

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0057b.10
(306 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
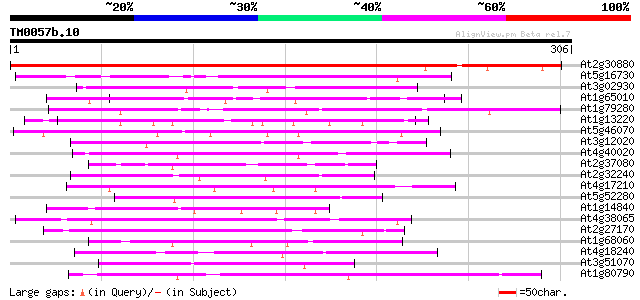
Score E
Sequences producing significant alignments: (bits) Value
At2g30880 unknown protein 340 7e-94
At5g16730 putative protein 62 4e-10
At3g02930 unknown protein 59 4e-09
At1g65010 hypothetical protein 50 2e-06
At1g79280 hypothetical protein 49 3e-06
At1g13220 putative nuclear matrix constituent protein 48 6e-06
At5g46070 putative protein 47 1e-05
At3g12020 unknown protein 47 2e-05
At4g40020 putative protein 46 2e-05
At2g37080 putative myosin heavy chain 46 3e-05
At2g32240 putative myosin heavy chain 45 4e-05
At4g17210 putative protein 45 6e-05
At5g52280 hyaluronan mediated motility receptor-like protein 44 8e-05
At1g14840 unknown protein 44 1e-04
At4g38065 hypothetical protein 44 1e-04
At2g27170 putative chromosome associated protein 44 1e-04
At1g68060 unknown protein 44 1e-04
At4g18240 starch synthase-like protein 43 2e-04
At3g51070 putative protein 43 2e-04
At1g80790 hypothetical protein 43 2e-04
>At2g30880 unknown protein
Length = 504
Score = 340 bits (871), Expect = 7e-94
Identities = 193/310 (62%), Positives = 238/310 (76%), Gaps = 11/310 (3%)
Query: 1 MQISLRNTLGMMTNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLS 60
MQISLRN L + NK DGP+DDLTIMKETL VKDEEL N AR+LR+RDS I++IA+KLS
Sbjct: 180 MQISLRNALKITPNKPIDGPLDDLTIMKETLRVKDEELHNLARELRSRDSMIKEIADKLS 239
Query: 61 ETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKE 120
ETAEAA AAASAAHT+DEQ + C E E L DS++Q E + KLKE E K +LSKEK+
Sbjct: 240 ETAEAAVAAASAAHTMDEQRKIVCVEFERLTTDSQRQQEATKLKLKELEEKTFTLSKEKD 299
Query: 121 QLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGES 180
QL K+R+AA QEA+MWRSELGKARE VILEGA+ RAEEKVRVAEA+ EAK KEA Q E+
Sbjct: 300 QLVKERDAALQEAHMWRSELGKARERVVILEGAVVRAEEKVRVAEASGEAKSKEASQREA 359
Query: 181 AAVSEKQELLAYVDKLKAQLQRQHIDSTEVF-EKTESCSDTKHVDL---TEENVDKACLS 236
A +EKQELLAYV+ L+ QLQRQ +++ +V EKTES + + + TE+NVDKACLS
Sbjct: 360 TAWTEKQELLAYVNMLQTQLQRQQLETKQVCEEKTESTNGEASLPMTKETEKNVDKACLS 419
Query: 237 VSRSVPDPAAAECVVQMPTDQVI--IQPVGDNEWSDIQATEARVADVREVASESE---VS 291
+SR+ P E VV M +QV+ PVG+NEW+DIQATEARV+DVRE+++E+E +
Sbjct: 420 ISRTASIP--GENVVHMSEEQVVNAQPPVGENEWNDIQATEARVSDVREISAETERDRRN 477
Query: 292 SLDIPVISQQ 301
SLDIPV+S +
Sbjct: 478 SLDIPVVSPE 487
>At5g16730 putative protein
Length = 853
Score = 62.0 bits (149), Expect = 4e-10
Identities = 59/242 (24%), Positives = 111/242 (45%), Gaps = 26/242 (10%)
Query: 4 SLRNTLGMMTNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETA 63
S R + N+ D++ ++K L E + F +++ ++ I EKL+
Sbjct: 254 STREKTAISDNEMVAKLEDEIVVLKRDL----ESARGFEAEVKEKEM----IVEKLNVDL 305
Query: 64 EAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLS 123
EAA+ A S AH++ +W++ +E+ E+QLE A KL+ S S E +
Sbjct: 306 EAAKMAESNAHSLSNEWQSKAKEL-------EEQLE-EANKLER------SASVSLESVM 351
Query: 124 KQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAV 183
KQ E + + + +E+ +E V LE +A+ +E + V+E + +E + E
Sbjct: 352 KQLEGSNDKLHDTETEITDLKERIVTLETTVAKQKEDLEVSEQRLGSVEEEVSKNEKEVE 411
Query: 184 SEKQELLAYVDKLKAQLQRQHIDSTEV----FEKTESCSDTKHVDLTEENVDKACLSVSR 239
K EL ++ L+++ ++ V EK++ SD + EE KA S++
Sbjct: 412 KLKSELETVKEEKNRALKKEQDATSRVQRLSEEKSKLLSDLESSKEEEEKSKKAMESLAS 471
Query: 240 SV 241
++
Sbjct: 472 AL 473
>At3g02930 unknown protein
Length = 806
Score = 58.5 bits (140), Expect = 4e-09
Identities = 53/202 (26%), Positives = 95/202 (46%), Gaps = 23/202 (11%)
Query: 37 ELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSE- 95
+L+N AR L + + I E+L+ EAA+ A S AH ++W+N +E+E +++
Sbjct: 269 DLEN-ARSLEAKVKELEMIIEQLNVDLEAAKMAESYAHGFADEWQNKAKELEKRLEEANK 327
Query: 96 -------------KQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSE--L 140
KQLE+S +L + E +I L ++ E L + A+Q+ ++ +SE L
Sbjct: 328 LEKCASVSLVSVTKQLEVSNSRLHDMESEITDLKEKIELL--EMTVASQKVDLEKSEQKL 385
Query: 141 GKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQL 200
G A E E + EK++ + +A++ E A S Q LL K+ ++L
Sbjct: 386 GIAEEESSKSE----KEAEKLKNELETVNEEKTQALKKEQDATSSVQRLLEEKKKILSEL 441
Query: 201 QRQHIDSTEVFEKTESCSDTKH 222
+ + + + ES + H
Sbjct: 442 ESSKEEEEKSKKAMESLASALH 463
Score = 38.5 bits (88), Expect = 0.005
Identities = 42/189 (22%), Positives = 85/189 (44%), Gaps = 16/189 (8%)
Query: 23 DLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRN 82
++T +KE + + + + + DL + + E+ S++ + AE + T++E
Sbjct: 356 EITDLKEKIELLEMTVASQKVDLEKSEQKLGIAEEESSKSEKEAEKLKNELETVNE---- 411
Query: 83 ACEEIESLKKDSEKQLELSAQKLKEYEVKIIS-LSKEKEQLSKQREAATQEANMWRSELG 141
E+ ++LKK E+ S Q+L E + KI+S L KE+ K ++A A+
Sbjct: 412 --EKTQALKK--EQDATSSVQRLLEEKKKILSELESSKEEEEKSKKAMESLASALHEVSS 467
Query: 142 KARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQE----LLAYVDKLK 197
++RE + E L+R ++ + + IK + E + L+ V++ K
Sbjct: 468 ESRE---LKEKLLSRGDQNYETQIEDLKLVIKATNNKYENMLDEARHEIDVLVNAVEQTK 524
Query: 198 AQLQRQHID 206
Q + +D
Sbjct: 525 KQFESAMVD 533
Score = 34.7 bits (78), Expect = 0.066
Identities = 46/219 (21%), Positives = 87/219 (39%), Gaps = 21/219 (9%)
Query: 27 MKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEE 86
+KE LL + + QN+ + I+ K + A EQ + ++
Sbjct: 472 LKEKLLSRGD--QNYETQIEDLKLVIKATNNKYENMLDEARHEIDVLVNAVEQTK---KQ 526
Query: 87 IESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARES 146
ES D E + +KE++ ++ S+ KE +L + +EA+ + + R+
Sbjct: 527 FESAMVDWEMREAGLVNHVKEFDEEVSSMGKEMNRLGNLVKRTKEEADASWEKESQMRDC 586
Query: 147 GVILEGALARAEEKVRVAEANA---EAKIKEAVQGESAAVSEKQELLAYVDK-------- 195
+E + +E +R A+A + K+ + + V E EL D
Sbjct: 587 LKEVEDEVIYLQETLREAKAETLKLKGKMLDKETEFQSIVHENDELRVKQDDSLKKIKEL 646
Query: 196 ---LKAQLQRQHIDSTEVFEKTESCSD--TKHVDLTEEN 229
L+ L ++HI+ ++E D K V+ +EEN
Sbjct: 647 SELLEEALAKKHIEENGELSESEKDYDLLPKVVEFSEEN 685
Score = 34.3 bits (77), Expect = 0.087
Identities = 37/178 (20%), Positives = 75/178 (41%), Gaps = 21/178 (11%)
Query: 27 MKETLLVKDEELQNFARD---LRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNA 83
+K +L K+ E Q+ + LR + D +K+ E +E E A + H + +
Sbjct: 611 LKGKMLDKETEFQSIVHENDELRVKQD---DSLKKIKELSELLEEALAKKHIEENGELSE 667
Query: 84 CEE--------IESLKKDSEKQLELSAQKLKEYEVKIISLS-----KEKEQLSKQREAAT 130
E+ +E +++ + E + K++ + + L KEK++ S + E
Sbjct: 668 SEKDYDLLPKVVEFSEENGYRSAEEKSSKVETLDGMNMKLEEDTEKKEKKERSPEDETVE 727
Query: 131 QEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQE 188
E MW S + +E V + + EE + V + + + + GE + EK++
Sbjct: 728 VEFKMWESCQIEKKE--VFHKESAKEEEEDLNVVDQSQKTSPVNGLTGEDELLKEKEK 783
Score = 32.3 bits (72), Expect = 0.33
Identities = 53/258 (20%), Positives = 105/258 (40%), Gaps = 34/258 (13%)
Query: 36 EELQNFARDLRTRDSTIRDIAEKLS----ETAEAAEAAASAAHTIDEQWRNACEEIESLK 91
+EL+N + +T+ + ++L E A A +A + A D+ + A E ++
Sbjct: 169 KELENVKNQHASESATLLLVTQELENVNQELANAKDAKSKALCRADDASKMAAIHAEKVE 228
Query: 92 KDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILE 151
S + + L A ++ ++EKE +SK A A + +L + E+ LE
Sbjct: 229 ILSSELIRLKA---------LLDSTREKEIISKNEIALKLGAEI--VDLKRDLENARSLE 277
Query: 152 GALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHIDSTEVF 211
+ E + + EA A ES A E +L+ +L+ +
Sbjct: 278 AKVKELEMIIEQLNVDLEA----AKMAESYAHGFADEWQNKAKELEKRLEEAN------- 326
Query: 212 EKTESCSDTKHVDLTEENVDKACLSVSRSVPDPAAAECVVQMPTDQVIIQPVGDNEWSDI 271
K E C+ V +T++ L VS S +E + + +++ ++ D+
Sbjct: 327 -KLEKCASVSLVSVTKQ------LEVSNSRLHDMESE-ITDLKEKIELLEMTVASQKVDL 378
Query: 272 QATEARVADVREVASESE 289
+ +E ++ E +S+SE
Sbjct: 379 EKSEQKLGIAEEESSKSE 396
>At1g65010 hypothetical protein
Length = 1318
Score = 50.1 bits (118), Expect = 2e-06
Identities = 47/197 (23%), Positives = 89/197 (44%), Gaps = 18/197 (9%)
Query: 55 IAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIIS 114
+ E+L EAA+ A S ++ E+W+N E+E ++S + +++ ++ ++
Sbjct: 260 LVEQLKVDLEAAKMAESCTNSSVEEWKNKVHELEKEVEESNRSKSSASESMESVMKQLAE 319
Query: 115 LS-------------KEK-EQLSKQREAATQEANMWRSELGKARESGVILEGALARAEEK 160
L+ KEK E L K EA + + ++ A+E LE + + +
Sbjct: 320 LNHVLHETKSDNAAQKEKIELLEKTIEAQRTDLEEYGRQVCIAKEEASKLENLVESIKSE 379
Query: 161 VRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHIDSTEVFEKTESCSDT 220
+ +++ E K + A+ E AA S Q LL +L +L+R ++ + + ES +
Sbjct: 380 LEISQ---EEKTR-ALDNEKAATSNIQNLLDQRTELSIELERCKVEEEKSKKDMESLTLA 435
Query: 221 KHVDLTEENVDKACLSV 237
TE + KA L V
Sbjct: 436 LQEASTESSEAKATLLV 452
Score = 47.4 bits (111), Expect = 1e-05
Identities = 62/233 (26%), Positives = 110/233 (46%), Gaps = 17/233 (7%)
Query: 21 MDDLTIMKETLLVKDEELQNFA---RDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTID 77
+D+L+ TL LQN + ++LR R++T+ AE+LSE E+ AS T+
Sbjct: 809 IDELSTANGTLADNVTNLQNISEENKELRERETTLLKKAEELSELNESLVDKASKLQTVV 868
Query: 78 EQWRNACE-EIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMW 136
++ E E LKK E L + L + E K+ + EKE+L K+RE A +
Sbjct: 869 QENEELRERETAYLKKIEE--LSKLHEILSDQETKLQISNHEKEEL-KERETAYLKK--- 922
Query: 137 RSELGKARESGVILEGAL---ARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYV 193
EL K +E + E L E +R ++ A+ KI+E ++ + ++ EL A V
Sbjct: 923 IEELSKVQEDLLNKENELHGMVVEIEDLRSKDSLAQKKIEELSNFNASLLIKENELQAVV 982
Query: 194 DKLKAQLQRQHIDSTEVFEKTESCSDTKHVDLTEENVDKACLSVSRSVPDPAA 246
+ +L+ + + + + ++ SD K + +E +A + + + AA
Sbjct: 983 CE-NEELKSKQVSTLKTIDE---LSDLKQSLIHKEKELQAAIVENEKLKAEAA 1031
Score = 40.8 bits (94), Expect = 0.001
Identities = 46/221 (20%), Positives = 89/221 (39%), Gaps = 28/221 (12%)
Query: 28 KETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAA----EAAASAAHTIDEQWRNA 83
K TLLV EEL+N + + ++ EK + E A ++ S +I ++ N+
Sbjct: 447 KATLLVCQEELKNCESQVDSLKLASKETNEKYEKMLEDARNEIDSLKSTVDSIQNEFENS 506
Query: 84 --------------CEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAA 129
++ E S++++ LKE E + +E+ L + A
Sbjct: 507 KAGWEQKELHLMGCVKKSEEENSSSQEEVSRLVNLLKESEEDACARKEEEASLKNNLKVA 566
Query: 130 TQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQEL 189
E + LG+A+ + L+ +L EE ++ N A+I + E + + + +EL
Sbjct: 567 EGEVKYLQETLGEAKAESMKLKESLLDKEEDLK----NVTAEISSLREWEGSVLEKIEEL 622
Query: 190 LAYVDKLKAQLQRQHIDSTEVFEKTESCS--DTKHVDLTEE 228
K+K L + + ++ E + H+ EE
Sbjct: 623 ----SKVKESLVDKETKLQSITQEAEELKGREAAHMKQIEE 659
Score = 38.5 bits (88), Expect = 0.005
Identities = 48/208 (23%), Positives = 91/208 (43%), Gaps = 21/208 (10%)
Query: 27 MKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEE 86
+KE+LL K+E+L+N ++ + + EK+ E ++ E+ + + A EE
Sbjct: 587 LKESLLDKEEDLKNVTAEISSLREWEGSVLEKIEELSKVKESLVDKETKLQSITQEA-EE 645
Query: 87 IESLKKDSEKQLE---LSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKA 143
++ + KQ+E + L + K+ S+ +E E L K++EA L K
Sbjct: 646 LKGREAAHMKQIEELSTANASLVDEATKLQSIVQESEDL-KEKEA---------GYLKKI 695
Query: 144 RESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQEL-LAYVDKLKAQLQR 202
E V E + + + + + K +E A + + +EL +A + + +
Sbjct: 696 EELSVANESLADNVTDLQSIVQESKDLKEREV-----AYLKKIEELSVANESLVDKETKL 750
Query: 203 QHID-STEVFEKTESCSDTKHVDLTEEN 229
QHID E E+ K +L++EN
Sbjct: 751 QHIDQEAEELRGREASHLKKIEELSKEN 778
Score = 38.1 bits (87), Expect = 0.006
Identities = 54/203 (26%), Positives = 86/203 (41%), Gaps = 34/203 (16%)
Query: 47 TRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKD----------SEK 96
TR S R K+ + + I E + A E+IE LKKD SEK
Sbjct: 34 TRLSLDRSPPTKVHSRLVKGTELQTQLNQIQEDLKKADEQIELLKKDKAKAIDDLKESEK 93
Query: 97 QLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANM---------WRSELGKARESG 147
+E + +KLKE + ++E ++ K R ++A + ++EL R
Sbjct: 94 LVEEANEKLKE-ALAAQKRAEESFEVEKFRAVELEQAGLEAVQKKDVTSKNELESIRSQH 152
Query: 148 VI-LEGALARAEEKVRV---AEANAEAKIKEAVQGESAAV-----SEKQELLA-YVDKLK 197
+ + L+ EE RV A+AK K E A +EK E+LA + +LK
Sbjct: 153 ALDISALLSTTEELQRVKHELSMTADAKNKALSHAEEATKIAEIHAEKAEILASELGRLK 212
Query: 198 AQL----QRQHIDSTEVFEKTES 216
A L +++ I+ E+ K +S
Sbjct: 213 ALLGSKEEKEAIEGNEIVSKLKS 235
Score = 33.1 bits (74), Expect = 0.19
Identities = 50/235 (21%), Positives = 96/235 (40%), Gaps = 37/235 (15%)
Query: 21 MDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQW 80
+++LT +K+TL+ K ELQ + + +K+ E +++ W
Sbjct: 1037 IEELTNLKQTLIDKQNELQGVFHENEELKAKEASSLKKIDELLH-----------LEQSW 1085
Query: 81 RNACEEIESLKKDS---EKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQE----- 132
E + + +++ + Q L+A+K++E K+ EKE K REAA E
Sbjct: 1086 LEKESEFQRVTQENLELKTQDALAAKKIEELS-KLKESLLEKETELKCREAAALEKMEEP 1144
Query: 133 ---ANMWRSELGKARESGVILE-GALARAEEKVR------------VAEANAEAKIKEAV 176
N + +GK + E + +EK + + E+ EA I + +
Sbjct: 1145 SKHGNSELNSIGKDYDLVQFSEVNGASNGDEKTKTDHYQQRSREHMIQESPMEA-IDKHL 1203
Query: 177 QGESAAVSEKQELLAYVDKLKAQLQRQHIDSTEVFEKTESCSDTKHVDLTEENVD 231
GE AA+ + + ++ + + + DS ++ + S +D EE VD
Sbjct: 1204 MGERAAIHKVAHRVEGERNVEKESEFKMWDSYKIEKSEVSPERETELDSVEEEVD 1258
Score = 30.8 bits (68), Expect = 0.96
Identities = 35/180 (19%), Positives = 78/180 (42%), Gaps = 9/180 (5%)
Query: 18 DGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTID 77
D ++L + + L K EEL +L + +++IAE+ + E A +
Sbjct: 754 DQEAEELRGREASHLKKIEELSKENENLVDNVANMQNIAEESKDLREREVAYLKKIDELS 813
Query: 78 EQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWR 137
+ + +L+ SE+ EL ++ +L K+ E+LS+ E+ +A+ +
Sbjct: 814 TANGTLADNVTNLQNISEENKELRERE--------TTLLKKAEELSELNESLVDKASKLQ 865
Query: 138 SELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLK 197
+ + + E L + EE ++ E ++ + K + +++E AY+ K++
Sbjct: 866 TVVQENEELRERETAYLKKIEELSKLHEILSDQETKLQISNHEKEELKERE-TAYLKKIE 924
>At1g79280 hypothetical protein
Length = 2111
Score = 49.3 bits (116), Expect = 3e-06
Identities = 60/292 (20%), Positives = 130/292 (43%), Gaps = 27/292 (9%)
Query: 22 DDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKL-------SETAEAAEAAASAAH 74
D++ ++E L KD ++ + L + + I + ++L SE + + A A
Sbjct: 1382 DEVRQLEEKLKAKDAHAEDCKKVLLEKQNKISLLEKELTNCKKDLSEREKRLDDAQQAQA 1441
Query: 75 TIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEAN 134
T+ ++ +E+E KK L ++ +K YE KEK++LSKQ ++ ++
Sbjct: 1442 TMQSEFNKQKQELEKNKK-IHYTLNMTKRK---YE-------KEKDELSKQNQSLAKQLE 1490
Query: 135 MWRSELGKARESGVILEGALARAEEK---VRVAEANAEAKIKEAVQGESAAVSEKQELLA 191
+ E GK + ++E ++ EEK +++ + ++K+ V+ ++ + +K E L
Sbjct: 1491 EAKEEAGKRTTTDAVVEQSVKEREEKEKRIQILDKYVH-QLKDEVRKKTEDLKKKDEELT 1549
Query: 192 YVDKLKAQLQRQHIDSTEVFEKTESCSDTKHVDLTEENVDKACLSVSRSVPDPAAAECVV 251
+ ++++ DS +K ++ D + L E A +S + A+ +
Sbjct: 1550 KERSERKSVEKEVGDSLTKIKKEKTKVDEELAKL--ERYQTALTHLSEELEKLKHADGNL 1607
Query: 252 QMPTDQVII---QPVGDNEWSDIQATEARVADVREVASESEVSSLDIPVISQ 300
T V + + D + + A E R +AS S+VS+ ++++
Sbjct: 1608 PEGTSAVQVLSGSILNDQAAAYVSAVEYFERVARSIASNSQVSTKPTDMVTE 1659
Score = 35.8 bits (81), Expect = 0.030
Identities = 47/225 (20%), Positives = 93/225 (40%), Gaps = 47/225 (20%)
Query: 14 NKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAA 73
N T +D + + +T K+ L +L + + +LS+ E E A
Sbjct: 142 NSTIKSYLDKIVKLTDTSSEKEARLAEATAELARSQA----MCSRLSQEKELTERHAK-- 195
Query: 74 HTIDEQWRNACEEIESLKK-DSEKQLELSA--------------------QKLKEYEVKI 112
+DE+ + L++ S+ + E+SA ++L+E E KI
Sbjct: 196 -WLDEELTAKVDSYAELRRRHSDLESEMSAKLVDVEKNYIECSSSLNWHKERLRELETKI 254
Query: 113 ISLSKEKEQLSKQREAATQEANMWRSELGKA---------------RESGVILEGALARA 157
SL +E LS ++AAT + +EL A R++G LEG +
Sbjct: 255 GSL---QEDLSSCKDAATTTEEQYTAELFTANKLVDLYKESSEEWSRKAGE-LEGVIKAL 310
Query: 158 EEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQR 202
E ++ E++ + ++ + V + E +L ++K +A++++
Sbjct: 311 EARLSQVESSYKERLDKEVSTKQLLEKENGDLKQKLEKCEAEIEK 355
Score = 31.6 bits (70), Expect = 0.56
Identities = 39/147 (26%), Positives = 72/147 (48%), Gaps = 23/147 (15%)
Query: 86 EIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSK--QREAATQEANMWRSELGKA 143
++ +LK D E + E + YE ++I LS+ ++L+K Q AA QE SEL K
Sbjct: 1086 QMSTLKNDLETEHEKWRVAQRNYERQVILLSETIQELTKTSQALAALQEE---ASELRKL 1142
Query: 144 RESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQ--ELLAYVDKLKAQLQ 201
+ AR E +E NA+ ++ + + ++EK+ EL L ++L+
Sbjct: 1143 AD---------ARGIEN---SELNAKWSEEKLMLEQQKNLAEKKYHELNEQNKLLHSRLE 1190
Query: 202 RQHIDSTEVFEKTESCS----DTKHVD 224
+H++S E ++ + S D+ H++
Sbjct: 1191 AKHLNSAEKNSRSGTISSGSTDSDHLE 1217
Score = 31.2 bits (69), Expect = 0.73
Identities = 28/101 (27%), Positives = 51/101 (49%), Gaps = 6/101 (5%)
Query: 114 SLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVR-VAEANAE-AK 171
SL ++ L+K E + + R E +E+ +IL+ L+ EEK + E E +
Sbjct: 405 SLLRDGWSLAKIYEKYQEAVDAMRHEQLGRKEAEMILQRVLSELEEKAGFIQEERGEYER 464
Query: 172 IKEAV----QGESAAVSEKQELLAYVDKLKAQLQRQHIDST 208
+ EA Q +VSE+ + ++ +LKA L+R+ ++T
Sbjct: 465 VVEAYCLVNQKLQDSVSEQSNMEKFIMELKADLRRRERENT 505
>At1g13220 putative nuclear matrix constituent protein
Length = 1128
Score = 48.1 bits (113), Expect = 6e-06
Identities = 51/242 (21%), Positives = 101/242 (41%), Gaps = 35/242 (14%)
Query: 9 LGMMTNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKL--SETAEAA 66
L M +K T+ +D+T E L K++E L +++ +R EKL E E
Sbjct: 316 LSMSKSKETE---EDITKRLEELTTKEKEAHTLQITLLAKENELRAFEEKLIAREGTEIQ 372
Query: 67 EAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQR 126
+ + + E E ++K +K+L+ +K++E E + + + +E+L K+
Sbjct: 373 KLIDDQKEVLGSKMLEFELECEEIRKSLDKELQ---RKIEELERQKVEIDHSEEKLEKRN 429
Query: 127 EAATQ------------EANMW----RSELGKARESGVILEGA-----------LARAEE 159
+A + EA + R ++ +A E + LE L + E
Sbjct: 430 QAMNKKFDRVNEKEMDLEAKLKTIKEREKIIQAEEKRLSLEKQQLLSDKESLEDLQQEIE 489
Query: 160 KVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHIDSTEVFEKTESCSD 219
K+R E I+E + E++E L +LK+Q+++ + + ++ E+
Sbjct: 490 KIRAEMTKKEEMIEEECKSLEIKKEEREEYLRLQSELKSQIEKSRVHEEFLSKEVENLKQ 549
Query: 220 TK 221
K
Sbjct: 550 EK 551
Score = 43.1 bits (100), Expect = 2e-04
Identities = 48/224 (21%), Positives = 103/224 (45%), Gaps = 27/224 (12%)
Query: 27 MKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTID------EQW 80
+++ L +K++EL+ + R + S ++ E +++ E AHT+ E
Sbjct: 296 IEKKLKLKEKELEEWNRKVDLSMSKSKETEEDITKRLEELTTKEKEAHTLQITLLAKENE 355
Query: 81 RNACEEI------ESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEAN 134
A EE ++K + Q E+ K+ E+E++ + K L K+ + +E
Sbjct: 356 LRAFEEKLIAREGTEIQKLIDDQKEVLGSKMLEFELECEEIRK---SLDKELQRKIEELE 412
Query: 135 MWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIK------EAVQGESAAVS-EKQ 187
+ E+ + E A+ + ++V E + EAK+K + +Q E +S EKQ
Sbjct: 413 RQKVEIDHSEEKLEKRNQAMNKKFDRVNEKEMDLEAKLKTIKEREKIIQAEEKRLSLEKQ 472
Query: 188 ELLA---YVDKLKAQLQRQHIDSTEVFEKTESCSDTKHVDLTEE 228
+LL+ ++ L+ ++++ + T+ E E + K +++ +E
Sbjct: 473 QLLSDKESLEDLQQEIEKIRAEMTKKEEMIE--EECKSLEIKKE 514
Score = 40.0 bits (92), Expect = 0.002
Identities = 40/148 (27%), Positives = 66/148 (44%), Gaps = 14/148 (9%)
Query: 36 EELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEE--IESLKKD 93
+EL+ R+++ +S IR +E A A A+ + + E + E E+ +K
Sbjct: 164 QELEKALREIQEENSKIRLSSEAKLVEANALVASVNGRSSDVENKIYSAESKLAEATRKS 223
Query: 94 SEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREA-------ATQEANMWRSELGKARES 146
SE +L +LKE E + L +E+ +K+RE+ + N W +L ES
Sbjct: 224 SELKL-----RLKEVETRESVLQQERLSFTKERESYEGTFQKQREYLNEWEKKLQGKEES 278
Query: 147 GVILEGALARAEEKVRVAEANAEAKIKE 174
+ L + EEKV E + K KE
Sbjct: 279 ITEQKRNLNQREEKVNEIEKKLKLKEKE 306
Score = 39.3 bits (90), Expect = 0.003
Identities = 47/248 (18%), Positives = 111/248 (43%), Gaps = 30/248 (12%)
Query: 28 KETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNA---- 83
KE+L +E++ ++ ++ I + + L E E + Q +
Sbjct: 478 KESLEDLQQEIEKIRAEMTKKEEMIEEECKSLEIKKEEREEYLRLQSELKSQIEKSRVHE 537
Query: 84 ---CEEIESLKKDSE---KQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWR 137
+E+E+LK++ E K+ E+ +K Y + I +S+EKE+ + +
Sbjct: 538 EFLSKEVENLKQEKERFEKEWEILDEKQAVYNKERIRISEEKEKF--------ERFQLLE 589
Query: 138 SELGKARESGVILEGALARAEEKVRVA----EANAE---AKIKEAVQGESAAVSEKQELL 190
E K ES + ++ + + + +R+ EAN E + ++E V+ E + V + E++
Sbjct: 590 GERLKKEESALRVQ--IMQELDDIRLQRESFEANMEHERSALQEKVKLEQSKVIDDLEMM 647
Query: 191 AYVDKLKAQLQRQHIDSTEVFEKTESCSDTKHVDLTEENVDKACLSVSRSVPDPAAAECV 250
+++ Q +R+ D ++ ++ D + +L++ N K L +R + + +
Sbjct: 648 RRNLEIELQ-ERKEQDEKDLLDRMAQFEDKRMAELSDINHQKQAL--NREMEEMMSKRSA 704
Query: 251 VQMPTDQV 258
+Q ++++
Sbjct: 705 LQKESEEI 712
Score = 39.3 bits (90), Expect = 0.003
Identities = 52/231 (22%), Positives = 100/231 (42%), Gaps = 40/231 (17%)
Query: 34 KDEELQNFARDLRTRDSTIRDIAEKLSETAEA-----------------AEAAASAAHTI 76
K EL+ +++ TR+S ++ E+LS T E + +I
Sbjct: 222 KSSELKLRLKEVETRESVLQQ--ERLSFTKERESYEGTFQKQREYLNEWEKKLQGKEESI 279
Query: 77 DEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKI-ISLSKEKEQ-----------LSK 124
EQ RN + E + + EK+L+L ++L+E+ K+ +S+SK KE +K
Sbjct: 280 TEQKRNLNQREEKV-NEIEKKLKLKEKELEEWNRKVDLSMSKSKETEEDITKRLEELTTK 338
Query: 125 QREAATQEANMWRSELGKARESGVILEGALARAEEKV-RVAEANAEAKIKEAVQGESAAV 183
++EA T + + L K E E +AR ++ ++ + E + ++ E
Sbjct: 339 EKEAHTLQITL----LAKENELRAFEEKLIAREGTEIQKLIDDQKEVLGSKMLEFELECE 394
Query: 184 SEKQELLAYVDKLKAQLQRQHID---STEVFEKTESCSDTKHVDLTEENVD 231
++ L + + +L+RQ ++ S E EK + K + E+ +D
Sbjct: 395 EIRKSLDKELQRKIEELERQKVEIDHSEEKLEKRNQAMNKKFDRVNEKEMD 445
Score = 37.7 bits (86), Expect = 0.008
Identities = 59/298 (19%), Positives = 127/298 (41%), Gaps = 20/298 (6%)
Query: 23 DLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRN 82
D + E + ++EL + ++ +++ K + +A + A
Sbjct: 81 DQEALLEKISTLEKELYGYQHNMGLLLMENKELVSKHEQLNQAFQEAQEILKREQSSHLY 140
Query: 83 ACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGK 142
A +E +++ K L L Q ++E E + + +E ++ EA EAN + +
Sbjct: 141 ALTTVEQREENLRKALGLEKQCVQELEKALREIQEENSKIRLSSEAKLVEANALVASVN- 199
Query: 143 ARESGVILEGALARAEEKVRVA---EANAEAKIKEAVQGESAAVSEKQELLAYVDKLKA- 198
R S V E + AE K+ A + + ++KE ES +QE L++ + ++
Sbjct: 200 GRSSDV--ENKIYSAESKLAEATRKSSELKLRLKEVETRESVL---QQERLSFTKERESY 254
Query: 199 ----QLQRQHIDSTE--VFEKTESCSDTK-HVDLTEENVDKA--CLSVSRSVPDPAAAEC 249
Q QR++++ E + K ES ++ K +++ EE V++ L + + +
Sbjct: 255 EGTFQKQREYLNEWEKKLQGKEESITEQKRNLNQREEKVNEIEKKLKLKEKELEEWNRKV 314
Query: 250 VVQMPTDQVIIQPVGDN-EWSDIQATEARVADVREVASESEVSSLDIPVISQQGTNHQ 306
+ M + + + E + EA + +A E+E+ + + +I+++GT Q
Sbjct: 315 DLSMSKSKETEEDITKRLEELTTKEKEAHTLQITLLAKENELRAFEEKLIAREGTEIQ 372
>At5g46070 putative protein
Length = 1060
Score = 47.0 bits (110), Expect = 1e-05
Identities = 56/255 (21%), Positives = 115/255 (44%), Gaps = 23/255 (9%)
Query: 3 ISLRNTLGMMTNKTT--DGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLS 60
I+L N +T K T + LT+++ L V + +L++F +L + T+ ++ +KL
Sbjct: 669 ITLANKDEEITEKATKLEKAEQSLTVLRSDLKVAESKLESFEVELASLRLTLSEMTDKLD 728
Query: 61 ETAEAAEAAASAAHTIDEQ-------WRNACEEIESLKKDSEKQLELSAQKLKEY--EVK 111
+ A A A+ ++++ +R+ + + +K+ K E+ A++ E + +
Sbjct: 729 SANKKALAYEKEANKLEQEKIRMEQKYRSEFQRFDEVKERC-KAAEIEAKRATELADKAR 787
Query: 112 IISLSKEKEQLSKQREAATQEANMWRSE--LGKARESGVILEGALAR-------AEEKVR 162
+++ +KE+ QR A + A + R+E + LE L R A KV
Sbjct: 788 TDAVTSQKEKSESQRLAMERLAQIERAERQVENLERQKTDLEDELDRLRVSEMEAVSKVT 847
Query: 163 VAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHIDSTEVFE--KTESCSDT 220
+ EA E + KE +++ + ++KL + ++ HI + E E +
Sbjct: 848 ILEARVEEREKEIGSLIKETNAQRAHNVKSLEKLLDEERKAHIAANRRAEALSLELQAAQ 907
Query: 221 KHVDLTEENVDKACL 235
HVD ++ + +A L
Sbjct: 908 AHVDNLQQELAQARL 922
Score = 36.6 bits (83), Expect = 0.017
Identities = 57/241 (23%), Positives = 98/241 (40%), Gaps = 42/241 (17%)
Query: 4 SLRNTLGMMTNKTTDGPMDDLTIMKETLLVKDEELQ-------NFAR--DLRTRDSTIRD 54
SLR TL MT+K L KE ++ E+++ F R +++ R
Sbjct: 715 SLRLTLSEMTDKLDSANKKALAYEKEANKLEQEKIRMEQKYRSEFQRFDEVKERCKAAEI 774
Query: 55 IAEKLSETAEAAEA-AASAAHTIDEQWRNACEEIESLKKDSEKQLE------------LS 101
A++ +E A+ A A ++ E R A E + +++ +E+Q+E L
Sbjct: 775 EAKRATELADKARTDAVTSQKEKSESQRLAMERLAQIER-AERQVENLERQKTDLEDELD 833
Query: 102 AQKLKEYEV--KIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGALARAEE 159
++ E E K+ L E+ K+ + +E N R+ K+ E L E
Sbjct: 834 RLRVSEMEAVSKVTILEARVEEREKEIGSLIKETNAQRAHNVKSLEK-------LLDEER 886
Query: 160 KVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHIDSTEVFEKTESCSD 219
K +A AN A+ A E Q A+VD L+ +L + + T + K + S
Sbjct: 887 KAHIA-ANRRAE---------ALSLELQAAQAHVDNLQQELAQARLKETALDNKIRAASS 936
Query: 220 T 220
+
Sbjct: 937 S 937
Score = 28.1 bits (61), Expect = 6.2
Identities = 33/198 (16%), Positives = 78/198 (38%), Gaps = 22/198 (11%)
Query: 9 LGMMTNKTTDGPMDDLT--------IMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLS 60
L + ++ +GP+ DLT I K +L +K +++ + L+ + + D
Sbjct: 463 LSVFLQQSLEGPIYDLTKRLIDSIAIEKNSLAMKFRSVEDAMKHLKQQ---LDDSERYKL 519
Query: 61 ETAEAAEAAASAAHTIDEQWRNACEEIE-----------SLKKDSEKQLELSAQKLKEYE 109
E + + + + +++ +R +++ +L K E + E + ++ Y+
Sbjct: 520 EYQKRYDESNNDKKKLEDIYRERITKLQGENSSLNERCSTLVKTVESKKEEIKEWIRNYD 579
Query: 110 VKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAE 169
++ +EQLS + E + + + ARE + K A A
Sbjct: 580 QIVLKQKAVQEQLSSEMEVLRTRSTTSEARVAAAREQAKSAAEETKEWKRKYDYAVGEAR 639
Query: 170 AKIKEAVQGESAAVSEKQ 187
+ +++A + + E Q
Sbjct: 640 SALQKAASVQERSGKETQ 657
>At3g12020 unknown protein
Length = 956
Score = 46.6 bits (109), Expect = 2e-05
Identities = 51/198 (25%), Positives = 89/198 (44%), Gaps = 16/198 (8%)
Query: 34 KDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAA----HTIDEQWRNACEEIES 89
K +EL+ A++L + KL+E + A+ ASAA + E+ + E
Sbjct: 768 KIKELKQDAKELSESKEQLELRNRKLAEESSYAKGLASAAAVELKALSEEVAKLMNQNER 827
Query: 90 LKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVI 149
L + Q AQ+ K + + +E L+K++E + + R EL ++E +
Sbjct: 828 LAAELATQKSPIAQRNKTGTTTNVRNNGRRESLAKRQEHDSPSMELKR-ELRMSKERELS 886
Query: 150 LEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHIDSTE 209
E AL E++ EA E ++E Q E+ +E + V KL+ + Q DS
Sbjct: 887 YEAALGEKEQR----EAELERILEETKQREAYLENELANMWVLVSKLR---RSQGADS-- 937
Query: 210 VFEKTESCSDTKHVDLTE 227
E ++S S+T+ + TE
Sbjct: 938 --EISDSISETRQTEQTE 953
>At4g40020 putative protein
Length = 615
Score = 46.2 bits (108), Expect = 2e-05
Identities = 46/213 (21%), Positives = 89/213 (41%), Gaps = 13/213 (6%)
Query: 35 DEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESL---K 91
D +Q+F D+ + + + E L++ EAA+A++ + E+ ++ E++S +
Sbjct: 115 DSSVQDF--DIESLKTEMESTKESLAQAHEAAQASSLKVSELLEEMKSVKNELKSATDAE 172
Query: 92 KDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILE 151
+EK ++ A LKE ++ + + EAA E+ W+ + + R+ +L+
Sbjct: 173 MTNEKAMDDLALALKEVATDCSQTKEKLVIVETELEAARIESQQWKDKYEEVRKDAELLK 232
Query: 152 GALAR----AEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHIDS 207
R AEE + + +GE EK LL ++L L S
Sbjct: 233 NTSERLRIEAEESLLAWNGKESVFVTCIKRGE----DEKNSLLDENNRLLEALVAAENLS 288
Query: 208 TEVFEKTESCSDTKHVDLTEENVDKACLSVSRS 240
+ E+ D + E NV K ++R+
Sbjct: 289 KKAKEENHKVRDILKQAINEANVAKEAAGIARA 321
Score = 40.8 bits (94), Expect = 0.001
Identities = 61/242 (25%), Positives = 101/242 (41%), Gaps = 24/242 (9%)
Query: 4 SLRNTLGMMTNK--TTDGPMDDLTIM-----------KETLLVKDEELQNF---ARDLRT 47
S++N L T+ T + MDDL + KE L++ + EL+ ++ +
Sbjct: 160 SVKNELKSATDAEMTNEKAMDDLALALKEVATDCSQTKEKLVIVETELEAARIESQQWKD 219
Query: 48 RDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKE 107
+ +R AE L T+E A + + +D + L +L E
Sbjct: 220 KYEEVRKDAELLKNTSERLRIEAEESLLAWNGKESVFVTCIKRGEDEKNSLLDENNRLLE 279
Query: 108 YEVKIISLSKE-KEQLSKQREAATQ---EANMWRSELGKARESGVILEGALARAEEKVRV 163
V +LSK+ KE+ K R+ Q EAN+ + G AR L+ AL EE+++
Sbjct: 280 ALVAAENLSKKAKEENHKVRDILKQAINEANVAKEAAGIARAENSNLKDALLDKEEELQF 339
Query: 164 AEANAE-AKIKEAVQGESAAVSEK---QELLAYVDKLKAQLQRQHIDSTEVFEKTESCSD 219
A E K+ EAV ++ +K + +A ++ + L RQ EV E E +
Sbjct: 340 ALKEIERVKVNEAVANDNIKKLKKMLSEIEVAMEEEKQRSLNRQESMPKEVVEVVEKKIE 399
Query: 220 TK 221
K
Sbjct: 400 EK 401
Score = 33.5 bits (75), Expect = 0.15
Identities = 24/116 (20%), Positives = 59/116 (50%), Gaps = 9/116 (7%)
Query: 27 MKETLLVKDEELQNFARDL---RTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNA 83
+K+ LL K+EELQ +++ + ++ D +KL + E A ++E+ + +
Sbjct: 326 LKDALLDKEEELQFALKEIERVKVNEAVANDNIKKLKKMLSEIEVA------MEEEKQRS 379
Query: 84 CEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSE 139
ES+ K+ + +E ++ ++ E K + ++KE +++E + ++ + + E
Sbjct: 380 LNRQESMPKEVVEVVEKKIEEKEKKEEKKENKKEKKESKKEKKEHSEKKEDKEKKE 435
>At2g37080 putative myosin heavy chain
Length = 583
Score = 45.8 bits (107), Expect = 3e-05
Identities = 42/160 (26%), Positives = 81/160 (50%), Gaps = 17/160 (10%)
Query: 44 DLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEI---ESLKKDSEKQLEL 100
D R+ + + +I +K T + E A+ + + E+ + A E++ E+LKK+++ Q E
Sbjct: 57 DRRSPRTPVNEIQKK--RTGKTPELASQISQ-LQEELKKAKEQLSASEALKKEAQDQAEE 113
Query: 101 SAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGALARAEEK 160
+ Q+L E S E +LS++R+ A W+SEL + + AL+ +
Sbjct: 114 TKQQLMEINASEDSRIDELRKLSQERDKA------WQSELEAMQRQHAMDSAALSSTMNE 167
Query: 161 VRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQL 200
V+ +A++ E+ E+ + E E L+ V+KL+ +L
Sbjct: 168 VQ----KLKAQLSESENVENLRM-ELNETLSLVEKLRGEL 202
>At2g32240 putative myosin heavy chain
Length = 1333
Score = 45.4 bits (106), Expect = 4e-05
Identities = 46/175 (26%), Positives = 79/175 (44%), Gaps = 13/175 (7%)
Query: 34 KDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKD 93
KD+EL + +KL E E + +A A +E + + +S +
Sbjct: 175 KDKELTEVKEAFDALGIELESSRKKLIELEEGLKRSAEEAQKFEELHKQSASHADS---E 231
Query: 94 SEKQLELSA------QKLKEYEVKIISLSKEKEQLS-KQREAATQEANMWRS--ELGKAR 144
S+K LE S + KE E K+ SL +E ++L+ K E EA + S EL +
Sbjct: 232 SQKALEFSELLKSTKESAKEMEEKMASLQQEIKELNEKMSENEKVEAALKSSAGELAAVQ 291
Query: 145 ESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQ 199
E + + L E+KV EA + ++ + ++ + A+ S +E L+ + L AQ
Sbjct: 292 EELALSKSRLLETEQKVSSTEALID-ELTQELEQKKASESRFKEELSVLQDLDAQ 345
Score = 40.4 bits (93), Expect = 0.001
Identities = 63/284 (22%), Positives = 119/284 (41%), Gaps = 32/284 (11%)
Query: 27 MKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEE 86
+ E L + +++++ L + EKL +T AA S + +++ A E+
Sbjct: 825 LTEKLRDLEGKIKSYEEQLAEASGKSSSLKEKLEQTLGRLAAAESVNEKLKQEFDQAQEK 884
Query: 87 IESLKKDSEKQLELSAQ---KLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKA 143
+SE E + Q K++E E I S S EKE K+ E A + N K
Sbjct: 885 SLQSSSESELLAETNNQLKIKIQELEGLIGSGSVEKETALKRLEEAIERFNQ------KE 938
Query: 144 RESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQ 203
ES ++ EK++ E E K A + A + K EL + KLK
Sbjct: 939 TESSDLV--------EKLKTHENQIEEYKKLAHEASGVADTRKVELEDALSKLK------ 984
Query: 204 HIDSTEVFEKTESCSDTKHVDLTEENVDKACLSVSRSVPDPAAAECVVQMPTDQVIIQPV 263
+++ST + E C L +E+ D A +++ ++ ++ T ++
Sbjct: 985 NLEST-IEELGAKCQ-----GLEKESGDLAEVNLKLNLELANHGSEANELQTKLSALEAE 1038
Query: 264 GDNEWSDIQATEARVADV-REVASESEVSSLDIPVISQQGTNHQ 306
+ ++++A++ + D+ +++ SE E L + S N+Q
Sbjct: 1039 KEQTANELEASKTTIEDLTKQLTSEGE--KLQSQISSHTEENNQ 1080
Score = 39.7 bits (91), Expect = 0.002
Identities = 45/211 (21%), Positives = 82/211 (38%), Gaps = 24/211 (11%)
Query: 34 KDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKD 93
K EL+ R+S + + + E A+ H + C+ +S +D
Sbjct: 565 KASELELSLTQSSARNSELEEDLRIALQKGAEHEDRANTTHQRSIELEGLCQSSQSKHED 624
Query: 94 SE---KQLELSAQ----KLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARES 146
+E K LEL Q +++E E ++ SL K+ + + + +S L +
Sbjct: 625 AEGRLKDLELLLQTEKYRIQELEEQVSSLEKKHGETEADSKGYLGQVAELQSTLEAFQVK 684
Query: 147 GVILEGALARAEEKVRVAEANAEA------KIKEAVQGESAAVSEKQELLAYV------- 193
LE AL A E + N A K++ V S +SE + LL +
Sbjct: 685 SSSLEAALNIATENEKELTENLNAVTSEKKKLEATVDEYSVKISESENLLESIRNELNVT 744
Query: 194 ----DKLKAQLQRQHIDSTEVFEKTESCSDT 220
+ ++ L+ + +EV EK +S ++
Sbjct: 745 QGKLESIENDLKAAGLQESEVMEKLKSAEES 775
Score = 39.7 bits (91), Expect = 0.002
Identities = 38/176 (21%), Positives = 73/176 (40%), Gaps = 4/176 (2%)
Query: 27 MKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEE 86
++E L DE L S ++ +KL E A SAA + +N E
Sbjct: 422 LEEKLKTSDENFSKTDALLSQALSNNSELEQKLKSLEELHSEAGSAAAAATQ--KNL--E 477
Query: 87 IESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARES 146
+E + + S + E + ++KE E K + ++ +L +Q +++ EL + E
Sbjct: 478 LEDVVRSSSQAAEEAKSQIKELETKFTAAEQKNAELEQQLNLLQLKSSDAERELKELSEK 537
Query: 147 GVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQR 202
L+ A+ AEE+ + A + ++A + E + + L+ LQ+
Sbjct: 538 SSELQTAIEVAEEEKKQATTQMQEYKQKASELELSLTQSSARNSELEEDLRIALQK 593
Score = 38.5 bits (88), Expect = 0.005
Identities = 54/225 (24%), Positives = 99/225 (44%), Gaps = 40/225 (17%)
Query: 24 LTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAE---------------- 67
L ++ L V +L++ DL+ ++ EKL E+ E
Sbjct: 734 LESIRNELNVTQGKLESIENDLKAAGLQESEVMEKLKSAEESLEQKGREIDEATTKRMEL 793
Query: 68 AAASAAHTIDEQWR--NACEEIESLKKDSE-----KQLELSAQKLKEYEVKIISLS---- 116
A + +ID + R A EE S +DSE ++L K+K YE ++ S
Sbjct: 794 EALHQSLSIDSEHRLQKAMEEFTS--RDSEASSLTEKLRDLEGKIKSYEEQLAEASGKSS 851
Query: 117 --KEKEQLSKQREAATQEAN-MWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIK 173
KEK + + R AA + N + E +A+E +L + E +AE N + KIK
Sbjct: 852 SLKEKLEQTLGRLAAAESVNEKLKQEFDQAQEK------SLQSSSESELLAETNNQLKIK 905
Query: 174 -EAVQGESAAVS-EKQELLAYVDKLKAQLQRQHIDSTEVFEKTES 216
+ ++G + S EK+ L +++ + ++ +S+++ EK ++
Sbjct: 906 IQELEGLIGSGSVEKETALKRLEEAIERFNQKETESSDLVEKLKT 950
Score = 36.6 bits (83), Expect = 0.017
Identities = 57/302 (18%), Positives = 123/302 (39%), Gaps = 24/302 (7%)
Query: 16 TTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHT 75
+T+ +D+LT E + + L+ D+ + + KLSE + A
Sbjct: 310 STEALIDELTQELEQKKASESRFKEELSVLQDLDAQTKGLQAKLSEQEGIN---SKLAEE 366
Query: 76 IDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANM 135
+ E+ E +ESL KD E++L + +KL E + +L +++ T+ N
Sbjct: 367 LKEK-----ELLESLSKDQEEKLRTANEKLAEVLKEKEALEANVAEVTSNVATVTEVCNE 421
Query: 136 WRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDK 195
+L + E+ + L++A E ++ + + SAA + Q+ L D
Sbjct: 422 LEEKLKTSDENFSKTDALLSQALSNNSELEQKLKSLEELHSEAGSAAAAATQKNLELEDV 481
Query: 196 LKAQLQRQHIDSTEV--FEKTESCSDTKHVDLTEE---------NVDKACLSVSRSVPDP 244
+++ Q +++ E + ++ K+ +L ++ + ++ +S +
Sbjct: 482 VRSSSQAAEEAKSQIKELETKFTAAEQKNAELEQQLNLLQLKSSDAERELKELSEKSSEL 541
Query: 245 AAAECVVQMPTDQVIIQPVGDNEWSDIQATEARVADVREVASESEVSSLDIPVISQQGTN 304
A V + Q Q E+ +A+E ++ + A SE+ D+ + Q+G
Sbjct: 542 QTAIEVAEEEKKQATTQM---QEYKQ-KASELELSLTQSSARNSELEE-DLRIALQKGAE 596
Query: 305 HQ 306
H+
Sbjct: 597 HE 598
Score = 35.8 bits (81), Expect = 0.030
Identities = 37/177 (20%), Positives = 70/177 (38%), Gaps = 19/177 (10%)
Query: 52 IRDIAEKLSETAEAA-------EAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQK 104
+ D+ S+ AE A E +AA + + ++ D+E++L+ ++K
Sbjct: 478 LEDVVRSSSQAAEEAKSQIKELETKFTAAEQKNAELEQQLNLLQLKSSDAERELKELSEK 537
Query: 105 LKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVA 164
E + I +EK+Q + Q + Q+A+ L ++ LE L A +K
Sbjct: 538 SSELQTAIEVAEEEKKQATTQMQEYKQKASELELSLTQSSARNSELEEDLRIALQKGAEH 597
Query: 165 EANAEAKIKEAVQGESAAVSEKQ------------ELLAYVDKLKAQLQRQHIDSTE 209
E A + +++ E S + ELL +K + Q + + S E
Sbjct: 598 EDRANTTHQRSIELEGLCQSSQSKHEDAEGRLKDLELLLQTEKYRIQELEEQVSSLE 654
Score = 34.3 bits (77), Expect = 0.087
Identities = 39/220 (17%), Positives = 87/220 (38%), Gaps = 8/220 (3%)
Query: 15 KTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAH 74
KT+D + L + EL+ + L S A ++ E ++
Sbjct: 427 KTSDENFSKTDALLSQALSNNSELEQKLKSLEELHSEAGSAAAAATQKNLELEDVVRSSS 486
Query: 75 TIDEQWRNACEEIES-------LKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQRE 127
E+ ++ +E+E+ + E+QL L K + E ++ LS++ +L E
Sbjct: 487 QAAEEAKSQIKELETKFTAAEQKNAELEQQLNLLQLKSSDAERELKELSEKSSELQTAIE 546
Query: 128 AATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQ 187
A +E +++ + ++ LE +L ++ + E + +++ + E A + Q
Sbjct: 547 VAEEEKKQATTQMQEYKQKASELELSLTQSSARNSELEEDLRIALQKGAEHEDRANTTHQ 606
Query: 188 ELLAYVDKLKAQLQRQHIDSTEVFEKTESCSDTKHVDLTE 227
+ ++ L Q +H D+ + E T+ + E
Sbjct: 607 RSIE-LEGLCQSSQSKHEDAEGRLKDLELLLQTEKYRIQE 645
Score = 29.6 bits (65), Expect = 2.1
Identities = 28/123 (22%), Positives = 55/123 (43%), Gaps = 21/123 (17%)
Query: 27 MKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAA--------------EAAASA 72
+++TL +L+ + T + ++ KL E A + +A
Sbjct: 1137 LEKTLSEVKAQLKENVENAATASVKVAELTSKLQEHEHIAGERDVLNEQVLQLQKELQAA 1196
Query: 73 AHTIDEQWRNACE---EIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAA 129
+IDEQ + + E+ES K S++++E + + E+E S+ K+ EQ + +A
Sbjct: 1197 QSSIDEQKQAHSQKQSELESALKKSQEEIEAKKKAVTEFE----SMVKDLEQKVQLADAK 1252
Query: 130 TQE 132
T+E
Sbjct: 1253 TKE 1255
Score = 28.9 bits (63), Expect = 3.6
Identities = 45/193 (23%), Positives = 73/193 (37%), Gaps = 32/193 (16%)
Query: 61 ETAEAAEAAASAAHTIDEQWRNACE-----------EIESLKKDSEKQLELSAQKLKEYE 109
E +A + A A H E+ + E E + K+ E +LE A +LK YE
Sbjct: 54 EAFDAKDDAEKADHVPVEEQKEVIERSSSGSQRELHESQEKAKELELELERVAGELKRYE 113
Query: 110 VKIISLSKE----KEQLSKQRE-----AATQEANMWRSELGKARESGVI--LEGAL---- 154
+ L E KE+L + + Q+ + G+ R S + LE AL
Sbjct: 114 SENTHLKDELLSAKEKLEETEKKHGDLEVVQKKQQEKIVEGEERHSSQLKSLEDALQSHD 173
Query: 155 ARAEEKVRVAEA------NAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHIDST 208
A+ +E V EA E+ K+ ++ E +E + + K +S
Sbjct: 174 AKDKELTEVKEAFDALGIELESSRKKLIELEEGLKRSAEEAQKFEELHKQSASHADSESQ 233
Query: 209 EVFEKTESCSDTK 221
+ E +E TK
Sbjct: 234 KALEFSELLKSTK 246
Score = 28.5 bits (62), Expect = 4.8
Identities = 30/117 (25%), Positives = 53/117 (44%), Gaps = 4/117 (3%)
Query: 99 ELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGALARAE 158
E+ ++ +E + + I + KE E A + E+ + SG E L ++
Sbjct: 35 EVPKEEKEEEDGEFIKVEKEAFDAKDDAEKADHVPVEEQKEVIERSSSGSQRE--LHESQ 92
Query: 159 EKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHIDSTEVFEKTE 215
EK + E E E + ES K ELL+ +KL+ + +++H D EV +K +
Sbjct: 93 EKAKELELELERVAGELKRYESENTHLKDELLSAKEKLE-ETEKKHGD-LEVVQKKQ 147
>At4g17210 putative protein
Length = 527
Score = 44.7 bits (104), Expect = 6e-05
Identities = 49/229 (21%), Positives = 95/229 (41%), Gaps = 26/229 (11%)
Query: 32 LVKDEELQNFARDL-RTRDSTIR---DIAEKLSETAEAAEAAASAAHTIDEQWRNACEEI 87
+V ++++ + D+ RD+ R D A K E + E + + T + A + +
Sbjct: 199 MVNSNKIKDMSHDIAEMRDAAERLNSDAARKKEEEEQIKEESIALRETYVCKKLEAKQRL 258
Query: 88 ESLKKDSEKQLELSAQKLKEYEV------KIISLSKEKEQLSKQREAATQEANMWRSELG 141
E LK+D + +L+ ++L E + I LS E ++ + E + ++S +G
Sbjct: 259 EDLKRDCDPELKKDIEELMEISTENERLQEEIKLSGELKEAKSAMQEIYDEESSYKSLVG 318
Query: 142 K--ARESGVILEGALARAEEKVRVAE-----ANAEAKIKEAVQGESAAVSEKQELLAYVD 194
GV E + +EK R A K+ E ++ E +E+ VD
Sbjct: 319 SLTVELDGVQRENRELKGKEKERQEAEEGEWVEASRKVDEIMREAEKTRKEAEEMRMNVD 378
Query: 195 KLKAQLQRQHIDSTEVFEKTESCSDTKHVDLTEENVDKACLSVSRSVPD 243
+L+ + +H+ E K +++ V+KA + R+V D
Sbjct: 379 ELRREAAAKHMVMGEA---------VKQLEIVGRAVEKAKTAEKRAVED 418
>At5g52280 hyaluronan mediated motility receptor-like protein
Length = 853
Score = 44.3 bits (103), Expect = 8e-05
Identities = 37/154 (24%), Positives = 73/154 (47%), Gaps = 9/154 (5%)
Query: 58 KLSETAEAAEAAASAAHTIDEQWRNACEEIE-------SLKKDSEKQLELSAQKLKEYEV 110
KL++ + + AAA+ I ++WR +E E + K ++K+L L+ + E
Sbjct: 663 KLTKLRDESSAAATETEKIIQEWRKERDEFERKLSLAKEVAKTAQKELTLTKSSNDDKET 722
Query: 111 KIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKV-RVAEANAE 169
++ +L E E LS Q + E + R+ L+ + R EE++ ++ +A E
Sbjct: 723 RLRNLKTEVEGLSLQYSELQNSFVQEKMENDELRKQVSNLKVDIRRKEEEMTKILDARME 782
Query: 170 AKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQ 203
A+ +E E +S+ + LAY + ++R+
Sbjct: 783 ARSQENGHKEE-NLSKLSDELAYCKNKNSSMERE 815
Score = 32.7 bits (73), Expect = 0.25
Identities = 44/194 (22%), Positives = 92/194 (46%), Gaps = 15/194 (7%)
Query: 31 LLVKD--EELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIE 88
L V+D E L+ ++ + +S + + A+KL E + ++ + T+ +Q + E++
Sbjct: 384 LAVRDLNEMLEQKNNEISSLNSLLEE-AKKLEEH-KGMDSGNNEIDTLKQQIEDLDWELD 441
Query: 89 SLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSE--LGKARES 146
S KK +E+Q L + +EYE K +Q+E + E S+ + + +
Sbjct: 442 SYKKKNEEQEILLDELTQEYESLKEENYKNVSSKLEQQECSNAEDEYLDSKDIIDELKSQ 501
Query: 147 GVILEGALAR-----AEEKVRVAEANAEAK-IKEAVQGESAAVSEKQELLAYVDKLKAQL 200
ILEG L + +E + V E ++ K +K+ ++ ++ A E + + + K +
Sbjct: 502 IEILEGKLKQQSLEYSECLITVNELESQVKELKKELEDQAQAYDEDIDTMM---REKTEQ 558
Query: 201 QRQHIDSTEVFEKT 214
+++ I + E KT
Sbjct: 559 EQRAIKAEENLRKT 572
>At1g14840 unknown protein
Length = 604
Score = 43.9 bits (102), Expect = 1e-04
Identities = 43/177 (24%), Positives = 77/177 (43%), Gaps = 27/177 (15%)
Query: 21 MDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQW 80
+D LT KE L+ E A + S + D+ K E + E ID+
Sbjct: 182 LDRLTKSKEAALLDAERTVQSAL---AKASMVDDLQNKNQELMKQIEICQEENRIIDKMH 238
Query: 81 RNACEEIESLKKDSEKQLE-------LSAQKLKEYEVKIISLSKEKEQLSKQ-------- 125
R E+E L + S ++LE +A +++Y+ K +++E++ L ++
Sbjct: 239 RQKVAEVEKLMQ-SVRELEEAVLAGGAAANAVRDYQRKFQEMNEERKILERELARAKVNA 297
Query: 126 REAATQEANMWRSELGKAR------ESGVILEGALARAEEKVRVAE--ANAEAKIKE 174
AT AN W+ K E L+G + + +K+ +A+ A +EA++KE
Sbjct: 298 NRVATVVANEWKDSNDKVMPVRQWLEERRFLQGEMQQLRDKLAIADRAAKSEAQLKE 354
>At4g38065 hypothetical protein
Length = 1050
Score = 43.5 bits (101), Expect = 1e-04
Identities = 42/221 (19%), Positives = 99/221 (44%), Gaps = 15/221 (6%)
Query: 4 SLRNTLGMMTNKTTDGPMDDLTIMKETLLVKDEELQNFAR--DLRTRDSTIRDIAEKLSE 61
SL+ + ++ + ++D+ + ++ + ELQN L+ +D + ++ +L
Sbjct: 802 SLQREVELLEQDSLRRELEDVVLAH---MIGERELQNEREICALQQKDQDLCEVKHELEG 858
Query: 62 TAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQ 121
+ ++ R E++ + + + + E + E E +I SLS++ E
Sbjct: 859 SLKSVSLLLQQKQNEVNMLRKTWEKLTARQILTAVETESKKMMIIELEGEISSLSQKLET 918
Query: 122 LSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESA 181
++ QEA R+EL + L+ + +EK+R +EA +KE ++
Sbjct: 919 SNESVSCFRQEATKSRAELETKQTE---LKEVTTQMQEKLRTSEAEKTELVKEV----AS 971
Query: 182 AVSEKQELLAYVDKLKAQLQRQHIDSTEV---FEKTESCSD 219
+EK+ LL+++ +++ + + + T++ E+ C D
Sbjct: 972 LSTEKRNLLSFISEMEDGMLKLYDGDTKLMKTLERVTQCCD 1012
Score = 32.7 bits (73), Expect = 0.25
Identities = 44/180 (24%), Positives = 82/180 (45%), Gaps = 23/180 (12%)
Query: 54 DIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKK---DSEKQLELSAQ-KLKEYE 109
+I EK E AE A +E+ + E +S+ K D +L + + K +E+E
Sbjct: 56 EIEEKSREIAELKRA--------NEELQRCLREKDSVVKRVNDVNDKLRANGEDKYREFE 107
Query: 110 VKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAE 169
+ ++ ++ S++ Q+ N++R+E+ L+G LA AE K AE +
Sbjct: 108 EEKRNMMSGLDEASEKNIDLEQKNNVYRAEIEG-------LKGLLAVAETKRIEAEKTVK 160
Query: 170 AKIKEAVQGESAAVSEKQELLAYVDKLK-AQLQRQHIDSTEVFEKTESCSDTKHVDLTEE 228
+KE + V ++E +KLK + Q +H++ E +EK ++ + EE
Sbjct: 161 G-MKEMRGRDDVVVKMEEEKSQVEEKLKWKKEQFKHLE--EAYEKLKNLFKDSKKEWEEE 217
>At2g27170 putative chromosome associated protein
Length = 1163
Score = 43.5 bits (101), Expect = 1e-04
Identities = 42/202 (20%), Positives = 84/202 (40%), Gaps = 14/202 (6%)
Query: 19 GPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDE 78
G + LT+MK+ ++ +L R + R+ + ET + H +DE
Sbjct: 142 GKIASLTLMKD---IERLDLLKEIGGTRVYEERRRESLRIMQETGNKRKQIIEVVHYLDE 198
Query: 79 QWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRS 138
+ R EE E L+K + + + + Y+ ++ ++ EQ+ R A++E+
Sbjct: 199 RLRELDEEKEELRKYQQLDKQRKSLEYTIYDKELHDAREKLEQVEVARTKASEESTKMYD 258
Query: 139 ELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLA-----YV 193
+ KA++ L+ +L K E K KE V+ + +K+ L +
Sbjct: 259 RVEKAQDDSKSLDESL-----KELTKELQTLYKEKETVEAQQTKALKKKTKLELDVKDFQ 313
Query: 194 DKLKAQLQRQHIDSTEVFEKTE 215
D++ +Q ++ D+ E E
Sbjct: 314 DRITGNIQSKN-DALEQLNTVE 334
Score = 37.7 bits (86), Expect = 0.008
Identities = 41/201 (20%), Positives = 97/201 (47%), Gaps = 25/201 (12%)
Query: 14 NKTTDGPMDDLTIMKETLLVKDEEL---QNFARDLRTR-DSTIRDIAEKLSETAEAAEAA 69
+K+ D + +LT +TL + E + Q A +T+ + ++D ++++ ++ A
Sbjct: 267 SKSLDESLKELTKELQTLYKEKETVEAQQTKALKKKTKLELDVKDFQDRITGNIQSKNDA 326
Query: 70 ASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAA 129
+T++ + +++ E+E++K E Q++ Q K +I L K L +++ A
Sbjct: 327 LEQLNTVEREMQDSLRELEAIKPLYESQVDKENQTSK----RINELEKTLSILYQKQGRA 382
Query: 130 TQEANMWRSELGKARESGVILEGALARAEEKVRVAEANA--EAKIKEAVQGESAAVSEKQ 187
TQ +N AR+ + E E+ RV ++N E K+++ + + ++E+
Sbjct: 383 TQFSNK------AARDKWLRKE-----IEDLKRVLDSNTVQEQKLQDEILRLNTDLTERD 431
Query: 188 ELL----AYVDKLKAQLQRQH 204
E + + +L++++ + H
Sbjct: 432 EHIKKHEVEIGELESRISKSH 452
>At1g68060 unknown protein
Length = 622
Score = 43.5 bits (101), Expect = 1e-04
Identities = 50/204 (24%), Positives = 84/204 (40%), Gaps = 39/204 (19%)
Query: 44 DLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEI--ESLKKDSEKQLELS 101
+L ++ +RD +LSE A A A + Q ACEE+ E K D + +L S
Sbjct: 71 ELNRLENEVRDKDRELSE----ANAEIKALRLSERQREKACEELTDELAKLDGKLKLTES 126
Query: 102 AQKLKEYEVKIISLSKEKEQLSKQREAAT------------------------QEANMWR 137
+ K E+K I+ K+ ++ AT E + R
Sbjct: 127 LLQSKNLEIKKINEEKKASMAAQFAAEATLRRVHAAQKDDDMPPIEAILAPLEAELKLAR 186
Query: 138 SELGKARESGVIL-------EGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELL 190
SE+GK +E L E AL AE V A A+A + + +Q ++ + ++ E+
Sbjct: 187 SEIGKLQEDNRALDRLTKSKEAALLDAERTVET--ALAKAALVDDLQNKNQELMKQIEIC 244
Query: 191 AYVDKLKAQLQRQHIDSTEVFEKT 214
+K+ ++ RQ + E +T
Sbjct: 245 QEENKILDRMHRQKVAEVEKLTQT 268
>At4g18240 starch synthase-like protein
Length = 1071
Score = 43.1 bits (100), Expect = 2e-04
Identities = 48/202 (23%), Positives = 85/202 (41%), Gaps = 17/202 (8%)
Query: 36 EELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSE 95
++L D I + KLSET E + AA ++ E++E L+ +
Sbjct: 211 DDLNKILSDKEALQGEINVLEMKLSETDERIKTAAQEKAHVELLE----EQLEKLRHEMI 266
Query: 96 KQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESG---VILEG 152
+E +++LSKE E L + + + M +SEL +++G V+LE
Sbjct: 267 SPIESDGY--------VLALSKELETLKLENLSLRNDIEMLKSELDSVKDTGERVVVLEK 318
Query: 153 ALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHIDSTEVFE 212
+ E V+ E+ +E V S E +L A V+ L+ L R + +
Sbjct: 319 ECSGLESSVKDLESKLSVS-QEDVSQLSTLKIECTDLWAKVETLQLLLDRATKQAEQAVI 377
Query: 213 KTESCSDTKH-VDLTEENVDKA 233
+ D ++ VD EE++ +A
Sbjct: 378 VLQQNQDLRNKVDKIEESLKEA 399
Score = 37.7 bits (86), Expect = 0.008
Identities = 42/204 (20%), Positives = 93/204 (45%), Gaps = 28/204 (13%)
Query: 3 ISLRNTLGMMTNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSET 62
+SLRN + M+ ++ +D + E ++V ++E +S+++D+ KLS +
Sbjct: 290 LSLRNDIEMLKSE-----LDSVKDTGERVVVLEKECSGL-------ESSVKDLESKLSVS 337
Query: 63 AEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQL 122
E ++ + W E ++ L + KQ E + L++ + + K +E L
Sbjct: 338 QEDVSQLSTLKIECTDLWAKV-ETLQLLLDRATKQAEQAVIVLQQNQDLRNKVDKIEESL 396
Query: 123 SKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAA 182
+EAN+++ K ++ +++ + EE++ ++A+I VQ +
Sbjct: 397 --------KEANVYKESSEKIQQYNELMQHKVTLLEERLE----KSDAEIFSYVQLYQES 444
Query: 183 VSEKQELLAYVDKLKAQLQRQHID 206
+ E QE L + LK + +++ D
Sbjct: 445 IKEFQETL---ESLKEESKKKSRD 465
>At3g51070 putative protein
Length = 895
Score = 43.1 bits (100), Expect = 2e-04
Identities = 36/145 (24%), Positives = 67/145 (45%), Gaps = 6/145 (4%)
Query: 49 DSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEY 108
D ++ E+ E +E +S T + Q N + E +KD+ K+ + + Q+ +E
Sbjct: 96 DDAVKSEDEQRKSAKEKSETTSSKTQTQETQQNNDDKISEEKEKDNGKENQ-TVQESEEG 154
Query: 109 EVKIISLSKEKEQLSKQRE-AATQEANMWRSELGKARESGVILEGALARAEE----KVRV 163
++K + EKEQ ++ E A TQ E G+ +E + +G E+ V
Sbjct: 155 QMKKVVKEFEKEQKQQRDEDAGTQPKGTQGQEQGQGKEQPDVEQGNKQGQEQDSNTDVTF 214
Query: 164 AEANAEAKIKEAVQGESAAVSEKQE 188
+A + + E QGE++ S+ +E
Sbjct: 215 TDATKQEQPMETGQGETSETSKNEE 239
>At1g80790 hypothetical protein
Length = 634
Score = 43.1 bits (100), Expect = 2e-04
Identities = 48/269 (17%), Positives = 110/269 (40%), Gaps = 26/269 (9%)
Query: 33 VKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESL-- 90
+ EE+QN + + D+A K++ T E + R E + L
Sbjct: 244 ITKEEIQN-------KSIVVDDLANKIAMTNEDLNKLQYMNNEKTLSLRRVLIEKDELDR 296
Query: 91 --KKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGV 148
K++++K ELS +K+ + +EKE+L+ + EA +W +L K +
Sbjct: 297 VYKQETKKMQELSREKINR-------IFREKERLTNELEAKMNNLKIWSKQLDKKQALTE 349
Query: 149 ILEGALARAEEKVRVAEANAEAKIKEAVQGESAAV-------SEKQELLAYVDKLKAQLQ 201
+ L ++K V ++ + E + + + +K+E L + +L+ +L
Sbjct: 350 LERQKLDEDKKKSDVMNSSLQLASLEQKKTDDRVLRLVDEHKRKKEETLNKILQLEKELD 409
Query: 202 RQHIDSTEVFEKTESCSDTKHVDLTEENVDKACLSVSRSVPDPAAAECVVQMPTDQVIIQ 261
+ E+ E KH D +E + K + + + + ++ ++++
Sbjct: 410 SKQKLQMEIQELKGKLKVMKHEDEDDEGIKKKMKKMKEELEEKCSELQDLEDTNSALMVK 469
Query: 262 PVGDNEWSDIQATEARVADVREVASESEV 290
N+ ++A + + ++RE+ S+ +
Sbjct: 470 ERKSND-EIVEARKFLITELRELVSDRNI 497
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.307 0.122 0.315
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,742,047
Number of Sequences: 26719
Number of extensions: 219676
Number of successful extensions: 1877
Number of sequences better than 10.0: 315
Number of HSP's better than 10.0 without gapping: 65
Number of HSP's successfully gapped in prelim test: 251
Number of HSP's that attempted gapping in prelim test: 1416
Number of HSP's gapped (non-prelim): 585
length of query: 306
length of database: 11,318,596
effective HSP length: 99
effective length of query: 207
effective length of database: 8,673,415
effective search space: 1795396905
effective search space used: 1795396905
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0057b.10


