

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0057b.8
(558 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
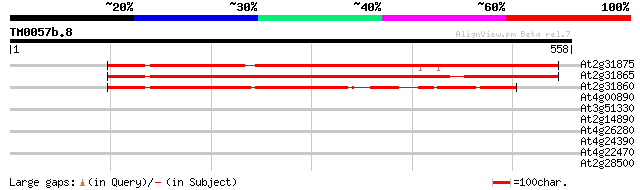
At2g31875 poly(ADP-ribose) like glycohydrolase 529 e-150
At2g31865 poly(ADP-ribose) like glycohydrolase 462 e-130
At2g31860 putative poly(ADP-ribose) glycohydrolase 309 2e-84
At4g00890 31 2.1
At3g51330 predicted GPI-anchored protein 30 2.7
At2g14890 arabinogalactan-protein AGP9 30 3.5
At4g26280 steroid sulfotransferase - like protein 29 7.9
At4g24390 transport inhibitor response-like protein 29 7.9
At4g22470 extensin - like protein 29 7.9
At2g28500 putative protein 29 7.9
>At2g31875 poly(ADP-ribose) like glycohydrolase
Length = 548
Score = 529 bits (1363), Expect = e-150
Identities = 274/466 (58%), Positives = 332/466 (70%), Gaps = 30/466 (6%)
Query: 98 EESRKWFQEVVPALGILLLRLPSLLEAHYQNADRVLDEDGGLVRTGLRLLDSQEPGIVFL 157
+ES++WF E++PAL LLL+ PSLLE H+QNAD ++ ++TGLRLL+SQ+ GIVFL
Sbjct: 86 KESKRWFDEIIPALASLLLQFPSLLEVHFQNADNIVSG----IKTGLRLLNSQQAGIVFL 141
Query: 158 SQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHYFQR 217
SQELI ALL CSFFCLFP + R KHL INFD LF SLY YSQ QESKI+CI+HYF+R
Sbjct: 142 SQELIGALLACSFFCLFPDDNRGAKHLPVINFDHLFASLYISYSQSQESKIRCIMHYFER 201
Query: 218 ITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLSEAV 277
S +P G+VSFERK+ + P+AD WS S + LC F+VHS GLIEDQ A+
Sbjct: 202 FCSCVPIGIVSFERKIT---------AAPDADFWSKSDVSLCAFKVHSFGLIEDQPDNAL 252
Query: 278 EVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSSYTG 337
EVDFAN+YLGGG+L RGCVQEEIRFMI+PELI MLFLP M DNEAIEIVG ERFS YTG
Sbjct: 253 EVDFANKYLGGGSLSRGCVQEEIRFMINPELIAGMLFLPRMDDNEAIEIVGAERFSCYTG 312
Query: 338 YASSFRFSGNYVDERDVDTLGRRKTRIVAIDALCSPGMRQYRPKFLLREINKAFCGFLYG 397
YASSFRF+G Y+D++ +D RR+TRIVAIDALC+P MR ++ LLREINKA CGFL
Sbjct: 313 YASSFRFAGEYIDKKAMDPFKRRRTRIVAIDALCTPKMRHFKDICLLREINKALCGFLNC 372
Query: 398 SKHQLYQKIL--------------QEKGCPSTLFDAATSTPM---ETSEGKCSNHEIRDS 440
SK +Q I ++ G T A+ TP+ E + K +N+ IRD
Sbjct: 373 SKAWEHQNIFMDEGDNEIQLVRNGRDSGLLRTETTASHRTPLNDVEMNREKPANNLIRDF 432
Query: 441 QNDYHRMEQCNNIGVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTFRLEALHN 500
+ E + GVATGNWGCG FGGDPE+K IQWLAASQ RPFI+YYTF +EAL N
Sbjct: 433 YVEGVDNEDHEDDGVATGNWGCGVFGGDPELKATIQWLAASQTRRPFISYYTFGVEALRN 492
Query: 501 IDKVAHWILSHRWTVGDLWNMLTEYSMNRSKGETNVGFLQWVLPSM 546
+D+V WILSH+WTVGDLWNM+ EYS R +T+VGF W+LPS+
Sbjct: 493 LDQVTKWILSHKWTVGDLWNMMLEYSAQRLYKQTSVGFFSWLLPSL 538
>At2g31865 poly(ADP-ribose) like glycohydrolase
Length = 522
Score = 462 bits (1188), Expect = e-130
Identities = 244/450 (54%), Positives = 304/450 (67%), Gaps = 18/450 (4%)
Query: 98 EESRKWFQEVVPALGILLLRLPSLLEAHYQNADRVLDEDGGLVRTGLRLLDSQEPGIVFL 157
EES +F EVVPAL LLL+LPS+LE HYQ AD VLD V++GLRLL QE GIV L
Sbjct: 88 EESANFFGEVVPALCRLLLQLPSMLEKHYQKADHVLDG----VKSGLRLLGPQEAGIVLL 143
Query: 158 SQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHYFQR 217
SQELIAALL CSFFCLFP +R K+LQ INF LF Y + KQE+KI+C+IHYF R
Sbjct: 144 SQELIAALLACSFFCLFPEVDRSLKNLQGINFSGLFSFPYMRHCTKQENKIKCLIHYFGR 203
Query: 218 ITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLSEAV 277
I MP G VSFERK+LPLE +SYP AD W+ S+ PLC E+H+SG IEDQ EA+
Sbjct: 204 ICRWMPTGFVSFERKILPLEYHPHFVSYPKADSWANSVTPLCSIEIHTSGAIEDQPCEAL 263
Query: 278 EVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSSYTG 337
EVDFA+EY GG L +QEEIRF+I+PELI M+FLP M NEAIEIVGVERFS YTG
Sbjct: 264 EVDFADEYFGGLTLSYDTLQEEIRFVINPELIAGMIFLPRMDANEAIEIVGVERFSGYTG 323
Query: 338 YASSFRFSGNYVDERDVDTLGRRKTRIVAIDALCSPGMRQYRPKFLLREINKAFCGFLYG 397
Y SF+++G+Y D +D+D RRKTR++AIDA+ PGM QY+ L+RE+NKAF G+++
Sbjct: 324 YGPSFQYAGDYTDNKDLDIFRRRKTRVIAIDAMPDPGMGQYKLDALIREVNKAFSGYMHQ 383
Query: 398 SKHQLYQKILQEKGCPST-LFDAATSTPMETSEGKCSNHEIRDSQNDYHRMEQCNNIGVA 456
K+ + K E L + S +E+S C +HE + IGVA
Sbjct: 384 CKYNIDVKHDPEASSSHVPLTSDSASQVIESSHRWCIDHEEK-------------KIGVA 430
Query: 457 TGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTFRLEALHNIDKVAHWILSHRWTVG 516
TGNWGCG FGGDPE+K ++QWLA SQ+ RPF++YYTF L+AL N+++V + TVG
Sbjct: 431 TGNWGCGVFGGDPELKIMLQWLAISQSGRPFMSYYTFGLQALQNLNQVIEMVALQEMTVG 490
Query: 517 DLWNMLTEYSMNRSKGETNVGFLQWVLPSM 546
DLW L EYS R T +GF W++ S+
Sbjct: 491 DLWKKLVEYSSERLSRRTWLGFFSWLMTSL 520
>At2g31860 putative poly(ADP-ribose) glycohydrolase
Length = 364
Score = 309 bits (792), Expect = 2e-84
Identities = 194/409 (47%), Positives = 250/409 (60%), Gaps = 50/409 (12%)
Query: 98 EESRKWFQEVVPALGILLLRLPSLLEAHYQNADRVLDEDGGLVRTGLRLLDSQEPGIVFL 157
EES +WF E +PA+ LLLR PSLLE+HY N+D +++ +TGLR+L + GIVFL
Sbjct: 4 EESSRWFNEFLPAMACLLLRFPSLLESHYLNSDNLING----TKTGLRVLVPNKAGIVFL 59
Query: 158 SQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDY-YSQKQESKIQCIIHYFQ 216
SQELI ALL CSFFCLFPV++R HL INFD+LFGSL + ++ QE+KI+CIIHYFQ
Sbjct: 60 SQELIGALLSCSFFCLFPVDDRGSNHLPIINFDKLFGSLINTGRNEHQENKIKCIIHYFQ 119
Query: 217 RITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLSEA 276
R++S++ G VSFERK+L LE D S + W S + LC EV +SGLIEDQ EA
Sbjct: 120 RLSSSISPGFVSFERKILSLEQDS---STLDEGFWGKSTVNLCPVEVRTSGLIEDQSVEA 176
Query: 277 VEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSSYT 336
+EVDFAN+ LGGGALR+GCVQEEIRFMI+PELIV MLFLP+M EAIE+VG ERFS YT
Sbjct: 177 LEVDFANKNLGGGALRKGCVQEEIRFMINPELIVGMLFLPTMEVTEAIEVVGAERFSLYT 236
Query: 337 GYASSFRFSGNYVDERDVDTLGRRKTRIVAIDALCSPGMRQYRPKFLLREINKAFCGFLY 396
G FR + KTRIVAIDAL PG+ QY+ + LL +
Sbjct: 237 G---CFR---------------KAKTRIVAIDALRHPGVSQYKLESLLSVL--------- 269
Query: 397 GSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRME-QCNNIGV 455
IL G P L+ + S ++ EI S ++ + +N+ +
Sbjct: 270 ---------ILSSSGRPIRLYMGSVS--LQGIGDVVLMVEILSSSLFFNGLRFHRSNLYL 318
Query: 456 ATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTFRLEALHNIDKV 504
+ C P V + L + QA RPF++YYTF EAL N+++V
Sbjct: 319 FSPLIACNY---QPMVVVWLTTLHSFQARRPFMSYYTFGFEALQNLNQV 364
>At4g00890
Length = 431
Score = 30.8 bits (68), Expect = 2.1
Identities = 23/90 (25%), Positives = 40/90 (43%), Gaps = 7/90 (7%)
Query: 38 SSISTPRDAFLLSVLAFPSRRITPATNCSAPFPTSEP-------PFPSLPNLYPLPLQTD 90
S + P +S+ + S +PAT + P P S P P PSLP + P +++
Sbjct: 282 SPMPPPSPTAQISLSSLKSPIPSPATITAPPPPFSSPLSQTTPSPKPSLPQIEPNQIKSP 341
Query: 91 TPSSSTIEESRKWFQEVVPALGILLLRLPS 120
+P + E Q +V L+ ++P+
Sbjct: 342 SPKLTNTESHASPEQNLVKPDANLMNKIPA 371
>At3g51330 predicted GPI-anchored protein
Length = 529
Score = 30.4 bits (67), Expect = 2.7
Identities = 23/63 (36%), Positives = 27/63 (42%), Gaps = 3/63 (4%)
Query: 61 PATNCSAPFPTSEPPFPSL--PNLYPLPLQTDTPSSSTIEESRKWFQEVVPALGILLLRL 118
P AP P++ P PSL P P Q D P +ST +VP LLL L
Sbjct: 465 PPPETEAPSPSASTPLPSLLPPPAAATPPQID-PRNSTRNSGTGTAANLVPLASQLLLLL 523
Query: 119 PSL 121
P L
Sbjct: 524 PLL 526
>At2g14890 arabinogalactan-protein AGP9
Length = 191
Score = 30.0 bits (66), Expect = 3.5
Identities = 12/31 (38%), Positives = 19/31 (60%)
Query: 60 TPATNCSAPFPTSEPPFPSLPNLYPLPLQTD 90
+P+ + S P P+S+ P PS ++ P P TD
Sbjct: 136 SPSPSSSPPLPSSDAPGPSTDSISPAPSPTD 166
>At4g26280 steroid sulfotransferase - like protein
Length = 314
Score = 28.9 bits (63), Expect = 7.9
Identities = 16/54 (29%), Positives = 26/54 (47%)
Query: 221 NMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLS 274
N QGV++F+R P + D I SYP + + + FE + +D +S
Sbjct: 45 NFLQGVLNFQRGFKPQDTDIIVASYPKSGTLWLKALTVALFERTKNPSHDDPMS 98
>At4g24390 transport inhibitor response-like protein
Length = 623
Score = 28.9 bits (63), Expect = 7.9
Identities = 23/93 (24%), Positives = 38/93 (40%), Gaps = 4/93 (4%)
Query: 295 CVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSSYTGYASSF---RFSGNYVDE 351
CV+ I F EL+V FL + N + + + R +S FS + V +
Sbjct: 238 CVESPINFKALEELVVRSPFLKKLRTNRFVSLEELHRLMVRAPQLTSLGTGSFSPDNVPQ 297
Query: 352 RDVDTLGRRKTRIVAIDALCSPGMRQYRPKFLL 384
+ R +C G R++RP++LL
Sbjct: 298 GEQQPDYAAAFR-ACKSIVCLSGFREFRPEYLL 329
>At4g22470 extensin - like protein
Length = 375
Score = 28.9 bits (63), Expect = 7.9
Identities = 25/87 (28%), Positives = 32/87 (36%), Gaps = 10/87 (11%)
Query: 57 RRITPATNCSAPFPTSEPPFPSLPNLYPLPLQTDTPSSSTIEESRKWFQEVVPALGILLL 116
RRI +C P PT PP L P P QT P I P L L+
Sbjct: 245 RRIPQGFSCPGPSPTISPPPLPPQTLKPPPPQTTPPPPPAI----------TPPLSPPLV 294
Query: 117 RLPSLLEAHYQNADRVLDEDGGLVRTG 143
+ S + + +L GL+ TG
Sbjct: 295 GICSKNDTELKICAGILAISDGLLTTG 321
>At2g28500 putative protein
Length = 229
Score = 28.9 bits (63), Expect = 7.9
Identities = 18/53 (33%), Positives = 26/53 (48%), Gaps = 7/53 (13%)
Query: 34 LEIDSSISTPRDAFLLSVLAFPSRRITPATNCSAPFPTSEPPFPSLPNLYPLP 86
+EI+ ++TP S +A + TP ++P PTS PP P P P P
Sbjct: 1 MEINGGVATPT----ASAVAKVTETTTPV---NSPSPTSSPPPPPSPQQPPQP 46
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.138 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,297,135
Number of Sequences: 26719
Number of extensions: 597990
Number of successful extensions: 1672
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 1647
Number of HSP's gapped (non-prelim): 20
length of query: 558
length of database: 11,318,596
effective HSP length: 104
effective length of query: 454
effective length of database: 8,539,820
effective search space: 3877078280
effective search space used: 3877078280
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0057b.8


