKFWDL 307
Query: 65 GQQRCLHSYAVHTDSVWAL 83
+ S T V L
Sbjct: 308 ETFELIGSGGTETTGVRCL 326
Score = 41.2 bits (95), Expect = 9e-05
Identities = 18/70 (25%), Positives = 37/70 (52%)
Query: 7 LLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQ 66
+LV+GG + V +W + + L GH+ I ++ D++ +G++ I+LWDL +
Sbjct: 124 VLVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDASEGLVAAGAASGTIKLWDLEE 183
Query: 67 QRCLHSYAVH 76
+ + + H
Sbjct: 184 AKVVRTLTGH 193
>At5g25150 transcription initiation factor IID-associated
factor-like protein
Length = 700
Score = 59.7 bits (143), Expect = 2e-10
Identities = 25/77 (32%), Positives = 40/77 (51%)
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQ 67
+ +G ++K VR+WD ++G GH + +L + GR+ SG D I +WDL
Sbjct: 514 IATGSSDKTVRLWDVQTGECVRIFIGHRSMVLSLAMSPDGRYMASGDEDGTIMMWDLSTA 573
Query: 68 RCLHSYAVHTDSVWALA 84
RC+ H VW+L+
Sbjct: 574 RCITPLMGHNSCVWSLS 590
Score = 40.4 bits (93), Expect = 1e-04
Identities = 30/118 (25%), Positives = 49/118 (41%), Gaps = 25/118 (21%)
Query: 1 MNEGGTLLVSGGTEKVVRVWDP----RSGSKTMK--------------------LKGHTD 36
++ G+L+ G ++ ++VWD ++GS ++ L GH+
Sbjct: 360 ISHDGSLVAGGFSDSSIKVWDMAKIGQAGSGALQAENDSSDQSIGPNGRRSYTLLLGHSG 419
Query: 37 NIRALLLDSTGRFCLSGSSDSMIRLWDLGQQRCLHSYAVHTDSVWALASTSTFSHVYS 94
+ + G F LS S+D+ IRLW L Y H VW A S F H ++
Sbjct: 420 PVYSATFSPPGDFVLSSSADTTIRLWSTKLNANLVCYKGHNYPVWD-AQFSPFGHYFA 476
Score = 37.7 bits (86), Expect = 0.001
Identities = 22/97 (22%), Positives = 40/97 (40%), Gaps = 3/97 (3%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G S ++ R+W + GH ++ + +GSSD +RLWD+
Sbjct: 472 GHYFASCSHDRTARIWSMDRIQPLRIMAGHLSDVD---WHPNCNYIATGSSDKTVRLWDV 528
Query: 65 GQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRDFSV 101
C+ + H V +LA + ++ SG D ++
Sbjct: 529 QTGECVRIFIGHRSMVLSLAMSPDGRYMASGDEDGTI 565
Score = 33.5 bits (75), Expect = 0.018
Identities = 17/76 (22%), Positives = 31/76 (40%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G ++S + +R+W + + + KGH + G + S S D R+W +
Sbjct: 430 GDFVLSSSADTTIRLWSTKLNANLVCYKGHNYPVWDAQFSPFGHYFASCSHDRTARIWSM 489
Query: 65 GQQRCLHSYAVHTDSV 80
+ + L A H V
Sbjct: 490 DRIQPLRIMAGHLSDV 505
Score = 29.6 bits (65), Expect = 0.26
Identities = 14/40 (35%), Positives = 25/40 (62%), Gaps = 3/40 (7%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLK---GHTDNIRAL 41
G+LL SG + V++WD S +K K + G+++ +R+L
Sbjct: 631 GSLLASGSADCTVKLWDVTSSTKLTKAEEKNGNSNRLRSL 670
>At5g23430 unknown protein
Length = 837
Score = 59.7 bits (143), Expect = 2e-10
Identities = 29/92 (31%), Positives = 45/92 (48%)
Query: 7 LLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQ 66
L+ +G +++WD L GH N ++ G F SGS D+ +++WD+ +
Sbjct: 73 LVAAGAASGTIKLWDLEEAKIVRTLTGHRSNCISVDFHPFGEFFASGSLDTNLKIWDIRK 132
Query: 67 QRCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
+ C+H+Y HT V L T V SGG D
Sbjct: 133 KGCIHTYKGHTRGVNVLRFTPDGRWVVSGGED 164
Score = 50.1 bits (118), Expect = 2e-07
Identities = 22/79 (27%), Positives = 38/79 (47%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G SG + +++WD R KGHT + L GR+ +SG D+++++WDL
Sbjct: 113 GEFFASGSLDTNLKIWDIRKKGCIHTYKGHTRGVNVLRFTPDGRWVVSGGEDNIVKVWDL 172
Query: 65 GQQRCLHSYAVHTDSVWAL 83
+ L + H + +L
Sbjct: 173 TAGKLLTEFKSHEGQIQSL 191
Score = 44.7 bits (104), Expect = 8e-06
Identities = 24/80 (30%), Positives = 38/80 (47%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G +VSGG + +V+VWD +G + K H I++L +GS+D ++ WDL
Sbjct: 155 GRWVVSGGEDNIVKVWDLTAGKLLTEFKSHEGQIQSLDFHPHEFLLATGSADRTVKFWDL 214
Query: 65 GQQRCLHSYAVHTDSVWALA 84
+ S T V L+
Sbjct: 215 ETFELIGSGGPETAGVRCLS 234
Score = 40.8 bits (94), Expect = 1e-04
Identities = 18/70 (25%), Positives = 37/70 (52%)
Query: 7 LLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQ 66
+LV+GG + V +W + + L GH+ I ++ D++ +G++ I+LWDL +
Sbjct: 31 VLVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDASEVLVAAGAASGTIKLWDLEE 90
Query: 67 QRCLHSYAVH 76
+ + + H
Sbjct: 91 AKIVRTLTGH 100
>At5g16750 WD40-repeat protein
Length = 876
Score = 56.6 bits (135), Expect = 2e-09
Identities = 32/100 (32%), Positives = 52/100 (52%), Gaps = 6/100 (6%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLL--DSTGRFCLSGSSDSMIRLW 62
G LL + G ++ V VWD G T +GH + ++L DS +SGS D+ +R+W
Sbjct: 114 GGLLATAGADRKVLVWDVDGGFCTHYFRGHKGVVSSILFHPDSNKNILISGSDDATVRVW 173
Query: 63 DLG----QQRCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
DL +++CL H +V ++A + ++S GRD
Sbjct: 174 DLNAKNTEKKCLAIMEKHFSAVTSIALSEDGLTLFSAGRD 213
Score = 56.2 bits (134), Expect = 3e-09
Identities = 25/91 (27%), Positives = 46/91 (50%)
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQ 67
+++ +K V++W GS +GHT ++ + G +S +D +++LW++
Sbjct: 556 VMTASGDKTVKIWAISDGSCLKTFEGHTSSVLRASFITDGTQFVSCGADGLLKLWNVNTS 615
Query: 68 RCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
C+ +Y H D VWALA + +GG D
Sbjct: 616 ECIATYDQHEDKVWALAVGKKTEMIATGGGD 646
Score = 42.4 bits (98), Expect = 4e-05
Identities = 22/86 (25%), Positives = 38/86 (43%)
Query: 16 VVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQRCLHSYAV 75
V+ + D S ++G +D + AL L + S IR+WDL +C+ S+
Sbjct: 41 VINIVDSTDSSVKSTIEGESDTLTALALSPDDKLLFSAGHSRQIRVWDLETLKCIRSWKG 100
Query: 76 HTDSVWALASTSTFSHVYSGGRDFSV 101
H V +A ++ + + G D V
Sbjct: 101 HEGPVMGMACHASGGLLATAGADRKV 126
Score = 40.8 bits (94), Expect = 1e-04
Identities = 20/93 (21%), Positives = 42/93 (44%)
Query: 6 TLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLG 65
+L+ +G ++ +W + LKGH I ++ + + ++ S D +++W +
Sbjct: 512 SLVCTGSEDRTASIWRLPDLVHVVTLKGHKRRIFSVEFSTVDQCVMTASGDKTVKIWAIS 571
Query: 66 QQRCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
CL ++ HT SV + + + S G D
Sbjct: 572 DGSCLKTFEGHTSSVLRASFITDGTQFVSCGAD 604
Score = 39.3 bits (90), Expect = 3e-04
Identities = 18/70 (25%), Positives = 31/70 (43%)
Query: 7 LLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQ 66
LL S G + +RVWD + KGH + + ++G + +D + +WD+
Sbjct: 74 LLFSAGHSRQIRVWDLETLKCIRSWKGHEGPVMGMACHASGGLLATAGADRKVLVWDVDG 133
Query: 67 QRCLHSYAVH 76
C H + H
Sbjct: 134 GFCTHYFRGH 143
Score = 38.9 bits (89), Expect = 4e-04
Identities = 21/64 (32%), Positives = 34/64 (52%), Gaps = 1/64 (1%)
Query: 2 NEGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRAL-LLDSTGRFCLSGSSDSMIR 60
+ G L+V+G +K VR+W+ S S GH +I A+ + F +SGS D ++
Sbjct: 413 SSGNVLIVTGSKDKTVRLWNATSKSCIGVGTGHNGDILAVAFAKKSFSFFVSGSGDRTLK 472
Query: 61 LWDL 64
+W L
Sbjct: 473 VWSL 476
Score = 38.1 bits (87), Expect = 7e-04
Identities = 26/86 (30%), Positives = 44/86 (50%), Gaps = 4/86 (4%)
Query: 17 VRVWDPRSGSKTMKLKGHTDNIRAL--LLDSTGRFCL-SGSSDSMIRLWDLGQQRCLHSY 73
VRV+D + S + L GH + + +L + S+G + +GS D +RLW+ + C+
Sbjct: 383 VRVYDVATMSCSYVLAGHKEVVLSLDTCVSSSGNVLIVTGSKDKTVRLWNATSKSCIGVG 442
Query: 74 AVHTDSVWALA-STSTFSHVYSGGRD 98
H + A+A + +FS SG D
Sbjct: 443 TGHNGDILAVAFAKKSFSFFVSGSGD 468
Score = 33.5 bits (75), Expect = 0.018
Identities = 15/58 (25%), Positives = 27/58 (45%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLW 62
GT VS G + ++++W+ + H D + AL + +G D++I LW
Sbjct: 595 GTQFVSCGADGLLKLWNVNTSECIATYDQHEDKVWALAVGKKTEMIATGGGDAVINLW 652
Score = 26.2 bits (56), Expect = 2.9
Identities = 10/21 (47%), Positives = 14/21 (66%)
Query: 1 MNEGGTLLVSGGTEKVVRVWD 21
++E G L S G +KVV +WD
Sbjct: 200 LSEDGLTLFSAGRDKVVNLWD 220
>At3g18130 protein kinase C-receptor/G-protein, putative
Length = 326
Score = 54.7 bits (130), Expect = 8e-09
Identities = 29/104 (27%), Positives = 51/104 (48%), Gaps = 5/104 (4%)
Query: 3 EGGTLLVSGGTEKVVRVW-----DPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDS 57
+ ++V+ +K + +W D G +L GH+ + ++L S G+F LSGS D
Sbjct: 26 DNSDIIVTASRDKSIILWKLTKDDKSYGVAQRRLTGHSHFVEDVVLSSDGQFALSGSWDG 85
Query: 58 MIRLWDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRDFSV 101
+RLWDL + HT V ++A ++ + S RD ++
Sbjct: 86 ELRLWDLATGETTRRFVGHTKDVLSVAFSTDNRQIVSASRDRTI 129
Score = 47.0 bits (110), Expect = 2e-06
Identities = 24/95 (25%), Positives = 42/95 (43%), Gaps = 9/95 (9%)
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQ 67
+VS +K V+VW+ ++ L GH+ + + + G C SG D +I LWDL +
Sbjct: 166 IVSASWDKTVKVWNLQNCKLRNSLVGHSGYLNTVAVSPDGSLCASGGKDGVILLWDLAEG 225
Query: 68 R---------CLHSYAVHTDSVWALASTSTFSHVY 93
+ +HS + W A+T ++
Sbjct: 226 KKLYSLEAGSIIHSLCFSPNRYWLCAATENSIRIW 260
Score = 42.7 bits (99), Expect = 3e-05
Identities = 18/63 (28%), Positives = 34/63 (53%)
Query: 1 MNEGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIR 60
++ G +SG + +R+WD +G T + GHT ++ ++ + R +S S D I+
Sbjct: 71 LSSDGQFALSGSWDGELRLWDLATGETTRRFVGHTKDVLSVAFSTDNRQIVSASRDRTIK 130
Query: 61 LWD 63
LW+
Sbjct: 131 LWN 133
Score = 39.7 bits (91), Expect = 3e-04
Identities = 23/75 (30%), Positives = 37/75 (48%), Gaps = 2/75 (2%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G+L SGG + V+ +WD G K L+ +I L S R+ L ++++ IR+WDL
Sbjct: 205 GSLCASGGKDGVILLWDLAEGKKLYSLE--AGSIIHSLCFSPNRYWLCAATENSIRIWDL 262
Query: 65 GQQRCLHSYAVHTDS 79
+ + V S
Sbjct: 263 ESKSVVEDLKVDLKS 277
>At5g13480 putative protein
Length = 679
Score = 53.9 bits (128), Expect = 1e-08
Identities = 27/87 (31%), Positives = 48/87 (55%), Gaps = 11/87 (12%)
Query: 6 TLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIR----- 60
+LLVSGG +++V++WD RSG + L GH + + ++ + G + L+ S D +I+
Sbjct: 253 SLLVSGGKDQLVKLWDTRSGRELCSLHGHKNIVLSVKWNQNGNWLLTASKDQIIKHLYDT 312
Query: 61 ------LWDLGQQRCLHSYAVHTDSVW 81
L+D+ + L S+ HT +W
Sbjct: 313 YACNLQLYDIRTMKELQSFRGHTKDLW 339
Score = 32.3 bits (72), Expect = 0.040
Identities = 15/76 (19%), Positives = 32/76 (41%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G L++G +W+ +S + M L+ H IR+++ + +SG ++ W
Sbjct: 135 GRRLITGSQSGEFTLWNGQSFNFEMILQAHDQPIRSMVWSHNENYMVSGDDGGTLKYWQN 194
Query: 65 GQQRCLHSYAVHTDSV 80
+ H +S+
Sbjct: 195 NMNNVKANKTAHKESI 210
Score = 27.7 bits (60), Expect = 0.98
Identities = 15/66 (22%), Positives = 31/66 (46%)
Query: 33 GHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQRCLHSYAVHTDSVWALASTSTFSHV 92
GH +++++ T +SG D +++LWD R L S H + V ++ + +
Sbjct: 238 GHGWDVKSVDWHPTKSLLVSGGKDQLVKLWDTRSGRELCSLHGHKNIVLSVKWNQNGNWL 297
Query: 93 YSGGRD 98
+ +D
Sbjct: 298 LTASKD 303
>At2g43770 putative splicing factor
Length = 343
Score = 53.9 bits (128), Expect = 1e-08
Identities = 25/73 (34%), Positives = 39/73 (53%), Gaps = 4/73 (5%)
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL--- 64
+ +GG + V+VWD R G TM L+GH D I + L G + L+ D+ + +WD+
Sbjct: 195 IFTGGVDNDVKVWDLRKGEATMTLEGHQDTITGMSLSPDGSYLLTNGMDNKLCVWDMRPY 254
Query: 65 -GQQRCLHSYAVH 76
Q RC+ + H
Sbjct: 255 APQNRCVKIFEGH 267
Score = 44.3 bits (103), Expect = 1e-05
Identities = 29/98 (29%), Positives = 43/98 (43%), Gaps = 1/98 (1%)
Query: 2 NEGGTLLVSGGTEKVVRVWDPRSGSKT-MKLKGHTDNIRALLLDSTGRFCLSGSSDSMIR 60
N GTL+ SG ++ + +W K M LKGH + I L S G +S S D +R
Sbjct: 62 NPAGTLIASGSHDREIFLWRVHGDCKNFMVLKGHKNAILDLHWTSDGSQIVSASPDKTVR 121
Query: 61 LWDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
WD+ + + A H+ V + T + G D
Sbjct: 122 AWDVETGKQIKKMAEHSSFVNSCCPTRRGPPLIISGSD 159
Score = 38.1 bits (87), Expect = 7e-04
Identities = 19/60 (31%), Positives = 31/60 (51%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
GT + +G ++++V +WD S KL GHT ++ + T S SSD I L ++
Sbjct: 284 GTKVTAGSSDRMVHIWDTTSRRTIYKLPGHTGSVNECVFHPTEPIIGSCSSDKNIYLGEI 343
Score = 34.7 bits (78), Expect = 0.008
Identities = 19/95 (20%), Positives = 40/95 (42%), Gaps = 1/95 (1%)
Query: 4 GGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWD 63
G L++SG + ++WD R I A+ +G D+ +++WD
Sbjct: 150 GPPLIISGSDDGTAKLWDMRQRGAIQTFPDKYQ-ITAVSFSDAADKIFTGGVDNDVKVWD 208
Query: 64 LGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
L + + H D++ ++ + S++ + G D
Sbjct: 209 LRKGEATMTLEGHQDTITGMSLSPDGSYLLTNGMD 243
Score = 29.6 bits (65), Expect = 0.26
Identities = 22/76 (28%), Positives = 32/76 (41%), Gaps = 1/76 (1%)
Query: 29 MKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL-GQQRCLHSYAVHTDSVWALASTS 87
M L GH + + + G SGS D I LW + G + H +++ L TS
Sbjct: 47 MLLSGHPSAVYTMKFNPAGTLIASGSHDREIFLWRVHGDCKNFMVLKGHKNAILDLHWTS 106
Query: 88 TFSHVYSGGRDFSVSA 103
S + S D +V A
Sbjct: 107 DGSQIVSASPDKTVRA 122
>At1g48630 unknown protein
Length = 326
Score = 53.9 bits (128), Expect = 1e-08
Identities = 28/104 (26%), Positives = 52/104 (49%), Gaps = 5/104 (4%)
Query: 3 EGGTLLVSGGTEKVVRVW-----DPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDS 57
+ ++V+ +K + +W D G ++ GH+ ++ ++L S G+F LSGS D
Sbjct: 26 DNSDVIVTSSRDKSIILWKLTKEDKSYGVAQRRMTGHSHFVQDVVLSSDGQFALSGSWDG 85
Query: 58 MIRLWDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRDFSV 101
+RLWDL + HT V ++A ++ + S RD ++
Sbjct: 86 ELRLWDLATGESTRRFVGHTKDVLSVAFSTDNRQIVSASRDRTI 129
Score = 47.0 bits (110), Expect = 2e-06
Identities = 24/95 (25%), Positives = 42/95 (43%), Gaps = 9/95 (9%)
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQ 67
+VS +K V+VW+ ++ L GH+ + + + G C SG D +I LWDL +
Sbjct: 166 IVSASWDKTVKVWNLQNCKLRNTLAGHSGYLNTVAVSPDGSLCASGGKDGVILLWDLAEG 225
Query: 68 R---------CLHSYAVHTDSVWALASTSTFSHVY 93
+ +HS + W A+T ++
Sbjct: 226 KKLYSLEAGSIIHSLCFSPNRYWLCAATENSIRIW 260
Score = 43.1 bits (100), Expect = 2e-05
Identities = 18/63 (28%), Positives = 34/63 (53%)
Query: 1 MNEGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIR 60
++ G +SG + +R+WD +G T + GHT ++ ++ + R +S S D I+
Sbjct: 71 LSSDGQFALSGSWDGELRLWDLATGESTRRFVGHTKDVLSVAFSTDNRQIVSASRDRTIK 130
Query: 61 LWD 63
LW+
Sbjct: 131 LWN 133
Score = 39.3 bits (90), Expect = 3e-04
Identities = 21/60 (35%), Positives = 33/60 (55%), Gaps = 2/60 (3%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G+L SGG + V+ +WD G K L+ +I L S R+ L ++++ IR+WDL
Sbjct: 205 GSLCASGGKDGVILLWDLAEGKKLYSLE--AGSIIHSLCFSPNRYWLCAATENSIRIWDL 262
Score = 26.6 bits (57), Expect = 2.2
Identities = 19/73 (26%), Positives = 30/73 (41%), Gaps = 19/73 (26%)
Query: 13 TEKVVRVWDPRSGSKTMKLK----------------GHTDNI---RALLLDSTGRFCLSG 53
TE +R+WD S S LK G+ + +L + G SG
Sbjct: 253 TENSIRIWDLESKSVVEDLKVDLKAEAEKTDGSTGIGNKTKVIYCTSLNWSADGNTLFSG 312
Query: 54 SSDSMIRLWDLGQ 66
+D +IR+W +G+
Sbjct: 313 YTDGVIRVWGIGR 325
>At1g18080 putative guanine nucleotide-binding protein beta
subunit
Length = 327
Score = 53.5 bits (127), Expect = 2e-08
Identities = 30/104 (28%), Positives = 50/104 (47%), Gaps = 5/104 (4%)
Query: 3 EGGTLLVSGGTEKVVRVW-----DPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDS 57
+ ++VS +K + +W D G +L GH+ + ++L S G+F LSGS D
Sbjct: 26 DNADIIVSASRDKSIILWKLTKDDKAYGVAQRRLTGHSHFVEDVVLSSDGQFALSGSWDG 85
Query: 58 MIRLWDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRDFSV 101
+RLWDL + HT V ++A + + S RD ++
Sbjct: 86 ELRLWDLAAGVSTRRFVGHTKDVLSVAFSLDNRQIVSASRDRTI 129
Score = 47.4 bits (111), Expect = 1e-06
Identities = 21/65 (32%), Positives = 34/65 (52%)
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQ 67
+VS +K V+VW+ + L GHT + + + G C SG D ++ LWDL +
Sbjct: 167 IVSASWDKTVKVWNLSNCKLRSTLAGHTGYVSTVAVSPDGSLCASGGKDGVVLLWDLAEG 226
Query: 68 RCLHS 72
+ L+S
Sbjct: 227 KKLYS 231
Score = 41.2 bits (95), Expect = 9e-05
Identities = 18/63 (28%), Positives = 33/63 (51%)
Query: 1 MNEGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIR 60
++ G +SG + +R+WD +G T + GHT ++ ++ R +S S D I+
Sbjct: 71 LSSDGQFALSGSWDGELRLWDLAAGVSTRRFVGHTKDVLSVAFSLDNRQIVSASRDRTIK 130
Query: 61 LWD 63
LW+
Sbjct: 131 LWN 133
Score = 37.7 bits (86), Expect = 0.001
Identities = 20/60 (33%), Positives = 33/60 (54%), Gaps = 2/60 (3%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G+L SGG + VV +WD G K L+ +++ L S R+ L +++ I++WDL
Sbjct: 206 GSLCASGGKDGVVLLWDLAEGKKLYSLE--ANSVIHALCFSPNRYWLCAATEHGIKIWDL 263
Score = 30.8 bits (68), Expect = 0.12
Identities = 22/96 (22%), Positives = 45/96 (45%), Gaps = 5/96 (5%)
Query: 8 LVSGGTEKVVRVWDPRSGSK---TMKLKGHTDNIRALLL--DSTGRFCLSGSSDSMIRLW 62
+VS ++ +++W+ K + +GH D + + ++ +S S D +++W
Sbjct: 120 IVSASRDRTIKLWNTLGECKYTISEGGEGHRDWVSCVRFSPNTLQPTIVSASWDKTVKVW 179
Query: 63 DLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
+L + + A HT V +A + S SGG+D
Sbjct: 180 NLSNCKLRSTLAGHTGYVSTVAVSPDGSLCASGGKD 215
>At3g49660 putative WD-40 repeat - protein
Length = 317
Score = 53.1 bits (126), Expect = 2e-08
Identities = 27/91 (29%), Positives = 44/91 (47%)
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQ 67
+VS +K +++WD +GS L GHT+ + + +SGS D +R+WD+
Sbjct: 86 IVSASDDKTLKLWDVETGSLIKTLIGHTNYAFCVNFNPQSNMIVSGSFDETVRIWDVTTG 145
Query: 68 RCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
+CL H+D V A+ S + S D
Sbjct: 146 KCLKVLPAHSDPVTAVDFNRDGSLIVSSSYD 176
Score = 50.4 bits (119), Expect = 1e-07
Identities = 22/69 (31%), Positives = 36/69 (51%)
Query: 2 NEGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRL 61
N ++VSG ++ VR+WD +G L H+D + A+ + G +S S D + R+
Sbjct: 122 NPQSNMIVSGSFDETVRIWDVTTGKCLKVLPAHSDPVTAVDFNRDGSLIVSSSYDGLCRI 181
Query: 62 WDLGQQRCL 70
WD G C+
Sbjct: 182 WDSGTGHCV 190
Score = 44.7 bits (104), Expect = 8e-06
Identities = 27/107 (25%), Positives = 50/107 (46%), Gaps = 3/107 (2%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDN---IRALLLDSTGRFCLSGSSDSMIRL 61
G ++ G + +R+W+ S GH + I + + G+ +SGS D+ + +
Sbjct: 210 GKFILVGTLDNTLRLWNISSAKFLKTYTGHVNAQYCISSAFSVTNGKRIVSGSEDNCVHM 269
Query: 62 WDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRDFSVSAVFLKK 108
W+L ++ L HT++V +A T + + SG D +V KK
Sbjct: 270 WELNSKKLLQKLEGHTETVMNVACHPTENLIASGSLDKTVRIWTQKK 316
Score = 41.2 bits (95), Expect = 9e-05
Identities = 26/103 (25%), Positives = 45/103 (43%), Gaps = 7/103 (6%)
Query: 5 GTLLVSGGTEKVVRVW------DPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSM 58
G LL S +K +R + DP + + GH + I + S RF +S S D
Sbjct: 36 GRLLASASADKTIRTYTINTINDPIA-EPVQEFTGHENGISDVAFSSDARFIVSASDDKT 94
Query: 59 IRLWDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRDFSV 101
++LWD+ + + HT+ + + + + SG D +V
Sbjct: 95 LKLWDVETGSLIKTLIGHTNYAFCVNFNPQSNMIVSGSFDETV 137
>At5g56130 unknown protein (At5g56130)
Length = 315
Score = 52.8 bits (125), Expect = 3e-08
Identities = 31/112 (27%), Positives = 54/112 (47%), Gaps = 6/112 (5%)
Query: 2 NEGGTLLVSGGTEKVVRVWD----PRSGSKTMKLKGHTDNIRALLLD-STGRFCLSGSSD 56
N GT L SG ++ R+W+ S +K ++LKGHTD++ L D + S D
Sbjct: 29 NSNGTKLASGSVDQTARIWNIEPHGHSKAKDLELKGHTDSVDQLCWDPKHSDLVATASGD 88
Query: 57 SMIRLWDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRDFSVSAVFLKK 108
+RLWD +C + +++ + +HV G RD ++ + ++K
Sbjct: 89 KSVRLWDARSGKCTQQVELSGENI-NITYKPDGTHVAVGNRDDELTILDVRK 139
Score = 40.8 bits (94), Expect = 1e-04
Identities = 22/65 (33%), Positives = 32/65 (48%), Gaps = 6/65 (9%)
Query: 31 LKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQRCLHSYAVHTDSVWALASTSTFS 90
L HT + +D GR+ GS+DS++ LWD+ CL ++ T W + T S
Sbjct: 187 LTAHTAGCYCIAIDPKGRYFAVGSADSLVSLWDISDMLCLRTF---TKLEWPV---RTIS 240
Query: 91 HVYSG 95
YSG
Sbjct: 241 FNYSG 245
Score = 31.6 bits (70), Expect = 0.068
Identities = 18/65 (27%), Positives = 33/65 (50%), Gaps = 1/65 (1%)
Query: 7 LLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQ 66
L+ + +K VR+WD RSG T +++ +NI + G G+ D + + D+ +
Sbjct: 81 LVATASGDKSVRLWDARSGKCTQQVELSGENIN-ITYKPDGTHVAVGNRDDELTILDVRK 139
Query: 67 QRCLH 71
+ LH
Sbjct: 140 FKPLH 144
Score = 28.5 bits (62), Expect = 0.58
Identities = 19/65 (29%), Positives = 29/65 (44%), Gaps = 8/65 (12%)
Query: 22 PRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQRCLHSYAV------ 75
P + + +GH + ++ +S G SGS D R+W++ HS A
Sbjct: 7 PFKSLHSREYQGHKKKVHSVAWNSNGTKLASGSVDQTARIWNIEPHG--HSKAKDLELKG 64
Query: 76 HTDSV 80
HTDSV
Sbjct: 65 HTDSV 69
>At5g52820 Notchless protein homolog
Length = 473
Score = 52.8 bits (125), Expect = 3e-08
Identities = 26/94 (27%), Positives = 46/94 (48%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G + S +K VR+W+ +G +GH + + + R LSGS DS +++W++
Sbjct: 372 GKWIASASFDKSVRLWNGITGQFVTVFRGHVGPVYQVSWSADSRLLLSGSKDSTLKIWEI 431
Query: 65 GQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
++ H D V+A+ + V SGG+D
Sbjct: 432 RTKKLKQDLPGHADEVFAVDWSPDGEKVVSGGKD 465
Score = 40.8 bits (94), Expect = 1e-04
Identities = 17/56 (30%), Positives = 30/56 (53%)
Query: 7 LLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLW 62
LL+SG + +++W+ R+ L GH D + A+ G +SG D +++LW
Sbjct: 416 LLLSGSKDSTLKIWEIRTKKLKQDLPGHADEVFAVDWSPDGEKVVSGGKDRVLKLW 471
Score = 37.7 bits (86), Expect = 0.001
Identities = 19/67 (28%), Positives = 33/67 (48%)
Query: 31 LKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQRCLHSYAVHTDSVWALASTSTFS 90
+ GH + + + G+ SGS D+ +RLWDL + L + H + V +A +
Sbjct: 105 IAGHAEAVLCVSFSPDGKQLASGSGDTTVRLWDLYTETPLFTCKGHKNWVLTVAWSPDGK 164
Query: 91 HVYSGGR 97
H+ SG +
Sbjct: 165 HLVSGSK 171
Score = 37.4 bits (85), Expect = 0.001
Identities = 22/92 (23%), Positives = 43/92 (45%), Gaps = 1/92 (1%)
Query: 8 LVSGGTEKVVRVWDPRSGSKTMK-LKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQ 66
LVSG + + +W+P + K L GH + + G++ S S D +RLW+
Sbjct: 332 LVSGSDDFTMFLWEPSVSKQPKKRLTGHQQLVNHVYFSPDGKWIASASFDKSVRLWNGIT 391
Query: 67 QRCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
+ + + H V+ ++ ++ + SG +D
Sbjct: 392 GQFVTVFRGHVGPVYQVSWSADSRLLLSGSKD 423
Score = 34.3 bits (77), Expect = 0.011
Identities = 17/59 (28%), Positives = 27/59 (44%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWD 63
G L SG + VR+WD + + KGH + + + G+ +SGS I W+
Sbjct: 121 GKQLASGSGDTTVRLWDLYTETPLFTCKGHKNWVLTVAWSPDGKHLVSGSKSGEICCWN 179
Score = 32.3 bits (72), Expect = 0.040
Identities = 26/103 (25%), Positives = 45/103 (43%), Gaps = 7/103 (6%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGS-KTMKLKGHTDNIRALL-----LDSTGRFCLSGSSDSM 58
G LVSG + W+P+ G + L GH I + L S R ++ S D
Sbjct: 163 GKHLVSGSKSGEICCWNPKKGELEGSPLTGHKKWITGISWEPVHLSSPCRRFVTSSKDGD 222
Query: 59 IRLWDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRDFSV 101
R+WD+ ++ + + HT +V +Y+G +D ++
Sbjct: 223 ARIWDITLKKSIICLSGHTLAV-TCVKWGGDGIIYTGSQDCTI 264
Score = 30.4 bits (67), Expect = 0.15
Identities = 18/81 (22%), Positives = 35/81 (42%), Gaps = 1/81 (1%)
Query: 9 VSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQR 68
V+ + R+WD + L GHT + + G +GS D I++W+ Q +
Sbjct: 215 VTSSKDGDARIWDITLKKSIICLSGHTLAVTCVKWGGDG-IIYTGSQDCTIKMWETTQGK 273
Query: 69 CLHSYAVHTDSVWALASTSTF 89
+ H + +LA ++ +
Sbjct: 274 LIRELKGHGHWINSLALSTEY 294
Score = 28.1 bits (61), Expect = 0.75
Identities = 15/55 (27%), Positives = 27/55 (48%), Gaps = 5/55 (9%)
Query: 4 GGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDS-----TGRFCLSG 53
G ++ +G + +++W+ G +LKGH I +L L + TG F +G
Sbjct: 251 GDGIIYTGSQDCTIKMWETTQGKLIRELKGHGHWINSLALSTEYVLRTGAFDHTG 305
>At5g50230 putative protein
Length = 515
Score = 52.0 bits (123), Expect = 5e-08
Identities = 31/103 (30%), Positives = 51/103 (49%), Gaps = 3/103 (2%)
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQ 67
L +GG ++ V++WD SG+ L G NI + + + ++ +S + + +WD+
Sbjct: 246 LFTGGQDRAVKMWDTNSGTLIKSLYGSLGNILDMAVTHDNKSVIAATSSNNLFVWDVSSG 305
Query: 68 RCLHSYAVHTDSVWALASTSTFS--HVYSGGRDFSVSAVFLKK 108
R H+ HTD V A+ S FS HV S D ++ L K
Sbjct: 306 RVRHTLTGHTDKVCAV-DVSKFSSRHVVSAAYDRTIKLWDLHK 347
Score = 46.6 bits (109), Expect = 2e-06
Identities = 25/91 (27%), Positives = 45/91 (48%), Gaps = 1/91 (1%)
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQ 67
+VS ++ +++WD G T + T N A+ L G SG D +RLWD+
Sbjct: 331 VVSAAYDRTIKLWDLHKGYCTNTVL-FTSNCNAICLSIDGLTVFSGHMDGNLRLWDIQTG 389
Query: 68 RCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
+ L A H+ +V +++ + + + + GRD
Sbjct: 390 KLLSEVAGHSSAVTSVSLSRNGNRILTSGRD 420
Score = 32.0 bits (71), Expect = 0.052
Identities = 14/59 (23%), Positives = 31/59 (51%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWD 63
G + SG + +R+WD ++G ++ GH+ + ++ L G L+ D++ ++D
Sbjct: 369 GLTVFSGHMDGNLRLWDIQTGKLLSEVAGHSSAVTSVSLSRNGNRILTSGRDNVHNVFD 427
>At2g41500 putative U4/U6 small nuclear ribonucleoprotein
Length = 554
Score = 52.0 bits (123), Expect = 5e-08
Identities = 32/111 (28%), Positives = 51/111 (45%), Gaps = 13/111 (11%)
Query: 3 EGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLW 62
+ G L S G + + RVWD R+G + +GH + ++ G SG D+ R+W
Sbjct: 391 QDGALAASCGLDSLARVWDLRTGRSILVFQGHIKPVFSVNFSPNGYHLASGGEDNQCRIW 450
Query: 63 DLGQQRCLHSYAVHTDSV------------WALASTSTFSHVYSGGRDFSV 101
DL ++ L+ H + V A AS +++S GRDFS+
Sbjct: 451 DLRMRKSLYIIPAHANLVSQVKYEPQEGYFLATASYDMKVNIWS-GRDFSL 500
Score = 49.3 bits (116), Expect = 3e-07
Identities = 25/94 (26%), Positives = 46/94 (48%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G L + +K R+WD +G++ + +GH+ ++ + G S DS+ R+WDL
Sbjct: 351 GKYLGTTSYDKTWRLWDINTGAELLLQEGHSRSVYGIAFQQDGALAASCGLDSLARVWDL 410
Query: 65 GQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
R + + H V+++ + H+ SGG D
Sbjct: 411 RTGRSILVFQGHIKPVFSVNFSPNGYHLASGGED 444
Score = 34.3 bits (77), Expect = 0.011
Identities = 18/77 (23%), Positives = 35/77 (45%), Gaps = 1/77 (1%)
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQ 67
L + ++ ++W G+ +GH D + + +G++ + S D RLWD+
Sbjct: 313 LATASADRTAKLWKT-DGTLLQTFEGHLDRLARVAFHPSGKYLGTTSYDKTWRLWDINTG 371
Query: 68 RCLHSYAVHTDSVWALA 84
L H+ SV+ +A
Sbjct: 372 AELLLQEGHSRSVYGIA 388
Score = 30.0 bits (66), Expect = 0.20
Identities = 15/58 (25%), Positives = 24/58 (40%)
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLW 62
G L + + V +W R S L GH + +L + + + S D I+LW
Sbjct: 478 GYFLATASYDMKVNIWSGRDFSLVKSLAGHESKVASLDITADSSCIATVSHDRTIKLW 535
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.132 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,546,439
Number of Sequences: 26719
Number of extensions: 92115
Number of successful extensions: 888
Number of sequences better than 10.0: 195
Number of HSP's better than 10.0 without gapping: 171
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 215
Number of HSP's gapped (non-prelim): 613
length of query: 108
length of database: 11,318,596
effective HSP length: 84
effective length of query: 24
effective length of database: 9,074,200
effective search space: 217780800
effective search space used: 217780800
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0062.3


