

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
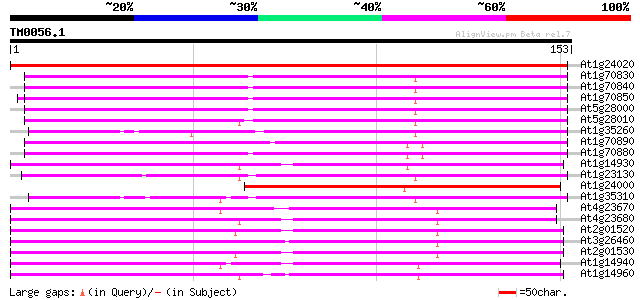
Query= TM0056.1
(153 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g24020 pollen allergen-like protein 192 6e-50
At1g70830 unknown protein 89 9e-19
At1g70840 unknown protein (At1g70840) 88 2e-18
At1g70850 putative protein 87 4e-18
At5g28000 putative protein 86 1e-17
At5g28010 major latex protein homolog - like 83 7e-17
At1g35260 unknown protein 82 9e-17
At1g70890 unknown protein 81 3e-16
At1g70880 unknown protein 72 2e-13
At1g14930 major latex homologue type2 70 4e-13
At1g23130 unknown protein 69 8e-13
At1g24000 hypothetical protein 67 4e-12
At1g35310 unknown protein 64 4e-11
At4g23670 putative major latex protein 63 7e-11
At4g23680 putative major latex protein 60 4e-10
At2g01520 unknown protein 60 4e-10
At3g26460 major latex protein, putative 59 1e-09
At2g01530 unknown protein 58 2e-09
At1g14940 major latex protein type3 58 2e-09
At1g14960 major latex protein, putative 58 2e-09
>At1g24020 pollen allergen-like protein
Length = 155
Score = 192 bits (488), Expect = 6e-50
Identities = 89/152 (58%), Positives = 119/152 (77%)
Query: 1 MGTRGKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITY 60
MG G L V++++KS A+K+W+ L D +FPKAFP+DYK+I++L GDG A GS+R ITY
Sbjct: 1 MGLSGVLHVEVEVKSPAEKFWVALGDGINLFPKAFPNDYKTIQVLAGDGNAPGSIRLITY 60
Query: 61 AEGSPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAVGEGSEVKWSAE 120
EGSPLVK S E+I+A D E K++SY++I GE+L+YYK FKGTI+VI GS +KWS E
Sbjct: 61 GEGSPLVKISAERIEAVDLENKSMSYSIIGGEMLEYYKTFKGTITVIPKDGGSLLKWSGE 120
Query: 121 YEKASTDIPDPSVVKDFAVKNFLEVDEYVLQQ 152
+EK + +I DP V+KDFAVKNF E+DEY+L+Q
Sbjct: 121 FEKTAHEIDDPHVIKDFAVKNFKEIDEYLLKQ 152
>At1g70830 unknown protein
Length = 335
Score = 89.0 bits (219), Expect = 9e-19
Identities = 50/150 (33%), Positives = 81/150 (53%), Gaps = 3/150 (2%)
Query: 5 GKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITYAEGS 64
GKLE D+++K++ADK+ KA P + + ++ EGD GS+ Y
Sbjct: 24 GKLETDVEIKASADKFHHMFAGKPHHVSKASPGNIQGCDLHEGDWGTVGSIVFWNYVHDG 83
Query: 65 PLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAV--GEGSEVKWSAEYE 122
K + E+I+A + +K +++ VI+G+L++ YK F TI V G GS V W EYE
Sbjct: 84 E-AKVAKERIEAVEPDKNLITFRVIEGDLMKEYKSFLLTIQVTPKPGGPGSIVHWHLEYE 142
Query: 123 KASTDIPDPSVVKDFAVKNFLEVDEYVLQQ 152
K S ++ P + F V+ E+DE++L +
Sbjct: 143 KISEEVAHPETLLQFCVEVSKEIDEHLLAE 172
Score = 87.4 bits (215), Expect = 3e-18
Identities = 50/150 (33%), Positives = 80/150 (53%), Gaps = 3/150 (2%)
Query: 5 GKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITYAEGS 64
GKLE D+++K++A+K+ KA P + + ++ EGD GS+ Y
Sbjct: 186 GKLETDVEIKASAEKFHHMFAGKPHHVSKASPGNIQGCDLHEGDWGQVGSIVFWNYVHDR 245
Query: 65 PLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAV--GEGSEVKWSAEYE 122
K + E+I+A + K +++ VIDG+L++ YK F TI V G GS V W EYE
Sbjct: 246 E-AKVAKERIEAVEPNKNLITFRVIDGDLMKEYKSFLLTIQVTPKLGGPGSIVHWHLEYE 304
Query: 123 KASTDIPDPSVVKDFAVKNFLEVDEYVLQQ 152
K S ++ P + F V+ E+DE++L +
Sbjct: 305 KISEEVAHPETLLQFCVEVSKEIDEHLLAE 334
>At1g70840 unknown protein (At1g70840)
Length = 171
Score = 88.2 bits (217), Expect = 2e-18
Identities = 53/150 (35%), Positives = 77/150 (51%), Gaps = 3/150 (2%)
Query: 5 GKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITYAEGS 64
GKLE DI++K++A K+ KA P + E+ EGD GS+ Y
Sbjct: 22 GKLETDIEIKASAGKFHHMFAGRPHHVSKATPGKIQGCELHEGDWGKVGSIVFWNYVHDG 81
Query: 65 PLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAV--GEGSEVKWSAEYE 122
K + E+I+A + EK +++ VI+G+LL+ YK F TI V G GS V W EYE
Sbjct: 82 E-AKVAKERIEAVEPEKNLITFRVIEGDLLKEYKSFVITIQVTPKRGGPGSVVHWHVEYE 140
Query: 123 KASTDIPDPSVVKDFAVKNFLEVDEYVLQQ 152
K + P DF V+ E+DE++L +
Sbjct: 141 KIDDKVAHPETFLDFCVEVSKEIDEHLLNE 170
>At1g70850 putative protein
Length = 316
Score = 87.0 bits (214), Expect = 4e-18
Identities = 50/152 (32%), Positives = 82/152 (53%), Gaps = 3/152 (1%)
Query: 3 TRGKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITYAE 62
T LE ++++K++A+K+ KA P + +S ++ EGD GS+ Y
Sbjct: 165 TTETLETEVEIKASAEKFHHMFAGKPHHVSKATPGNIQSCDLHEGDWGTVGSIVFWNYVH 224
Query: 63 GSPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAV--GEGSEVKWSAE 120
K + E+I+A D EK +++ VI+G+L++ YK F TI V G GS V W E
Sbjct: 225 DGE-AKVAKERIEAVDPEKNLITFRVIEGDLMKEYKSFVITIQVTPKHGGSGSVVHWHFE 283
Query: 121 YEKASTDIPDPSVVKDFAVKNFLEVDEYVLQQ 152
YEK + ++ P + FAV+ E+DE++L +
Sbjct: 284 YEKINEEVAHPETLLQFAVEVSKEIDEHLLAE 315
Score = 86.3 bits (212), Expect = 6e-18
Identities = 49/150 (32%), Positives = 81/150 (53%), Gaps = 3/150 (2%)
Query: 5 GKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITYAEGS 64
GKLE ++++K++A ++ KA P + +S ++ EGD GS+ Y
Sbjct: 11 GKLETEVEIKASAGQFHHMFAGKPHHVSKASPGNIQSCDLHEGDWGTVGSIVFWNYVHDG 70
Query: 65 PLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAV--GEGSEVKWSAEYE 122
K + E+I+A + EK +++ VI+G+L++ YK F TI V G GS V W EYE
Sbjct: 71 E-AKVAKERIEAVEPEKNLITFRVIEGDLMKEYKSFLITIQVTPKHGGPGSIVHWHLEYE 129
Query: 123 KASTDIPDPSVVKDFAVKNFLEVDEYVLQQ 152
K S ++ P + F V+ E+DE++L +
Sbjct: 130 KISDEVAHPETLLQFCVEVSQEIDEHLLSE 159
>At5g28000 putative protein
Length = 164
Score = 85.5 bits (210), Expect = 1e-17
Identities = 49/150 (32%), Positives = 80/150 (52%), Gaps = 3/150 (2%)
Query: 5 GKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITYAEGS 64
G++EV++D+KS A+K++ KA ++ ++LEG+ S+ + Y
Sbjct: 15 GEVEVEVDIKSPAEKFYQVYVGRPDHVAKATSSKVQACDLLEGEWGTISSIVNWNYVYAG 74
Query: 65 PLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAV--GEGSEVKWSAEYE 122
K + E+I+ + EKK + + VI+G++L YK F TISV G GS KW EYE
Sbjct: 75 K-AKVAKERIELVEPEKKLIKFRVIEGDVLAVYKNFFITISVTPKEGGVGSVAKWHLEYE 133
Query: 123 KASTDIPDPSVVKDFAVKNFLEVDEYVLQQ 152
K ++PDP F + E+DE++L +
Sbjct: 134 KNDVNVPDPENFLPFLAEMTKEIDEHLLSE 163
>At5g28010 major latex protein homolog - like
Length = 166
Score = 82.8 bits (203), Expect = 7e-17
Identities = 49/151 (32%), Positives = 82/151 (53%), Gaps = 5/151 (3%)
Query: 5 GKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITYA-EG 63
GKLEV++++K+ A ++ KA P + +S ++ +G+ GS+ + Y EG
Sbjct: 17 GKLEVEVEIKAPAAIFYHIYAGRPHHVAKATPRNVQSCDLHDGEWGTVGSIVYWNYVHEG 76
Query: 64 SPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAV--GEGSEVKWSAEY 121
K + E+I+ + EKK + + VI+G+++ YK F TI V G GS VKW EY
Sbjct: 77 Q--AKVAKERIELVEPEKKLIKFRVIEGDVMAEYKSFLITIQVTPKEGGTGSVVKWHIEY 134
Query: 122 EKASTDIPDPSVVKDFAVKNFLEVDEYVLQQ 152
EK ++P P + F + E+DE++L +
Sbjct: 135 EKIDENVPHPENLLPFFAEMTKEIDEHLLSE 165
>At1g35260 unknown protein
Length = 152
Score = 82.4 bits (202), Expect = 9e-17
Identities = 51/151 (33%), Positives = 86/151 (56%), Gaps = 8/151 (5%)
Query: 6 KLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGD-GKAAGSVRHITYAEGS 64
++EVD+D+K+ ADK+ +R S + PKA H K ++LEG+ GK + +G
Sbjct: 5 EIEVDVDIKTRADKFHKFIRRSQHV-PKA-THYIKGCDLLEGEWGKVGSILLWKLVFDGE 62
Query: 65 PLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAV---GEGSEVKWSAEY 121
P V S + I+ D+EK + V++G L + YK F T+ V++ G GS VKW+ +Y
Sbjct: 63 PRV--SKDMIEVIDEEKNVIQLRVLEGPLKKEYKSFLKTMKVMSPKHGGPGSVVKWNMKY 120
Query: 122 EKASTDIPDPSVVKDFAVKNFLEVDEYVLQQ 152
E+ ++ P+ + F V+ E+D+Y+L +
Sbjct: 121 ERIDQNVDHPNRLLQFFVEVTKEIDQYLLSK 151
>At1g70890 unknown protein
Length = 158
Score = 80.9 bits (198), Expect = 3e-16
Identities = 49/150 (32%), Positives = 76/150 (50%), Gaps = 3/150 (2%)
Query: 5 GKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITYAEGS 64
GKLE ++++K++A K+ + KA P E+ EGD GS+ Y
Sbjct: 9 GKLETEVEIKASAKKFHHMFTERPHHVSKATPDKIHGCELHEGDWGKVGSIVIWKYVHDG 68
Query: 65 PLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVI-AVGE-GSEVKWSAEYE 122
L KI+A D EK +++ V++G+L+ YK F T+ V GE GS W EYE
Sbjct: 69 KLTVGKN-KIEAVDPEKNLITFKVLEGDLMNEYKSFAFTLQVTPKQGESGSIAHWHLEYE 127
Query: 123 KASTDIPDPSVVKDFAVKNFLEVDEYVLQQ 152
K S ++ P + F V+ E+DE++L +
Sbjct: 128 KISEEVAHPETLLQFCVEISKEIDEHLLAE 157
>At1g70880 unknown protein
Length = 159
Score = 71.6 bits (174), Expect = 2e-13
Identities = 45/150 (30%), Positives = 77/150 (51%), Gaps = 3/150 (2%)
Query: 5 GKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITYAEGS 64
G +E ++LKS+ +K+ L A P + +S E+ EG+ G+V Y
Sbjct: 10 GIVETTVELKSSVEKFHDLLVGRPHHMSNATPSNIQSAELQEGEMGQVGAVILWNYVHDG 69
Query: 65 PLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVI-AVGE-GSEVKWSAEYE 122
KS+ ++I++ D EK ++Y V++G+LL+ Y F T V GE GS W EYE
Sbjct: 70 E-AKSAKQRIESLDPEKNRITYRVVEGDLLKEYTSFVTTFQVTPKEGEPGSVAHWHFEYE 128
Query: 123 KASTDIPDPSVVKDFAVKNFLEVDEYVLQQ 152
K + ++ P + A + ++DE++L +
Sbjct: 129 KINEEVAHPETLLQLATEVSKDMDEHLLSE 158
>At1g14930 major latex homologue type2
Length = 155
Score = 70.1 bits (170), Expect = 4e-13
Identities = 40/153 (26%), Positives = 77/153 (50%), Gaps = 5/153 (3%)
Query: 1 MGTRGKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITY 60
M GK ++ LK +A+K++ R +FP A H + + + +GD + GS++ Y
Sbjct: 1 MAMSGKYVAEVPLKGSAEKHYRRWRSQNNLFPDAIGHHVQGVTVHDGDWDSHGSIKSWNY 60
Query: 61 A-EGSPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVI-AVGEGSEVKWS 118
+G P V +I DDEK +++ ++G +++ YKK++ + I EG K +
Sbjct: 61 TLDGKPEVIKEKREI---DDEKMALTFRGLEGHVMEKYKKYEVILQFIPKSNEGCVCKVT 117
Query: 119 AEYEKASTDIPDPSVVKDFAVKNFLEVDEYVLQ 151
+E + D P+P F ++D+++L+
Sbjct: 118 LIWENRNEDSPEPINYMKFVKSLVADMDDHILK 150
>At1g23130 unknown protein
Length = 160
Score = 69.3 bits (168), Expect = 8e-13
Identities = 51/152 (33%), Positives = 77/152 (50%), Gaps = 6/152 (3%)
Query: 4 RGKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITYA-E 62
+G+LEVDI++K++A K+ L KA P D + + EG+ GSV YA +
Sbjct: 11 QGELEVDIEIKASAKKFHHMLSGRPQDVAKATP-DIQGCALHEGEFGKVGSVIFWNYAID 69
Query: 63 GSPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAV--GEGSEVKWSAE 120
G P K + E+I+A D EK + VIDG+L + +K F TI G GS VK +
Sbjct: 70 GQP--KVARERIEAVDQEKNLLVLRVIDGDLTKEFKSFLVTIQATPKLRGPGSVVKCHLK 127
Query: 121 YEKASTDIPDPSVVKDFAVKNFLEVDEYVLQQ 152
YE+ + P + + K +DE +L +
Sbjct: 128 YERIDEKVAPPEKLLELFAKLTESMDEMLLSE 159
>At1g24000 hypothetical protein
Length = 122
Score = 67.0 bits (162), Expect = 4e-12
Identities = 35/88 (39%), Positives = 54/88 (60%), Gaps = 2/88 (2%)
Query: 65 PLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISV--IAVGEGSEVKWSAEYE 122
P K+ +I+A D KKT++ + E+ +Y+K KG+I+V I VG+GS V W+ +E
Sbjct: 31 PFEKNGKTEIEAVDLVKKTMTIQMSGSEIQKYFKTLKGSIAVTPIGVGDGSHVVWTFHFE 90
Query: 123 KASTDIPDPSVVKDFAVKNFLEVDEYVL 150
K DI DP + D +VK F ++DE +L
Sbjct: 91 KVHKDIDDPHSIIDESVKYFKKLDEAIL 118
>At1g35310 unknown protein
Length = 151
Score = 63.5 bits (153), Expect = 4e-11
Identities = 46/151 (30%), Positives = 81/151 (53%), Gaps = 9/151 (5%)
Query: 6 KLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVR--HITYAEG 63
++EVD+++KS ADK+++ R S KA + + ++LEG+ GS+ +T +G
Sbjct: 5 EVEVDVEIKSTADKFFMFSRRSQHA-SKATRY-VQGCDLLEGEWGEVGSILLWKLT-VDG 61
Query: 64 SPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAV--GEGSEVKWSAEY 121
P K S + I+A D + + + V++G L + Y F T+ V G GS VKW+ +Y
Sbjct: 62 EP--KVSKDMIEAIDMKMNMIQWRVLEGPLKEEYNIFSKTMKVSPKQGGSGSVVKWNLKY 119
Query: 122 EKASTDIPDPSVVKDFAVKNFLEVDEYVLQQ 152
E+ + + F V+ E+D+Y+L +
Sbjct: 120 ERIDEKVAHLERLLQFFVECVNEIDQYLLSE 150
>At4g23670 putative major latex protein
Length = 151
Score = 62.8 bits (151), Expect = 7e-11
Identities = 41/152 (26%), Positives = 75/152 (48%), Gaps = 7/152 (4%)
Query: 1 MGTRGKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVR--HI 58
M T G ++ LK +A+K++ + + +F A H +++ + EG+ + GS+R
Sbjct: 1 MATSGTYVTEVPLKGSAEKHYKSWKSENHVFADAIGHHIQNVVVHEGEHDSHGSIRSWDY 60
Query: 59 TYAEGSPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAVGEGSEV-KW 117
TY + K E DDE KT++ +DG ++++ K F I E S V K
Sbjct: 61 TYDGKKEMFKEKRE----IDDENKTLTKRGLDGHVMEHLKVFDIIYEFIPKSEDSCVCKI 116
Query: 118 SAEYEKASTDIPDPSVVKDFAVKNFLEVDEYV 149
+ +EK + D P+PS F + ++++ +V
Sbjct: 117 TMIWEKRNDDFPEPSGYMKFVKQMVVDIEGHV 148
>At4g23680 putative major latex protein
Length = 151
Score = 60.5 bits (145), Expect = 4e-10
Identities = 39/151 (25%), Positives = 73/151 (47%), Gaps = 5/151 (3%)
Query: 1 MGTRGKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITY 60
M T G ++ LK +A+KY+ ++ +FP A H +++ + EG+ + GS+R Y
Sbjct: 1 MATSGTYVTEVPLKGSAEKYYKRWKNENHVFPDAIGHHIQNVTVHEGEHDSHGSIRSWNY 60
Query: 61 A-EGSPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAVGEGSEV-KWS 118
+G V +I DDE KT++ ++G +++ K + I E + + K +
Sbjct: 61 TWDGKEEVFKERREI---DDETKTLTLRGLEGHVMEQLKVYDVVYQFIPKSEDTCIGKIT 117
Query: 119 AEYEKASTDIPDPSVVKDFAVKNFLEVDEYV 149
+EK + D P+PS F ++ +V
Sbjct: 118 LIWEKRNDDSPEPSGYMKFVKSLVADMGNHV 148
>At2g01520 unknown protein
Length = 151
Score = 60.5 bits (145), Expect = 4e-10
Identities = 38/153 (24%), Positives = 74/153 (47%), Gaps = 5/153 (3%)
Query: 1 MGTRGKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITY 60
M T G ++ LK +A+K++ R +FP A H + + I +G+ + G+++ Y
Sbjct: 1 MATSGTYVTEVPLKGSAEKHYKRWRSENHLFPDAIGHHIQGVTIHDGEWDSHGAIKIWNY 60
Query: 61 -AEGSPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAVGEGSEV-KWS 118
+G P V +I DDE V++ ++G +++ K + I + K +
Sbjct: 61 TCDGKPEVFKERREI---DDENMAVTFRGLEGHVMEQLKVYDVIFQFIQKSPDDIICKIT 117
Query: 119 AEYEKASTDIPDPSVVKDFAVKNFLEVDEYVLQ 151
+EK + D+P+PS F ++D++VL+
Sbjct: 118 MIWEKQNDDMPEPSNYMKFVKSLAADMDDHVLK 150
>At3g26460 major latex protein, putative
Length = 152
Score = 58.9 bits (141), Expect = 1e-09
Identities = 35/152 (23%), Positives = 72/152 (47%), Gaps = 2/152 (1%)
Query: 1 MGTRGKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITY 60
M T G ++ LK A+K++ R+ +FP A H + + + +G+ GS++ Y
Sbjct: 1 MATSGTYVTEVPLKGTAEKHFQRWRNENHLFPDAVGHHIQGVSVHDGEWDTHGSIKIWNY 60
Query: 61 AEGSPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAVGEGSEV-KWSA 119
G + E+ + DDE T+ ++G +++ K ++ I E + K +
Sbjct: 61 TLGDGKQEEFKERRE-MDDENNTMKVVGLEGHVMEQLKVYEIDFQFIPKSEEDCICKITM 119
Query: 120 EYEKASTDIPDPSVVKDFAVKNFLEVDEYVLQ 151
+EK + D P+PS ++++++VL+
Sbjct: 120 IWEKRNDDFPEPSSYMQLLKSMVIDMEDHVLK 151
>At2g01530 unknown protein
Length = 151
Score = 58.2 bits (139), Expect = 2e-09
Identities = 38/153 (24%), Positives = 73/153 (46%), Gaps = 5/153 (3%)
Query: 1 MGTRGKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITY 60
M T G ++ LK +ADK++ RD +FP A H + + + +G+ + +++ Y
Sbjct: 1 MATSGTYVTEVPLKGSADKHYKRWRDENHLFPDAIGHHIQGVTVHDGEWDSHEAIKIWNY 60
Query: 61 -AEGSPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAVGEGSEV-KWS 118
+G P V ++I DDE +++ ++G +++ K + V K +
Sbjct: 61 TCDGKPEVFKERKEI---DDENMVITFRGLEGHVMEQLKVYDLIYQFSQKSPDDIVCKIT 117
Query: 119 AEYEKASTDIPDPSVVKDFAVKNFLEVDEYVLQ 151
+EK + D P+PS F ++DE+VL+
Sbjct: 118 MIWEKRTDDSPEPSNYMKFLKSVVADMDEHVLK 150
>At1g14940 major latex protein type3
Length = 155
Score = 58.2 bits (139), Expect = 2e-09
Identities = 41/153 (26%), Positives = 75/153 (48%), Gaps = 7/153 (4%)
Query: 1 MGTRGKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVR--HI 58
M G ++ LK +A+K++ R+ +F A H + + +GD + GS+R +I
Sbjct: 1 MAMSGTYVAEVPLKGSAEKHYKKWRNENHVFQDAVGHHIQGCTMHDGDWDSHGSIRSWNI 60
Query: 59 TYAEGSPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAVG-EGSEVKW 117
T +G P V +I DDEK ++ ++G+ ++ YKK++ I EG K
Sbjct: 61 T-CDGKPEVFKEKREI---DDEKMALTLKGLEGQAMEKYKKYEVIYQFIPKSKEGCVCKI 116
Query: 118 SAEYEKASTDIPDPSVVKDFAVKNFLEVDEYVL 150
+ +EK + + P+P F ++D++VL
Sbjct: 117 TLIWEKRNENSPEPINYMKFVKSLVADMDDHVL 149
>At1g14960 major latex protein, putative
Length = 153
Score = 57.8 bits (138), Expect = 2e-09
Identities = 39/153 (25%), Positives = 74/153 (47%), Gaps = 5/153 (3%)
Query: 1 MGTRGKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITY 60
M G D+ LK +A+K++ R + P+A H + + + +GD + G+++ Y
Sbjct: 1 MAMSGTYVTDVPLKGSAEKHYKRWRSENHLVPEAIGHLIQGVTVHDGDWDSHGTIKIWNY 60
Query: 61 A-EGSPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAVG-EGSEVKWS 118
+G V E+I+ DDE V+ +DG +++ K + T+ I +G K +
Sbjct: 61 TRDGKEEVLK--ERIE-MDDENMAVTINGLDGHVMEVLKVYVTTLHFIPESKDGCVCKIT 117
Query: 119 AEYEKASTDIPDPSVVKDFAVKNFLEVDEYVLQ 151
+EK + D P+P F + +D+++LQ
Sbjct: 118 MVWEKRTEDSPEPIEFMKFVEQMIAHMDDHILQ 150
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.132 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,453,557
Number of Sequences: 26719
Number of extensions: 138640
Number of successful extensions: 371
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 287
Number of HSP's gapped (non-prelim): 56
length of query: 153
length of database: 11,318,596
effective HSP length: 90
effective length of query: 63
effective length of database: 8,913,886
effective search space: 561574818
effective search space used: 561574818
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0056.1


