

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0057b.1
(388 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
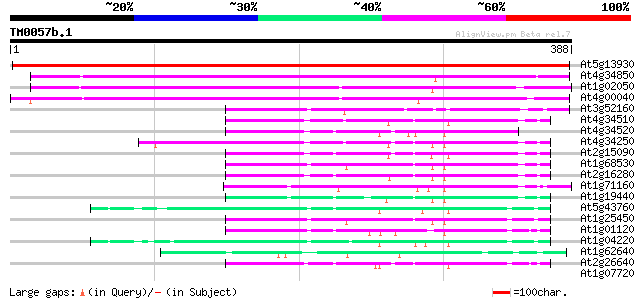
At5g13930 chalcone synthase (naringenin-chalcone synthase) (test... 644 0.0
At4g34850 chalcone synthase - like protein 282 3e-76
At1g02050 unknown protein 276 1e-74
At4g00040 chalcone synthase like protein 276 2e-74
At3g52160 beta-ketoacyl-CoA synthase like protein 64 1e-10
At4g34510 putative ketoacyl-CoA synthase 60 2e-09
At4g34520 fatty acid elongase 1 59 3e-09
At4g34250 fatty acid elongase - like protein 57 1e-08
At2g15090 putative fatty acid elongase 53 2e-07
At1g68530 very-long-chain fatty acid condensing enzyme (CUT1) 53 3e-07
At2g16280 putative beta-ketoacyl-CoA synthase 52 4e-07
At1g71160 putative ketoacyl-CoA synthase 52 6e-07
At1g19440 very-long-chain fatty acid condensing enzyme CUT1 like... 52 7e-07
At5g43760 beta-ketoacyl-CoA synthase 51 1e-06
At1g25450 unknown protein 50 2e-06
At1g01120 putative fatty acid elongase 3-ketoacyl-CoA synthase 1... 50 2e-06
At1g04220 beta-ketoacyl-CoA synthase like protein 49 4e-06
At1g62640 3-ketoacyl-acyl carrier protein synthase III (KAS III) 47 1e-05
At2g26640 putative beta-ketoacyl-CoA synthase 46 3e-05
At1g07720 fatty acid elongase 3-ketoacyl-CoA synthase, putative 38 0.011
>At5g13930 chalcone synthase (naringenin-chalcone synthase) (testa 4
protein) (sp|P13114)
Length = 395
Score = 644 bits (1660), Expect = 0.0
Identities = 319/386 (82%), Positives = 353/386 (90%), Gaps = 1/386 (0%)
Query: 3 SVADIRKAQRAKGPATIFAIGTANPPSCVDQSTYPDFYFRVTNSEHKAELKEKFQRMCDK 62
S+ +IR+AQRA GPA I AIGTANP + V Q+ YPD+YFR+TNSEH +LKEKF+RMCDK
Sbjct: 8 SLDEIRQAQRADGPAGILAIGTANPENHVLQAEYPDYYFRITNSEHMTDLKEKFKRMCDK 67
Query: 63 SMIKKRYMHLTEDLLKENPNMCAYMAPSLDARQDMVVVEVPRLGKEAAVKAIKEWGQPKS 122
S I+KR+MHLTE+ LKENP+MCAYMAPSLD RQD+VVVEVP+LGKEAAVKAIKEWGQPKS
Sbjct: 68 STIRKRHMHLTEEFLKENPHMCAYMAPSLDTRQDIVVVEVPKLGKEAAVKAIKEWGQPKS 127
Query: 123 KITQLIFCTTSGVDMPGADY*LTKLLGLRPYVKRYMMYQQGCFAGGTVLRLAKDLAENNK 182
KIT ++FCTTSGVDMPGADY LTKLLGLRP VKR MMYQQGCFAGGTVLR+AKDLAENN+
Sbjct: 128 KITHVVFCTTSGVDMPGADYQLTKLLGLRPSVKRLMMYQQGCFAGGTVLRIAKDLAENNR 187
Query: 183 GARVLVVCSELTAVTFRGPSDTHLDSLVGQALFGDGAAALIVGSDPIPEV-KKPLFELVW 241
GARVLVVCSE+TAVTFRGPSDTHLDSLVGQALF DGAAALIVGSDP V +KP+FE+V
Sbjct: 188 GARVLVVCSEITAVTFRGPSDTHLDSLVGQALFSDGAAALIVGSDPDTSVGEKPIFEMVS 247
Query: 242 TAQTIAPDSEGAIDGHLREVGLTFHLLKDVPGIVSKNIEKALIEAFQPLGISDYNSIFWI 301
AQTI PDS+GAIDGHLREVGLTFHLLKDVPG++SKNI K+L EAF+PLGISD+NS+FWI
Sbjct: 248 AAQTILPDSDGAIDGHLREVGLTFHLLKDVPGLISKNIVKSLDEAFKPLGISDWNSLFWI 307
Query: 302 AHPGGPAILDQVEQKLALKPEKMRATREVLSEYGNMSSACVLFILDEMRKKSAQDGLKTT 361
AHPGGPAILDQVE KL LK EKMRATR VLSEYGNMSSACVLFILDEMR+KSA+DG+ TT
Sbjct: 308 AHPGGPAILDQVEIKLGLKEEKMRATRHVLSEYGNMSSACVLFILDEMRRKSAKDGVATT 367
Query: 362 GEGLEWGVLFGFGPGLTIETVVLRSV 387
GEGLEWGVLFGFGPGLT+ETVVL SV
Sbjct: 368 GEGLEWGVLFGFGPGLTVETVVLHSV 393
>At4g34850 chalcone synthase - like protein
Length = 392
Score = 282 bits (721), Expect = 3e-76
Identities = 151/375 (40%), Positives = 224/375 (59%), Gaps = 4/375 (1%)
Query: 15 GPATIFAIGTANPPSCVDQSTYPDFYFRVTNSEHKAELKEKFQRMCDKSMIKKRYMHLTE 74
G ATI A+G A P V Q D YF+ T + ELK+K R+C + +K RY+ ++E
Sbjct: 18 GKATILALGKAFPHQLVMQEYLVDGYFKTTKCDDP-ELKQKLTRLCKTTTVKTRYVVMSE 76
Query: 75 DLLKENPNMCAYMAPSLDARQDMVVVEVPRLGKEAAVKAIKEWGQPKSKITQLIFCTTSG 134
++LK+ P + ++ R D+ V + EA+ IK WG+ S IT +++ ++S
Sbjct: 77 EILKKYPELAIEGGSTVTQRLDICNDAVTEMAVEASRACIKNWGRSISDITHVVYVSSSE 136
Query: 135 VDMPGADY*LTKLLGLRPYVKRYMMYQQGCFAGGTVLRLAKDLAENNKGARVLVVCSELT 194
+PG D L K LGL P R ++Y GC G LR+AKD+AENN G+RVL+ SE T
Sbjct: 137 ARLPGGDLYLAKGLGLSPDTHRVLLYFVGCSGGVAGLRVAKDIAENNPGSRVLLATSETT 196
Query: 195 AVTFRGPSDTHLDSLVGQALFGDGAAALIVGSDPIPEVKKPLFELVWTAQTIAPDSEGAI 254
+ F+ PS LVG ALFGDGA A+I+GSDP P +KPLFEL Q P++E I
Sbjct: 197 IIGFKPPSVDRPYDLVGVALFGDGAGAMIIGSDPDPICEKPLFELHTAIQNFLPETEKTI 256
Query: 255 DGHLREVGLTFHLLKDVPGIVSKNIEKALIEAFQPLGIS--DYNSIFWIAHPGGPAILDQ 312
DG L E G+ F L +++P I+ N+E + G++ +YN +FW HPGGPAIL++
Sbjct: 257 DGRLTEQGINFKLSRELPQIIEDNVENFCKKLIGKAGLAHKNYNQMFWAVHPGGPAILNR 316
Query: 313 VEQKLALKPEKMRATREVLSEYGNMSSACVLFILDEMRKKSAQDGLKTTGEGLEWGVLFG 372
+E++L L PEK+ +R L +YGN SS ++++L+ M ++S + E EWG++
Sbjct: 317 IEKRLNLSPEKLSPSRRALMDYGNASSNSIVYVLEYMLEESKKVRNMNEEEN-EWGLILA 375
Query: 373 FGPGLTIETVVLRSV 387
FGPG+T E ++ R++
Sbjct: 376 FGPGVTFEGIIARNL 390
>At1g02050 unknown protein
Length = 395
Score = 276 bits (707), Expect = 1e-74
Identities = 150/377 (39%), Positives = 228/377 (59%), Gaps = 10/377 (2%)
Query: 15 GPATIFAIGTANPPSCVDQSTYPDFYFRVTNSEHKAELKEKFQRMCDKSMIKKRYMHLTE 74
G AT+ A+G A P V Q + + R T + A +KEK + +C + +K RY LT
Sbjct: 24 GKATLLALGKAFPSQVVPQENLVEGFLRDTKCDD-AFIKEKLEHLCKTTTVKTRYTVLTR 82
Query: 75 DLLKENPNMCAYMAPSLDARQDMVVVEVPRLGKEAAVKAIKEWGQPKSKITQLIFCTTSG 134
++L + P + +P++ R ++ V + EA++ IKEWG+P IT +++ ++S
Sbjct: 83 EILAKYPELTTEGSPTIKQRLEIANEAVVEMALEASLGCIKEWGRPVEDITHIVYVSSSE 142
Query: 135 VDMPGADY*LTKLLGLRPYVKRYMMYQQGCFAGGTVLRLAKDLAENNKGARVLVVCSELT 194
+ +PG D L+ LGLR V R M+Y GC+ G T LR+AKD+AENN G+RVL+ SE T
Sbjct: 143 IRLPGGDLYLSAKLGLRNDVNRVMLYFLGCYGGVTGLRVAKDIAENNPGSRVLLTTSETT 202
Query: 195 AVTFRGPSDTHLDSLVGQALFGDGAAALIVGSDPIPEVKKPLFELVWTAQTIAPDSEGAI 254
+ FR P+ LVG ALFGDGAAA+I+G+DP E + P EL + Q P ++ I
Sbjct: 203 ILGFRPPNKARPYDLVGAALFGDGAAAVIIGADP-RECEAPFMELHYAVQQFLPGTQNVI 261
Query: 255 DGHLREVGLTFHLLKDVPGIVSKNIEKALIEAFQPLG--ISDYNSIFWIAHPGGPAILDQ 312
+G L E G+ F L +D+P + +NIE+ + G ++N +FW HPGGPAIL++
Sbjct: 262 EGRLTEEGINFKLGRDLPQKIEENIEEFCKKLMGKAGDESMEFNDMFWAVHPGGPAILNR 321
Query: 313 VEQKLALKPEKMRATREVLSEYGNMSSACVLFILDEMRKKSAQDGLKTTGEGL-EWGVLF 371
+E KL L+ EK+ ++R L +YGN+SS +L++++ MR D LK G+ EWG+
Sbjct: 322 LETKLKLEKEKLESSRRALVDYGNVSSNTILYVMEYMR-----DELKKKGDAAQEWGLGL 376
Query: 372 GFGPGLTIETVVLRSVT 388
FGPG+T E +++RS+T
Sbjct: 377 AFGPGITFEGLLIRSLT 393
>At4g00040 chalcone synthase like protein
Length = 385
Score = 276 bits (705), Expect = 2e-74
Identities = 152/391 (38%), Positives = 232/391 (58%), Gaps = 10/391 (2%)
Query: 1 MVSVADIRKAQRA--KGPATIFAIGTANPPSCVDQSTYPDFYFRVTNSEHKAELKEKFQR 58
M+ A + K +R +G AT+ A+G A P + V Q + Y R ++ + +K+K Q
Sbjct: 1 MLVSARVEKQKRVAYQGKATVLALGKALPSNVVSQENLVEEYLREIKCDNLS-IKDKLQH 59
Query: 59 MCDKSMIKKRYMHLTEDLLKENPNMCAYMAPSLDARQDMVVVEVPRLGKEAAVKAIKEWG 118
+C + +K RY ++ + L + P + +P++ R ++ V ++ EA++ IKEWG
Sbjct: 60 LCKSTTVKTRYTVMSRETLHKYPELATEGSPTIKQRLEIANDAVVQMAYEASLVCIKEWG 119
Query: 119 QPKSKITQLIFCTTSGVDMPGADY*LTKLLGLRPYVKRYMMYQQGCFAGGTVLRLAKDLA 178
+ IT L++ ++S +PG D L+ LGL V+R M+Y GC+ G + LR+AKD+A
Sbjct: 120 RAVEDITHLVYVSSSEFRLPGGDLYLSAQLGLSNEVQRVMLYFLGCYGGLSGLRVAKDIA 179
Query: 179 ENNKGARVLVVCSELTAVTFRGPSDTHLDSLVGQALFGDGAAALIVGSDPIPEVKKPLFE 238
ENN G+RVL+ SE T + FR P+ +LVG ALFGDGAAALI+G+DP E + P E
Sbjct: 180 ENNPGSRVLLTTSETTVLGFRPPNKARPYNLVGAALFGDGAAALIIGADP-TESESPFME 238
Query: 239 LVWTAQTIAPDSEGAIDGHLREVGLTFHLLKDVPGIVSKNIEK--ALIEAFQPLGISDYN 296
L Q P ++G IDG L E G+TF L +D+P + N+E+ + A G + N
Sbjct: 239 LHCAMQQFLPQTQGVIDGRLSEEGITFKLGRDLPQKIEDNVEEFCKKLVAKAGSGALELN 298
Query: 297 SIFWIAHPGGPAILDQVEQKLALKPEKMRATREVLSEYGNMSSACVLFILDEMRKKSAQD 356
+FW HPGGPAIL +E KL LKPEK+ +R L +YGN+SS + +I+D++R + +
Sbjct: 299 DLFWAVHPGGPAILSGLETKLKLKPEKLECSRRALMDYGNVSSNTIFYIMDKVRDELEKK 358
Query: 357 GLKTTGEGLEWGVLFGFGPGLTIETVVLRSV 387
G EG EWG+ FGPG+T E ++R++
Sbjct: 359 GT----EGEEWGLGLAFGPGITFEGFLMRNL 385
>At3g52160 beta-ketoacyl-CoA synthase like protein
Length = 451
Score = 63.9 bits (154), Expect = 1e-10
Identities = 69/241 (28%), Positives = 111/241 (45%), Gaps = 16/241 (6%)
Query: 150 LRPYVKRYMMYQQGCFAGGTVLRLAKDLAENNKGARVLVVCSELTAVTFRGPSDTHLDSL 209
LR + Y + GC AG + LAKDL ++G+ LVV +E+ + T+ +D L L
Sbjct: 210 LRHNTESYNLGGMGCSAGVIAIDLAKDLLNAHQGSYALVVSTEIVSFTWYSGNDVAL--L 267
Query: 210 VGQALFGDGAAALIVGSDPIP--EVKKPLFELVWTAQTIAPDSEGAID-GHLREVGLTFH 266
F GAAA+++ S I K L +LV T + + S +I+ R+ +
Sbjct: 268 PPNCFFRMGAAAVMLSSRRIDRWRAKYQLMQLVRTHKGMEDTSYKSIELREDRDGKQGLY 327
Query: 267 LLKDVPGIVSKNIEKALIEAFQPLGISDYNSIFWIAHPGGPAILDQVEQKLALKPEKMRA 326
+ +DV V ++ KA I L S + I +A +LD + + L L E M A
Sbjct: 328 VSRDVME-VGRHALKANIATLGRLEPS-FEHICVLA--SSKKVLDDIHKDLKLTEENMEA 383
Query: 327 TREVLSEYGNMSSACVLFILDEMRKKSAQDGLKTTGEGLEWGVLFGFGPGLTIETVVLRS 386
+R L +GN SS+ + + L + K+ K W + GFG G +VV ++
Sbjct: 384 SRRTLERFGNTSSSSIWYELAYLEHKA-----KMKRGDRVWQI--GFGSGFKCNSVVWKA 436
Query: 387 V 387
+
Sbjct: 437 L 437
>At4g34510 putative ketoacyl-CoA synthase
Length = 487
Score = 60.5 bits (145), Expect = 2e-09
Identities = 64/259 (24%), Positives = 109/259 (41%), Gaps = 44/259 (16%)
Query: 150 LRPYVKRYMMYQQGCFAGGTVLRLAKDLAENNKGARVLVVCSE-LTAVTFRGPSDTHLDS 208
LR +K + + GC AG + LA D+ + ++ LVV +E +T + G
Sbjct: 204 LRGNIKSFNLGGMGCSAGVIAVDLASDMLQIHRNTFALVVSTENITQNWYFGNKKA---M 260
Query: 209 LVGQALFGDGAAALIVGSDPIPEVKKPLFELVWTAQTIAPDSEGAIDGHLRE----VGLT 264
L+ LF G +A+++ + P+ K+ ++LV T +T E A + +E +
Sbjct: 261 LIPNCLFRVGGSAVLLSNKPLDR-KRSKYKLVHTVRTHKGSDENAFNCVYQEQDECLKTG 319
Query: 265 FHLLKDVPGIVSKNIEKALIEAFQPLGISDYNSIFWIA---------------------- 302
L KD+ I + + K I + PL + I + A
Sbjct: 320 VSLSKDLMAIAGEAL-KTNITSLGPLVLPISEQILFFATFVAKRLFNDKKKKPYIPDFKL 378
Query: 303 -------HPGGPAILDQVEQKLALKPEKMRATREVLSEYGNMSSACVLFILDEMRKKSAQ 355
H GG A++D++E+ L L P+ + A+R L +GN SS+ + + L K
Sbjct: 379 ALDHFCIHAGGRAVIDELEKSLKLSPKHVEASRMTLHRFGNTSSSSIWYELAYTEAK--- 435
Query: 356 DGLKTTGEGLEWGVLFGFG 374
G G + W + FG G
Sbjct: 436 -GRMRKGNRV-WQIAFGSG 452
>At4g34520 fatty acid elongase 1
Length = 506
Score = 59.3 bits (142), Expect = 3e-09
Identities = 59/238 (24%), Positives = 107/238 (44%), Gaps = 41/238 (17%)
Query: 150 LRPYVKRYMMYQQGCFAGGTVLRLAKDLAENNKGARVLVVCSE-LTAVTFRGPSDTHLDS 208
LR +K + + GC AG + LAKDL +K LVV +E +T + G + +
Sbjct: 209 LRSNIKSFNLGGMGCSAGVIAIDLAKDLLHVHKNTYALVVSTENITQGIYAGENRS---M 265
Query: 209 LVGQALFGDGAAALIVGSDPIPEVKKPLFELVWTAQTIAPDSEGAI------DGHLREVG 262
+V LF G AA+++ S+ + ++ ++LV T +T + + D ++G
Sbjct: 266 MVSNCLFRVGGAAILL-SNKSGDRRRSKYKLVHTVRTHTGADDKSFRCVQQEDDESGKIG 324
Query: 263 LTFHLLKDVPGI----VSKNI----------------------EKALIEAFQPLGISDYN 296
+ L KD+ + ++KNI +K L + + + D+
Sbjct: 325 VC--LSKDITNVAGTTLTKNIATLGPLILPLSEKFLFFATFVAKKLLKDKIKHYYVPDFK 382
Query: 297 SIF--WIAHPGGPAILDQVEQKLALKPEKMRATREVLSEYGNMSSACVLFILDEMRKK 352
+ H GG A++D++E+ L L P + A+R L +GN SS+ + + L + K
Sbjct: 383 LAVDHFCIHAGGRAVIDELEKNLGLSPIDVEASRSTLHRFGNTSSSSIWYELAYIEAK 440
>At4g34250 fatty acid elongase - like protein
Length = 493
Score = 57.4 bits (137), Expect = 1e-08
Identities = 77/322 (23%), Positives = 134/322 (40%), Gaps = 49/322 (15%)
Query: 90 SLDARQDMVV--VEVPRLGKEAAVKAIKEWGQPKSKITQLIFCTTSGVDMPGADY*LTKL 147
+L +Q++ V +E + A + G S I L+ +++ P L
Sbjct: 145 TLPLQQNLAVSRIETEEVIIGAVDNLFRNTGISPSDIGILVVNSSTFNPTPSLSSILVNK 204
Query: 148 LGLRPYVKRYMMYQQGCFAGGTVLRLAKDLAENNKGARVLVVCSE-LTAVTFRGPSDTHL 206
LR +K + GC AG + AK L + ++ LVV +E +T + G + +
Sbjct: 205 FKLRDNIKSLNLGGMGCSAGVIAIDAAKSLLQVHRNTYALVVSTENITQNLYMGNNKS-- 262
Query: 207 DSLVGQALFGDGAAALIVGSDPIPEVKKPLFELVWTAQTIAPDSEGAIDGHLRE------ 260
LV LF G AA+++ + I K+ +ELV T + + + + +E
Sbjct: 263 -MLVTNCLFRIGGAAILLSNRSIDR-KRAKYELVHTVRVHTGADDRSYECATQEEDEDGI 320
Query: 261 VGLTFHLLKDVPGIVSKNIEKALIEAFQPLG--------------------------ISD 294
VG++ L K++P + ++ + K I PL I D
Sbjct: 321 VGVS--LSKNLPMVAARTL-KINIATLGPLVLPISEKFHFFVRFVKKKFLNPKLKHYIPD 377
Query: 295 YNSIF--WIAHPGGPAILDQVEQKLALKPEKMRATREVLSEYGNMSSACVLFILDEMRKK 352
+ F + H GG A++D++E+ L L P + A+R L +GN SS+ + + L K
Sbjct: 378 FKLAFEHFCIHAGGRALIDEMEKNLHLTPLDVEASRMTLHRFGNTSSSSIWYELAYTEAK 437
Query: 353 SAQDGLKTTGEGLEWGVLFGFG 374
G T G+ + W + G G
Sbjct: 438 ----GRMTKGDRI-WQIALGSG 454
>At2g15090 putative fatty acid elongase
Length = 476
Score = 53.1 bits (126), Expect = 2e-07
Identities = 63/260 (24%), Positives = 113/260 (43%), Gaps = 47/260 (18%)
Query: 150 LRPYVKRYMMYQQGCFAGGTVLRLAKDLAENNKGARVLVVCSE-LTAVTFRGPSDTHLDS 208
LR +K + GC AG + +AK L + ++ +VV +E +T + G + +
Sbjct: 194 LRDNIKSLNLGGMGCSAGVIAVDVAKGLLQVHRNTYAIVVSTENITQNLYLGKNKS---M 250
Query: 209 LVGQALFGDGAAALIVGSDPIPEVKKPLFELVWTAQTIAPDSEGAIDGHLRE------VG 262
LV LF G AA+++ S+ + + +ELV T + + + + +E +G
Sbjct: 251 LVTNCLFRVGGAAVLL-SNRSRDRNRAKYELVHTVRIHTGSDDRSFECATQEEDEDGIIG 309
Query: 263 LTFHLLKDVPGIVSKNIEKALIEAFQPL----------------------GISDYNSIFW 300
+T L K++P + ++ + K I PL + +Y F
Sbjct: 310 VT--LTKNLPMVAARTL-KINIATLGPLVLPLKEKLAFFITFVKKKYFKPELRNYTPDFK 366
Query: 301 IA------HPGGPAILDQVEQKLALKPEKMRATREVLSEYGNMSSACVLFILDEMRKKSA 354
+A H GG A++D++E+ L L P + A+R L +GN SS+ + + L K
Sbjct: 367 LAFEHFCIHAGGRALIDELEKNLKLSPLHVEASRMTLHRFGNTSSSSIWYELAYTEAK-- 424
Query: 355 QDGLKTTGEGLEWGVLFGFG 374
G G+ + W + G G
Sbjct: 425 --GRMKEGDRI-WQIALGSG 441
>At1g68530 very-long-chain fatty acid condensing enzyme (CUT1)
Length = 497
Score = 52.8 bits (125), Expect = 3e-07
Identities = 62/256 (24%), Positives = 109/256 (42%), Gaps = 39/256 (15%)
Query: 150 LRPYVKRYMMYQQGCFAGGTVLRLAKDLAENNKGARVLVVCSELTAVTFRGPSDTHLDSL 209
LR +K + + GC AG + LA+DL + + + ++V +E+ + ++ + L
Sbjct: 211 LRSNIKSFNLSGMGCSAGLISVDLARDLLQVHPNSNAIIVSTEIITPNYYQGNERAM--L 268
Query: 210 VGQALFGDGAAALIVGSDPIPE--VKKPLFELVWTAQTIAPDSEGAI-DGHLREVGLTFH 266
+ LF GAAA+ + + K L LV T + S + + +E + +
Sbjct: 269 LPNCLFRMGAAAIHMSNRRSDRWRAKYKLSHLVRTHRGADDKSFYCVYEQEDKEGHVGIN 328
Query: 267 LLKDVPGIVSKNIEKALIEAFQPLG--------------------------ISDYNSIF- 299
L KD+ I + + KA I PL I D+ F
Sbjct: 329 LSKDLMAIAGEAL-KANITTIGPLVLPASEQLLFLTSLIGRKIFNPKWKPYIPDFKLAFE 387
Query: 300 -WIAHPGGPAILDQVEQKLALKPEKMRATREVLSEYGNMSSACVLFILDEMRKKSAQDGL 358
+ H GG A++D++++ L L E + A+R L +GN SS+ + + L + K G
Sbjct: 388 HFCIHAGGRAVIDELQKNLQLSGEHVEASRMTLHRFGNTSSSSLWYELSYIESK----GR 443
Query: 359 KTTGEGLEWGVLFGFG 374
G+ + W + FG G
Sbjct: 444 MRRGDRV-WQIAFGSG 458
>At2g16280 putative beta-ketoacyl-CoA synthase
Length = 512
Score = 52.4 bits (124), Expect = 4e-07
Identities = 67/258 (25%), Positives = 111/258 (42%), Gaps = 43/258 (16%)
Query: 150 LRPYVKRYMMYQQGCFAGGTVLRLAKDLAENNKGARVLVVCSE-LTAVTFRGPSDTHLDS 208
LR VK + + GC AG + LAKD+ + ++ +VV +E +T + G
Sbjct: 230 LRGNVKSFNLGGMGCSAGVISIDLAKDMLQVHRNTYAVVVSTENITQNWYFGNKKA---M 286
Query: 209 LVGQALFGDGAAALIVGSDPIPEVKKPLFELVWTAQTIAPDSEGAIDGHLREV---GLT- 264
L+ LF G +A+++ S+ + ++ ++LV T +T E A + +E G T
Sbjct: 287 LIPNCLFRVGGSAILL-SNKGKDRRRSKYKLVHTVRTHKGAVEKAFNCVYQEQDDNGKTG 345
Query: 265 FHLLKDVPGIVSKNIEKALIEAFQPLG--------------------------ISDYNSI 298
L KD+ I + + KA I PL I D+
Sbjct: 346 VSLSKDLMAIAGEAL-KANITTLGPLVLPISEQILFFMTLVTKKLFNSKLKPYIPDFKLA 404
Query: 299 F--WIAHPGGPAILDQVEQKLALKPEKMRATREVLSEYGNMSSACVLFILDEMRKKSAQD 356
F + H GG A++D++E+ L L + A+R L +GN SS+ + + L + K
Sbjct: 405 FDHFCIHAGGRAVIDELEKNLQLSQTHVEASRMTLHRFGNTSSSSIWYELAYIEAK---- 460
Query: 357 GLKTTGEGLEWGVLFGFG 374
G G + W + FG G
Sbjct: 461 GRMKKGNRV-WQIAFGSG 477
>At1g71160 putative ketoacyl-CoA synthase
Length = 460
Score = 52.0 bits (123), Expect = 6e-07
Identities = 64/269 (23%), Positives = 113/269 (41%), Gaps = 38/269 (14%)
Query: 149 GLRPYVKRYMMYQQGCFAGGTVLRLAKDLAENNKGARVLVVCSELTAVTFRGPSDTHLDS 208
G+R +K + + GC AG + L KDL + + + LV+ E AV+ G
Sbjct: 168 GMRSDIKSFSLSGMGCSAGILSVNLVKDLMKIHGDSLALVLSME--AVSPNGYRGKCKSM 225
Query: 209 LVGQALFGDGAAALIVGS--DPIPEVKKPLFELVWTAQTIAPDSEGAIDGHLREVG-LTF 265
L+ +F G AA+++ + + K L ++ T +S ++ + E G +
Sbjct: 226 LIANTIFRMGGAAILLSNRKQDSHKAKYKLQHIIRTHVGSDTESYESVMQQVDEEGKVGV 285
Query: 266 HLLKDVPGIVSKNIE-----------------KALIEAFQ-------PLGISDYNSIF-- 299
L K + + SK ++ K +I Q + ++ F
Sbjct: 286 ALSKQLVRVASKALKINVVQLGPRVLPYSEQLKYIISFIQRKWGMHKEIYTPNFKKAFEH 345
Query: 300 WIAHPGGPAILDQVEQKLALKPEKMRATREVLSEYGNMSSACVLFILDEMRKKSAQDGLK 359
+ H GG AI++ VE+ L L E + A+R L YGN SS+ + + L + K G
Sbjct: 346 FCIHAGGRAIIEGVEKHLKLDKEDVEASRSTLYRYGNTSSSSLWYELQYLEAK----GRM 401
Query: 360 TTGEGLEWGVLFGFGPGLTIETVVLRSVT 388
G+ + W + GFG G + V + ++
Sbjct: 402 KMGDKV-WQI--GFGSGFKANSAVWKCIS 427
>At1g19440 very-long-chain fatty acid condensing enzyme CUT1 like
protein
Length = 516
Score = 51.6 bits (122), Expect = 7e-07
Identities = 65/261 (24%), Positives = 106/261 (39%), Gaps = 49/261 (18%)
Query: 150 LRPYVKRYMMYQQGCFAGGTVLRLAKDLAENNKGARVLVVCSELTAVTFRGPSDTHLDSL 209
LR ++ Y + GC AG + LAKD+ ++ +VV +E +T L
Sbjct: 234 LRGNIRSYNLGGMGCSAGVIAVDLAKDMLLVHRNTYAVVVSTE--NITQNWYFGNKKSML 291
Query: 210 VGQALFGDGAAALIVGSDPIPEVKKPLFELVWTAQTIAPDSEGAIDGHLR-------EVG 262
+ LF G +A+++ S+ + ++ + LV +T GA D R + G
Sbjct: 292 IPNCLFRVGGSAVLL-SNKSRDKRRSKYRLVHVVRT----HRGADDKAFRCVYQEQDDTG 346
Query: 263 LT-FHLLKDVPGIVSKNIEKALIEAFQPLG--------------------------ISDY 295
T L KD+ I + + K I PL I D+
Sbjct: 347 RTGVSLSKDLMAIAGETL-KTNITTLGPLVLPISEQILFFMTLVVKKLFNGKVKPYIPDF 405
Query: 296 NSIF--WIAHPGGPAILDQVEQKLALKPEKMRATREVLSEYGNMSSACVLFILDEMRKKS 353
F + H GG A++D++E+ L L P + A+R L +GN SS+ + + L + K
Sbjct: 406 KLAFEHFCIHAGGRAVIDELEKNLQLSPVHVEASRMTLHRFGNTSSSSIWYELAYIEAK- 464
Query: 354 AQDGLKTTGEGLEWGVLFGFG 374
G G + W + FG G
Sbjct: 465 ---GRMRRGNRV-WQIAFGSG 481
>At5g43760 beta-ketoacyl-CoA synthase
Length = 529
Score = 50.8 bits (120), Expect = 1e-06
Identities = 77/355 (21%), Positives = 144/355 (39%), Gaps = 57/355 (16%)
Query: 57 QRMCDKSMIKKRYMHLTEDLLKENPNMCAYMAPSLDARQDMVVVEVPRLGKEAAVKAIKE 116
Q++ ++S + ++ + E LL+ PN C +AR++ V A+ A+ E
Sbjct: 151 QKILERSGLGQK-TYFPEALLRVPPNPCME-----EARKEAETVMF------GAIDAVLE 198
Query: 117 WGQPKSKITQLIFCTTSGVD-MPGADY*LTKLLGLRPYVKRYMMYQQGCFAGGTVLRLAK 175
K K ++ S + P + LR + Y + GC AG + LAK
Sbjct: 199 KTGVKPKDIGILVVNCSLFNPTPSLSAMIVNKYKLRGNILSYNLGGMGCSAGLISIDLAK 258
Query: 176 DLAENNKGARVLVVCSELTAVTFRGPSDTHLDSLVGQALFGDGAAALIVGSDPIPEVKKP 235
+ + + LVV +E + + +D + L+ +F G AA+++ S+ + +
Sbjct: 259 QMLQVQPNSYALVVSTENITLNWYLGNDRSM--LLSNCIFRMGGAAVLL-SNRSSDRSRS 315
Query: 236 LFELVWTAQTIAPDSEGAI-------DGHLREVG-LTFHLLKDVPGIVSKNIEKALI--- 284
++L+ T +T + A D + E G + L K++ I + ++ +
Sbjct: 316 KYQLIHTVRTHKGADDNAFGCVYQREDNNAEETGKIGVSLSKNLMAIAGEALKTNITTLG 375
Query: 285 -------------------EAFQPLGISDYNSIFWIA------HPGGPAILDQVEQKLAL 319
+ F+ I Y F +A H GG A+LD++E+ L L
Sbjct: 376 PLVLPMSEQLLFFATLVARKVFKVKKIKPYIPDFKLAFEHFCIHAGGRAVLDEIEKNLDL 435
Query: 320 KPEKMRATREVLSEYGNMSSACVLFILDEMRKKSAQDGLKTTGEGLEWGVLFGFG 374
M +R L+ +GN SS+ + + E+ A+ +K W + FG G
Sbjct: 436 SEWHMEPSRMTLNRFGNTSSSSLWY---ELAYSEAKGRIKRGDR--TWQIAFGSG 485
>At1g25450 unknown protein
Length = 492
Score = 50.1 bits (118), Expect = 2e-06
Identities = 58/256 (22%), Positives = 106/256 (40%), Gaps = 39/256 (15%)
Query: 150 LRPYVKRYMMYQQGCFAGGTVLRLAKDLAENNKGARVLVVCSELTAVTFRGPSDTHLDSL 209
LR +K Y + GC A + +A+DL + + + +++ +E+ + ++ + L
Sbjct: 206 LRSNIKSYNLSGMGCSASLISVDVARDLLQVHPNSNAIIISTEIITPNYYKGNERAM--L 263
Query: 210 VGQALFGDGAAALIVGSDPIPE--VKKPLFELVWTAQTIAPDSEGAI-DGHLREVGLTFH 266
+ LF G AA+++ + K L LV T + S + + + + +
Sbjct: 264 LPNCLFRMGGAAILLSNRRSDRWRAKYKLCHLVRTHRGADDKSYNCVMEQEDKNGNVGIN 323
Query: 267 LLKDVPGIVSKNIEKALIEAFQPLG--------------------------ISDYNSIF- 299
L KD+ I + + KA I PL I D+ F
Sbjct: 324 LSKDLMTIAGEAL-KANITTIGPLVLPASEQLLFLSSLIGRKIFNPKWKPYIPDFKQAFE 382
Query: 300 -WIAHPGGPAILDQVEQKLALKPEKMRATREVLSEYGNMSSACVLFILDEMRKKSAQDGL 358
+ H GG A++D++++ L L E + A+R L +GN SS+ + + E+ AQ +
Sbjct: 383 HFCIHAGGRAVIDELQKNLQLSGEHVEASRMTLHRFGNTSSSSLWY---ELSYIEAQGRM 439
Query: 359 KTTGEGLEWGVLFGFG 374
K W + FG G
Sbjct: 440 KRNDR--VWQIAFGSG 453
>At1g01120 putative fatty acid elongase 3-ketoacyl-CoA synthase 1
(At1g01120)
Length = 528
Score = 50.1 bits (118), Expect = 2e-06
Identities = 64/260 (24%), Positives = 110/260 (41%), Gaps = 47/260 (18%)
Query: 150 LRPYVKRYMMYQQGCFAGGTVLRLAKDLAENNKGARVLVVCSELTAVTFRGPSDTHLDSL 209
+R +K Y + GC AG + LA +L + N + +VV +E + + +D + L
Sbjct: 244 MREDIKSYNLGGMGCSAGLISIDLANNLLKANPNSYAVVVSTENITLNWYFGNDRSM--L 301
Query: 210 VGQALFGDGAAALIVGSDPIPEVKKPLFELVWTAQTIA-------------PDSEGAID- 255
+ +F G AA+++ S+ + KK + LV +T D G I
Sbjct: 302 LCNCIFRMGGAAILL-SNRRQDRKKSKYSLVNVVRTHKGSDDKNYNCVYQKEDERGTIGV 360
Query: 256 ----------GHLREVGLTF---------HLLKDVPGIVSKNIEKALIEAFQPLGISDYN 296
G + +T L + +V + + K ++ + P D+
Sbjct: 361 SLARELMSVAGDALKTNITTLGPMVLPLSEQLMFLISLVKRKMFKLKVKPYIP----DFK 416
Query: 297 SIF--WIAHPGGPAILDQVEQKLALKPEKMRATREVLSEYGNMSSACVLFILDEMRKKSA 354
F + H GG A+LD+V++ L LK M +R L +GN SS+ + + EM A
Sbjct: 417 LAFEHFCIHAGGRAVLDEVQKNLDLKDWHMEPSRMTLHRFGNTSSSSLWY---EMAYTEA 473
Query: 355 QDGLKTTGEGLEWGVLFGFG 374
+ +K G+ L W + FG G
Sbjct: 474 KGRVK-AGDRL-WQIAFGSG 491
>At1g04220 beta-ketoacyl-CoA synthase like protein
Length = 528
Score = 49.3 bits (116), Expect = 4e-06
Identities = 81/352 (23%), Positives = 140/352 (39%), Gaps = 53/352 (15%)
Query: 57 QRMCDKSMIKKRYMHLTEDLLKENPNMCAYMAPSLDARQDMVVVEVPRLGKEAAVKAIKE 116
Q++ ++S + ++ + E LL+ PN C +AR++ E G AV +++
Sbjct: 145 QKILERSGLGQK-TYFPEALLRVPPNPCMS-----EARKE---AETVMFGAIDAV--LEK 193
Query: 117 WGQPKSKITQLIFCTTSGVDMPGADY*LTKLLGLRPYVKRYMMYQQGCFAGGTVLRLAKD 176
G I L+ + P + LR V Y + GC AG + LAK
Sbjct: 194 TGVNPKDIGILVVNCSLFNPTPSLSAMIVNKYKLRGNVLSYNLGGMGCSAGLISIDLAKQ 253
Query: 177 LAENNKGARVLVVCSELTAVTFRGPSDTHLDSLVGQALFGDGAAALIVGSDPIPEVKKPL 236
L + + LVV +E + + +D + L+ +F G AA+++ + +
Sbjct: 254 LLQVQPNSYALVVSTENITLNWYLGNDRSM--LLSNCIFRMGGAAVLLSNRSSDRCRSK- 310
Query: 237 FELVWTAQTIAPDSEGAI----------DGHLREVGLTFHLLKDVPGIVSKNI------- 279
++L+ T +T + A D V L+ +L+ + NI
Sbjct: 311 YQLIHTVRTHKGSDDNAFNCVYQREDNDDNKQIGVSLSKNLMAIAGEALKTNITTLGPLV 370
Query: 280 ----EKALIEA-------FQPLGISDYNSIFWIA------HPGGPAILDQVEQKLALKPE 322
E+ L A F I Y F +A H GG A+LD++E+ L L
Sbjct: 371 LPMSEQLLFFATLVARKVFNVKKIKPYIPDFKLAFEHFCIHAGGRAVLDEIEKNLDLSEW 430
Query: 323 KMRATREVLSEYGNMSSACVLFILDEMRKKSAQDGLKTTGEGLEWGVLFGFG 374
M +R L+ +GN SS+ + + E+ A+ +K W + FG G
Sbjct: 431 HMEPSRMTLNRFGNTSSSSLWY---ELAYSEAKGRIKRGDR--TWQIAFGSG 477
>At1g62640 3-ketoacyl-acyl carrier protein synthase III (KAS III)
Length = 404
Score = 47.4 bits (111), Expect = 1e-05
Identities = 74/305 (24%), Positives = 118/305 (38%), Gaps = 47/305 (15%)
Query: 105 LGKEAAVKAIKEWGQPKSKITQLIFCTTSGVDMPGADY*LTKLLGLRPYVKRYMMYQQGC 164
L EAA KA++ I ++ CT++ D+ GA + K LG K + Y
Sbjct: 121 LAVEAATKALEMAEVVPEDIDLVLMCTSTPDDLFGAAPQIQKALGC---TKNPLAYDITA 177
Query: 165 FAGGTVLRLAKDLAENNKGA--RVLVV----CSELTAVTFRGPSDTHLDSLVGQALFGDG 218
G VL L G VLV+ S T RG LFGD
Sbjct: 178 ACSGFVLGLVSAACHIRGGGFKNVLVIGADSLSRFVDWTDRGTC----------ILFGDA 227
Query: 219 AAALIVGSDPIPEV-------------KKPLFELVWTAQTIAPDSE-GAIDGHLREVGLT 264
A A++V + I + ++ L V +Q S G++ G +
Sbjct: 228 AGAVVVQACDIEDDGLFSFDVHSDGDGRRHLNASVKESQNDGESSSNGSVFGDFPPKQSS 287
Query: 265 FHLL----KDVPGIVSKNIEKALIEAFQPLGISDYNSIFWIAHPGGPAILDQVEQKLALK 320
+ + K+V K + +++ A Q G+ + + H I+D V +L
Sbjct: 288 YSCIQMNGKEVFRFAVKCVPQSIESALQKAGLPASAIDWLLLHQANQRIIDSVATRLHFP 347
Query: 321 PEKMRATREVLSEYGNMSSACVLFILDEMRKKSAQDGLKTTGEGLEWGVLFGFGPGLTIE 380
PE++ + L+ YGN S+A + LDE + + G G + GFG GLT
Sbjct: 348 PERVISN---LANYGNTSAASIPLALDE----AVRSGKVKPGHTI---ATSGFGAGLTWG 397
Query: 381 TVVLR 385
+ ++R
Sbjct: 398 SAIMR 402
>At2g26640 putative beta-ketoacyl-CoA synthase
Length = 509
Score = 46.2 bits (108), Expect = 3e-05
Identities = 62/259 (23%), Positives = 107/259 (40%), Gaps = 45/259 (17%)
Query: 150 LRPYVKRYMMYQQGCFAGGTVLRLAKDLAENNKGARVLVVCSELTAVTFRGPSDTHLDSL 209
LR + Y + GC AG + LAK L + +V+ E + + +D L
Sbjct: 222 LRGNILSYNLGGMGCSAGLISIDLAKHLLHSIPNTYAMVISMENITLNWYFGNDR--SKL 279
Query: 210 VGQALFGDGAAALIVGSDPIPEVKKPLFELVWTAQTIAPDSE---GAI---DGHLREVGL 263
V LF G AA+++ S+ + ++ +ELV T +T + G I + ++G+
Sbjct: 280 VSNCLFRMGGAAILL-SNKRWDRRRSKYELVDTVRTHKGADDKCFGCITQEEDSASKIGV 338
Query: 264 TFHLLKDVPGIVSKNIEKALIEAFQPLGISDYNSIFWIA--------------------- 302
T L K++ + + K I PL + + + A
Sbjct: 339 T--LSKELMAVAGDAL-KTNITTLGPLVLPTSEQLLFFATLVGRKLFKMKIKPYIPDFKL 395
Query: 303 -------HPGGPAILDQVEQKLALKPEKMRATREVLSEYGNMSSACVLFILDEMRKKSAQ 355
H GG A+LD++E+ L L M +R L +GN SS+ + + E+ A+
Sbjct: 396 AFEHFCIHAGGRAVLDELEKNLKLTEWHMEPSRMTLYRFGNTSSSSLWY---ELAYSEAK 452
Query: 356 DGLKTTGEGLEWGVLFGFG 374
+K G+ + W + FG G
Sbjct: 453 GRIK-KGDRI-WQIAFGSG 469
>At1g07720 fatty acid elongase 3-ketoacyl-CoA synthase, putative
Length = 478
Score = 37.7 bits (86), Expect = 0.011
Identities = 24/86 (27%), Positives = 40/86 (45%), Gaps = 1/86 (1%)
Query: 268 LKDVPGIVSKNIEKALIEAFQPLGISDYNSI-FWIAHPGGPAILDQVEQKLALKPEKMRA 326
L+ P S N+ +A +A GI+ I + H GG A++D + L L +
Sbjct: 313 LRSSPSKGSTNVTQAAPKAGVKAGINFKTGIDHFCIHTGGKAVIDAIGYSLDLNEYDLEP 372
Query: 327 TREVLSEYGNMSSACVLFILDEMRKK 352
R L +GN S++ + ++L M K
Sbjct: 373 ARMTLHRFGNTSASSLWYVLGYMEAK 398
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.138 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,462,936
Number of Sequences: 26719
Number of extensions: 348797
Number of successful extensions: 837
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 779
Number of HSP's gapped (non-prelim): 49
length of query: 388
length of database: 11,318,596
effective HSP length: 101
effective length of query: 287
effective length of database: 8,619,977
effective search space: 2473933399
effective search space used: 2473933399
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0057b.1


