

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0055.8
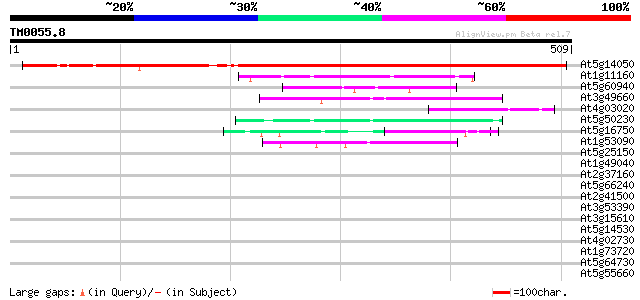
(509 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g14050 unknown protein 592 e-169
At1g11160 hypothetical protein 48 1e-05
At5g60940 cleavage stimulation factor 50K chain 46 4e-05
At3g49660 putative WD-40 repeat - protein 46 4e-05
At4g03020 putative WD-repeat protein 44 2e-04
At5g50230 putative protein 44 2e-04
At5g16750 WD40-repeat protein 44 3e-04
At1g53090 phytochrome A supressor spa1, putative 42 8e-04
At5g25150 transcription initiation factor IID-associated factor-... 41 0.002
At1g49040 hypothetical protein; similar to EST emb|Z26090.1 41 0.002
At2g37160 unknown protein 40 0.002
At5g66240 WD repeat protein-like 40 0.003
At2g41500 putative U4/U6 small nuclear ribonucleoprotein 40 0.003
At3g53390 putative protein 39 0.005
At3g15610 unknown protein 39 0.009
At5g14530 unknown protein 38 0.012
At4g02730 putative WD-repeat protein 38 0.012
At1g73720 unknown protein 38 0.012
At5g64730 unknown protein 37 0.020
At5g55660 putative protein 37 0.020
>At5g14050 unknown protein
Length = 546
Score = 592 bits (1525), Expect = e-169
Identities = 312/501 (62%), Positives = 391/501 (77%), Gaps = 23/501 (4%)
Query: 12 KIEEKEVVDREE-NSDVDTVKSKKRKRDRKREEHVVDMVEQVREMRKLESFLFGSLYSPL 70
K + ++V D EE SD D + K+RK ++++++ ++ E V EM+KLE+ +FGSLYSP+
Sbjct: 19 KKQYEDVEDEEEIGSDDDLTRGKRRKTEKEKQK--LEESELV-EMKKLENLIFGSLYSPV 75
Query: 71 ESGKEVDGEVEPSDLFFTDRSADSVLSVCDEDGEFSDGSGDGDDGL-----GRKPAWVDE 125
GKE E + S LF DRSA + ++DG+ + D ++G + AW DE
Sbjct: 76 TFGKEE--EEDGSALFHVDRSAVRQIPDYEDDGDDDEELSDEENGQVVAIRKGEAAWEDE 133
Query: 126 EEEKFTVNIAKVNRLRKLRKDEDESFISGSEYVARLRAHHVKLNRGTDWAQLDSRLKLDR 185
EE++ V+IA VNRLRKLRK+E+E ISGSEY+ARLRAHH KLN GTDWA+ DS++
Sbjct: 134 EEKQINVDIASVNRLRKLRKEENEGLISGSEYIARLRAHHAKLNPGTDWARPDSQI---- 189
Query: 186 SDYDGELTDDENEAVVRRGYENVDDILRTNEDLVANS-SSKLLPGHLEYSRLVDANIQDP 244
DGE +DD++ + G VDDILRTNEDLV S +KL G LEYS+LVDAN DP
Sbjct: 190 --VDGESSDDDD---TQDG--GVDDILRTNEDLVVKSRGNKLCAGRLEYSKLVDANAADP 242
Query: 245 SNGPVNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQ 304
SNGP+NSV FH+NAQLLLTAGLD++LRFFQIDGKRNTKIQSIFLEDCPIRKA+FLP+GSQ
Sbjct: 243 SNGPINSVHFHQNAQLLLTAGLDRRLRFFQIDGKRNTKIQSIFLEDCPIRKAAFLPNGSQ 302
Query: 305 VILSGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVST 364
VI+SGRRKFFYSFDL K +KIGPLVGREEKSLE FE+S DS IAFVGNEGYILLVST
Sbjct: 303 VIVSGRRKFFYSFDLEKAKFDKIGPLVGREEKSLEYFEVSQDSNTIAFVGNEGYILLVST 362
Query: 365 KTKQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTA 424
KTK+L+G+LKM+G+ RSLAFSEDG+ LLS+GGDGQVY WDLRT C++KGVDEG T+
Sbjct: 363 KTKELIGTLKMNGSVRSLAFSEDGKHLLSSGGDGQVYVWDLRTMKCLYKGVDEGSTCGTS 422
Query: 425 LCTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAIC 484
LC+S +G LFA+G+D GIVN+Y + +F+GGKRKPIKT++NL +K+DFM+FNHD+QILAI
Sbjct: 423 LCSSLNGALFASGTDRGIVNIYKKSEFVGGKRKPIKTVDNLTSKIDFMKFNHDAQILAIV 482
Query: 485 SSMKKSSLKLIHIPSYTVFSN 505
S+M K+S+KL+H+PS TVFSN
Sbjct: 483 STMNKNSVKLVHVPSLTVFSN 503
>At1g11160 hypothetical protein
Length = 974
Score = 47.8 bits (112), Expect = 1e-05
Identities = 61/226 (26%), Positives = 93/226 (40%), Gaps = 22/226 (9%)
Query: 208 VDDILRTNED---LVANSSSKLLPGHLEYSRLVDANIQDPSNGPVNSVQFHRNAQLLLTA 264
VD + +E+ L SS + LE S++V A SN ++V+FH + L +
Sbjct: 10 VDSVAFNSEEVLVLAGASSGVIKLWDLEESKMVRAFTGHRSN--CSAVEFHPFGEFLASG 67
Query: 265 GLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILSGRRKFFYSFDLVKGTL 324
D LR + D ++ IQ+ I F PDG V+ G +DL G L
Sbjct: 68 SSDTNLRVW--DTRKKGCIQTYKGHTRGISTIEFSPDGRWVVSGGLDNVVKVWDLTAGKL 125
Query: 325 EKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLVGSLKMSGT-TRSLA 383
+SL+ L ++A + + +T +L+G+ + T R++A
Sbjct: 126 LHEFKCHEGPIRSLDFHPLEF---LLATGSADRTVKFWDLETFELIGTTRPEATGVRAIA 182
Query: 384 FSEDGQKLLSAGGDG-QVYHWDLRTRTCIHKGVDEG-------CIN 421
F DGQ L DG +VY W+ GVD G CIN
Sbjct: 183 FHPDGQTLFCGLDDGLKVYSWE---PVICRDGVDMGWSTLGDFCIN 225
Score = 35.4 bits (80), Expect = 0.075
Identities = 49/189 (25%), Positives = 73/189 (37%), Gaps = 34/189 (17%)
Query: 318 DLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVS-------------- 363
D V E++ L G +++++L + AF G+ V
Sbjct: 11 DSVAFNSEEVLVLAGASSGVIKLWDLEESKMVRAFTGHRSNCSAVEFHPFGEFLASGSSD 70
Query: 364 -------TKTKQLVGSLKMSGTTRSLA---FSEDGQKLLSAGGDGQVYHWDLRTRTCIHK 413
T+ K + + K G TR ++ FS DG+ ++S G D V WDL +H+
Sbjct: 71 TNLRVWDTRKKGCIQTYK--GHTRGISTIEFSPDGRWVVSGGLDNVVKVWDLTAGKLLHE 128
Query: 414 -GVDEGCINSTALCTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFM 472
EG I S L P L A GS V ++ E F + I T T V +
Sbjct: 129 FKCHEGPIRS--LDFHPLEFLLATGSADRTVKFWDLETF-----ELIGTTRPEATGVRAI 181
Query: 473 RFNHDSQIL 481
F+ D Q L
Sbjct: 182 AFHPDGQTL 190
>At5g60940 cleavage stimulation factor 50K chain
Length = 429
Score = 46.2 bits (108), Expect = 4e-05
Identities = 38/164 (23%), Positives = 70/164 (42%), Gaps = 8/164 (4%)
Query: 248 PVNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVIL 307
P+N + FH + +L+++ D ++FF + +F + +R SF P G + +L
Sbjct: 178 PINDLDFHPRSTILISSAKDNCIKFFDFSKTTAKRAFKVFQDTHNVRSISFHPSG-EFLL 236
Query: 308 SGRR---KFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILL--- 361
+G Y + + L P G ++ SS + +G I L
Sbjct: 237 AGTDHPIPHLYDVNTYQCFLPSNFPDSG-VSGAINQVRYSSTGSIYITASKDGAIRLFDG 295
Query: 362 VSTKTKQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDL 405
VS K + +G+ S F++D + +LS+G D V W++
Sbjct: 296 VSAKCVRSIGNAHGKSEVTSAVFTKDQRFVLSSGKDSTVKLWEI 339
Score = 34.3 bits (77), Expect = 0.17
Identities = 54/272 (19%), Positives = 102/272 (36%), Gaps = 47/272 (17%)
Query: 220 ANSSSKLLPGHLEYSRLVDANIQDPSNGPVNSVQFHRNAQLLLTAGLDQKLRFFQI---- 275
A SSK +P H ++ V +F + T G D ++ F++
Sbjct: 104 AKGSSKTIPKH-------ESKTLSEHKSVVRCARFSPDGMFFATGGADTSIKLFEVPKVK 156
Query: 276 -----DGKRNTKIQSIFLEDCPIRKASFLPDGSQVILSGRRKFFYSFDLVKGTLEKIGPL 330
D + I++ + PI F P + +I S + FD K T ++
Sbjct: 157 QMISGDTQARPLIRTFYDHAEPINDLDFHPRSTILISSAKDNCIKFFDFSKTTAKR---- 212
Query: 331 VGREEKSLEVFELSSDSQMIAFVGNEGYIL---------LVSTKTKQLV-----GSLKMS 376
+ +VF+ + + + I+F + ++L L T Q +S
Sbjct: 213 ------AFKVFQDTHNVRSISFHPSGEFLLAGTDHPIPHLYDVNTYQCFLPSNFPDSGVS 266
Query: 377 GTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHK-GVDEGCINST-ALCTSPSGTLF 434
G + +S G ++A DG + +D + C+ G G T A+ T +
Sbjct: 267 GAINQVRYSSTGSIYITASKDGAIRLFDGVSAKCVRSIGNAHGKSEVTSAVFTKDQRFVL 326
Query: 435 AAGSDS-----GIVNVYNREDFLGGKRKPIKT 461
++G DS I + +++LG KR +++
Sbjct: 327 SSGKDSTVKLWEIGSGRMVKEYLGAKRVKLRS 358
>At3g49660 putative WD-40 repeat - protein
Length = 317
Score = 46.2 bits (108), Expect = 4e-05
Identities = 47/225 (20%), Positives = 94/225 (40%), Gaps = 7/225 (3%)
Query: 227 LPGHLEYSRLVDANIQDPSNGPVNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNT---KI 283
+P ++ V + N V+SV+F + +LL +A D+ +R + I+ + +
Sbjct: 5 IPATASFTPYVHSQTLTSHNRAVSSVKFSSDGRLLASASADKTIRTYTINTINDPIAEPV 64
Query: 284 QSIFLEDCPIRKASFLPDGSQVILSGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFEL 343
Q + I +F D ++ + K +D+ G+L K L+G + V
Sbjct: 65 QEFTGHENGISDVAFSSDARFIVSASDDKTLKLWDVETGSLIKT--LIGHTNYAFCV-NF 121
Query: 344 SSDSQMIAFVGNEGYILLVSTKTKQLVGSLKM-SGTTRSLAFSEDGQKLLSAGGDGQVYH 402
+ S MI + + + T + + L S ++ F+ DG ++S+ DG
Sbjct: 122 NPQSNMIVSGSFDETVRIWDVTTGKCLKVLPAHSDPVTAVDFNRDGSLIVSSSYDGLCRI 181
Query: 403 WDLRTRTCIHKGVDEGCINSTALCTSPSGTLFAAGSDSGIVNVYN 447
WD T C+ +D+ + + SP+G G+ + ++N
Sbjct: 182 WDSGTGHCVKTLIDDENPPVSFVRFSPNGKFILVGTLDNTLRLWN 226
Score = 29.3 bits (64), Expect = 5.4
Identities = 30/139 (21%), Positives = 58/139 (41%), Gaps = 24/139 (17%)
Query: 243 DPSNGPVNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDG 302
D N PV+ V+F N + +L LD LR + I + K + + A +
Sbjct: 195 DDENPPVSFVRFSPNGKFILVGTLDNTLRLWNISSAKFLKTYTGHV------NAQYCISS 248
Query: 303 SQVILSGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLV 362
+ + +G+R +V G+ E+ + ++EL+S + G+ ++ V
Sbjct: 249 AFSVTNGKR-------IVSGS----------EDNCVHMWELNSKKLLQKLEGHTETVMNV 291
Query: 363 ST-KTKQLVGSLKMSGTTR 380
+ T+ L+ S + T R
Sbjct: 292 ACHPTENLIASGSLDKTVR 310
>At4g03020 putative WD-repeat protein
Length = 493
Score = 44.3 bits (103), Expect = 2e-04
Identities = 30/114 (26%), Positives = 53/114 (46%), Gaps = 3/114 (2%)
Query: 381 SLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCTSPSGTLFAAGSDS 440
S+ FS DG+++++ D +Y +DL + V +T SG L +GSD
Sbjct: 234 SVKFSTDGREVVAGSSDDSIYVYDLEANRVSLRTVAHTSDVNTVCFADESGNLILSGSDD 293
Query: 441 GIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICSSMKKSSLKL 494
+ V++R F+ G+ KP + + V F+ D + S+ K ++KL
Sbjct: 294 NLCKVWDRRCFI-GRDKPAGVLVGHLEGVTFIDSRGDGRY--FISNGKDQTIKL 344
>At5g50230 putative protein
Length = 515
Score = 43.9 bits (102), Expect = 2e-04
Identities = 49/242 (20%), Positives = 89/242 (36%), Gaps = 15/242 (6%)
Query: 206 ENVDDILRTNEDLVANSSSKLLPGHLEYSRLVDANIQDPSNGPVNSVQFHRNAQLLLTAG 265
+ VD I+R NED + +P AN G S+ F N+ L T G
Sbjct: 198 QQVDGIVRRNEDGTDHFVESTIPSTC-------ANRIHAHEGGCGSIVFEYNSGTLFTGG 250
Query: 266 LDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILSGRRKFFYSFDLVKGTLE 325
D+ ++ + D T I+S++ I + D VI + + +D+ G +
Sbjct: 251 QDRAVKMW--DTNSGTLIKSLYGSLGNILDMAVTHDNKSVIAATSSNNLFVWDVSSGRVR 308
Query: 326 KIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLVGSLKMSGTTRSLAFS 385
L G +K V S+ + + I L ++ + ++ S
Sbjct: 309 HT--LTGHTDKVCAVDVSKFSSRHVVSAAYDRTIKLWDLHKGYCTNTVLFTSNCNAICLS 366
Query: 386 EDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCTSPSGTLFAAGSDSGIVNV 445
DG + S DG + WD++T + + ++ + + +G D NV
Sbjct: 367 IDGLTVFSGHMDGNLRLWDIQTGKLLSEVAGHSSAVTSVSLSRNGNRILTSGRD----NV 422
Query: 446 YN 447
+N
Sbjct: 423 HN 424
>At5g16750 WD40-repeat protein
Length = 876
Score = 43.5 bits (101), Expect = 3e-04
Identities = 63/250 (25%), Positives = 92/250 (36%), Gaps = 53/250 (21%)
Query: 195 DENEAVVRRGYENVDDILRTNEDLVANSSSKLL--PGHLEYSRLVDANIQD------PSN 246
D ++ V+ E D L L + KLL GH R+ D
Sbjct: 46 DSTDSSVKSTIEGESDTLTA---LALSPDDKLLFSAGHSRQIRVWDLETLKCIRSWKGHE 102
Query: 247 GPVNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVI 306
GPV + H + LL TAG D+K+ + +DG T + F PD ++ I
Sbjct: 103 GPVMGMACHASGGLLATAGADRKVLVWDVDGGFCTHY--FRGHKGVVSSILFHPDSNKNI 160
Query: 307 LSGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKT 366
L + G ++ ++ V++L++ + T+
Sbjct: 161 L----------------------ISGSDDATVRVWDLNAKN----------------TEK 182
Query: 367 KQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALC 426
K L K S+A SEDG L SAG D V WDL +C K A+
Sbjct: 183 KCLAIMEKHFSAVTSIALSEDGLTLFSAGRDKVVNLWDLHDYSC--KATVATYEVLEAVT 240
Query: 427 TSPSGTLFAA 436
T SGT FA+
Sbjct: 241 TVSSGTPFAS 250
Score = 43.1 bits (100), Expect = 4e-04
Identities = 36/105 (34%), Positives = 49/105 (46%), Gaps = 4/105 (3%)
Query: 341 FELSSDSQMIAFVGNEGYILLVSTKTKQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQV 400
F +SSD IA + ++ ST + S T +LA S D + L SAG Q+
Sbjct: 25 FIVSSDGSFIACACGDVINIVDSTDSSVKSTIEGESDTLTALALSPDDKLLFSAGHSRQI 84
Query: 401 YHWDLRTRTCIH--KGVDEGCINSTALCTSPSGTLFAAGSDSGIV 443
WDL T CI KG EG + A C + G L AG+D ++
Sbjct: 85 RVWDLETLKCIRSWKG-HEGPVMGMA-CHASGGLLATAGADRKVL 127
Score = 41.6 bits (96), Expect = 0.001
Identities = 32/133 (24%), Positives = 58/133 (43%), Gaps = 4/133 (3%)
Query: 318 DLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLVGSLK-MS 376
++V T + + E +L LS D +++ G+ I + +T + + S K
Sbjct: 43 NIVDSTDSSVKSTIEGESDTLTALALSPDDKLLFSAGHSRQIRVWDLETLKCIRSWKGHE 102
Query: 377 GTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIH--KGVDEGCINSTALCTSPSGTLF 434
G +A G L +AG D +V WD+ C H +G +G ++S + +
Sbjct: 103 GPVMGMACHASGGLLATAGADRKVLVWDVDGGFCTHYFRG-HKGVVSSILFHPDSNKNIL 161
Query: 435 AAGSDSGIVNVYN 447
+GSD V V++
Sbjct: 162 ISGSDDATVRVWD 174
Score = 35.4 bits (80), Expect = 0.075
Identities = 32/150 (21%), Positives = 63/150 (41%), Gaps = 25/150 (16%)
Query: 299 LPDGSQVI-LSGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEG 357
LPD V+ L G ++ +S + T+++ + +K+++++ +S S + F G+
Sbjct: 528 LPDLVHVVTLKGHKRRIFSVEF--STVDQC-VMTASGDKTVKIWAISDGSCLKTFEGHTS 584
Query: 358 YILLVSTKTKQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDE 417
+L S F DG + +S G DG + W++ T CI D+
Sbjct: 585 SVLRAS--------------------FITDGTQFVSCGADGLLKLWNVNTSECI-ATYDQ 623
Query: 418 GCINSTALCTSPSGTLFAAGSDSGIVNVYN 447
AL + A G ++N+++
Sbjct: 624 HEDKVWALAVGKKTEMIATGGGDAVINLWH 653
>At1g53090 phytochrome A supressor spa1, putative
Length = 794
Score = 42.0 bits (97), Expect = 8e-04
Identities = 45/189 (23%), Positives = 87/189 (45%), Gaps = 14/189 (7%)
Query: 230 HLEYSRL-VDANIQDP----SNGPVNSVQFHRNAQLLLTAGLDQKLRFFQIDG--KRNTK 282
+L +S+L V A+++ S+ V ++ F R+ + TAG+++K++ F+ + K
Sbjct: 463 YLSFSKLRVKADLKQGDLLNSSNLVCAIGFDRDGEFFATAGVNKKIKIFECESIIKDGRD 522
Query: 283 IQSIFLEDCPIRKASFLPDGS----QVILSGRRKFFYSFDLVKGTLEKIGPLVGREEKSL 338
I +E K S + S QV S +D+ + L + + E++
Sbjct: 523 IHYPVVELASRSKLSGICWNSYIKSQVASSNFEGVVQVWDVARNQL--VTEMKEHEKRVW 580
Query: 339 EVFELSSDSQMIAFVGNEGYILLVSTKTKQLVGSLKMSGTTRSLAF-SEDGQKLLSAGGD 397
+ S+D ++A ++G + L S +G++K + F SE G+ L D
Sbjct: 581 SIDYSSADPTLLASGSDDGSVKLWSINQGVSIGTIKTKANICCVQFPSETGRSLAFGSAD 640
Query: 398 GQVYHWDLR 406
+VY++DLR
Sbjct: 641 HKVYYYDLR 649
>At5g25150 transcription initiation factor IID-associated
factor-like protein
Length = 700
Score = 40.8 bits (94), Expect = 0.002
Identities = 40/181 (22%), Positives = 67/181 (36%), Gaps = 37/181 (20%)
Query: 246 NGPVNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQV 305
+GPV S F +L++ D +R + K N + + P+ A F P G
Sbjct: 418 SGPVYSATFSPPGDFVLSSSADTTIRLWST--KLNANLVCYKGHNYPVWDAQFSPFGHYF 475
Query: 306 ILSG--RRKFFYSFDLVKGTLEKIGPL-------------VGREEKSLEVFELSSDSQMI 350
R +S D ++ G L G +K++ ++++ + +
Sbjct: 476 ASCSHDRTARIWSMDRIQPLRIMAGHLSDVDWHPNCNYIATGSSDKTVRLWDVQTGECVR 535
Query: 351 AFVGNEGYILLVSTKTKQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTC 410
F+G+ +L SLA S DG+ + S DG + WDL T C
Sbjct: 536 IFIGHRSMVL--------------------SLAMSPDGRYMASGDEDGTIMMWDLSTARC 575
Query: 411 I 411
I
Sbjct: 576 I 576
>At1g49040 hypothetical protein; similar to EST emb|Z26090.1
Length = 1264
Score = 40.8 bits (94), Expect = 0.002
Identities = 34/117 (29%), Positives = 56/117 (47%), Gaps = 10/117 (8%)
Query: 343 LSSDSQMIAFVGNEGYILLVSTKTKQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYH 402
+SSD I ++ +++ +T QL+ LK + S G+++L+A DG V
Sbjct: 982 ISSDRGKIVSGSDDLSVIVWDKQTTQLLEELKGHDSQVSCVKMLSGERVLTAAHDGTVKM 1041
Query: 403 WDLRTRTCIHKGVDEGCINSTALC---TSPSGTLFAAGSDSGIVNVYNREDFLGGKR 456
WD+RT C+ G +S L +G L AAG D+ + N++ D GK+
Sbjct: 1042 WDVRTDMCV---ATVGRCSSAILSLEYDDSTGILAAAGRDT-VANIW---DIRSGKQ 1091
Score = 28.5 bits (62), Expect = 9.2
Identities = 23/94 (24%), Positives = 43/94 (45%), Gaps = 3/94 (3%)
Query: 321 KGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLVGSL-KMSGTT 379
K T + + L G + + V LS + + A ++G + + +T V ++ + S
Sbjct: 1003 KQTTQLLEELKGHDSQVSCVKMLSGERVLTA--AHDGTVKMWDVRTDMCVATVGRCSSAI 1060
Query: 380 RSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHK 413
SL + + L +AG D WD+R+ +HK
Sbjct: 1061 LSLEYDDSTGILAAAGRDTVANIWDIRSGKQMHK 1094
>At2g37160 unknown protein
Length = 444
Score = 40.4 bits (93), Expect = 0.002
Identities = 23/69 (33%), Positives = 36/69 (51%), Gaps = 1/69 (1%)
Query: 344 SSDSQMIAFVGNEGYILLVSTKTKQLVGSLKMS-GTTRSLAFSEDGQKLLSAGGDGQVYH 402
S+D +A VG +GY+ + T++LV +K G A+S DG+ LL+ G D V
Sbjct: 224 SNDGAYLATVGRDGYLRIFDFSTQKLVCGVKSYYGALLCCAWSMDGKYLLTGGEDDLVQV 283
Query: 403 WDLRTRTCI 411
W + R +
Sbjct: 284 WSMEDRKVV 292
Score = 33.1 bits (74), Expect = 0.37
Identities = 22/74 (29%), Positives = 32/74 (42%), Gaps = 1/74 (1%)
Query: 377 GTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCTSPSGTLFAA 436
G S+AFS DG L + G DG + +D T+ + GV S G
Sbjct: 216 GAINSIAFSNDGAYLATVGRDGYLRIFDFSTQKLV-CGVKSYYGALLCCAWSMDGKYLLT 274
Query: 437 GSDSGIVNVYNRED 450
G + +V V++ ED
Sbjct: 275 GGEDDLVQVWSMED 288
Score = 29.3 bits (64), Expect = 5.4
Identities = 33/162 (20%), Positives = 61/162 (37%), Gaps = 33/162 (20%)
Query: 247 GPVNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVI 306
G +NS+ F + L T G D LR F +Q +
Sbjct: 216 GAINSIAFSNDGAYLATVGRDGYLRIFDF--------------------------STQKL 249
Query: 307 LSGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILL----V 362
+ G + ++ + +++ L G E+ ++V+ + D +++A+ G+ ++ V
Sbjct: 250 VCGVKSYYGALLCCAWSMDGKYLLTGGEDDLVQVWSM-EDRKVVAWEGSGENVMYRFGSV 308
Query: 363 STKTKQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWD 404
T+ L+ L+M L G S G Q HWD
Sbjct: 309 GQDTQLLLWDLEMDEIVVPLRRPPGGSPTYSTG--SQSAHWD 348
>At5g66240 WD repeat protein-like
Length = 331
Score = 40.0 bits (92), Expect = 0.003
Identities = 25/112 (22%), Positives = 50/112 (44%), Gaps = 5/112 (4%)
Query: 331 VGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLVGSLKMSG----TTRSLAFSE 386
VG + V + S+D +++ +G+I ++ + L+ + + +T AFS
Sbjct: 201 VGGDLSEANVVKFSNDGRLMLLTTMDGFIHVLDSFRGTLLSTFSVKPVAGESTLDAAFSP 260
Query: 387 DGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCTSPSGTLFAAGS 438
+G ++S GDG + W +R+ +H + G + P +F GS
Sbjct: 261 EGMFVVSGSGDGSTHAWGVRSGKQVHSWMGLGS-EPPVIKWGPGSPMFVTGS 311
>At2g41500 putative U4/U6 small nuclear ribonucleoprotein
Length = 554
Score = 40.0 bits (92), Expect = 0.003
Identities = 53/230 (23%), Positives = 93/230 (40%), Gaps = 20/230 (8%)
Query: 261 LLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILSGRRKFFYSFDLV 320
L TA D+ + ++ DG T +Q+ + + +F P G + + K + +D+
Sbjct: 313 LATASADRTAKLWKTDG---TLLQTFEGHLDRLARVAFHPSGKYLGTTSYDKTWRLWDIN 369
Query: 321 KGTLEKIGPLVGREEKSLEVFELS--SDSQMIAFVGNEGYILLVSTKTKQLVGSLKMSGT 378
G L+ +E S V+ ++ D + A G + + +T + + L G
Sbjct: 370 TGA-----ELLLQEGHSRSVYGIAFQQDGALAASCGLDSLARVWDLRTGRSI--LVFQGH 422
Query: 379 TR---SLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCTSPSGTLFA 435
+ S+ FS +G L S G D Q WDLR R ++ + S G A
Sbjct: 423 IKPVFSVNFSPNGYHLASGGEDNQCRIWDLRMRKSLYIIPAHANLVSQVKYEPQEGYFLA 482
Query: 436 AGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICS 485
S VN+++ DF +K++ +KV + DS +A S
Sbjct: 483 TASYDMKVNIWSGRDF-----SLVKSLAGHESKVASLDITADSSCIATVS 527
>At3g53390 putative protein
Length = 556
Score = 39.3 bits (90), Expect = 0.005
Identities = 23/79 (29%), Positives = 38/79 (47%), Gaps = 1/79 (1%)
Query: 337 SLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLV-GSLKMSGTTRSLAFSEDGQKLLSAG 395
S+ S+D +A VG +GY+ + T++LV G G ++S DG+ +L+ G
Sbjct: 303 SINSIAFSNDGAHLATVGRDGYLRIFDFLTQKLVCGGKSYYGALLCCSWSMDGKYILTGG 362
Query: 396 GDGQVYHWDLRTRTCIHKG 414
D V W + R + G
Sbjct: 363 EDDLVQVWSMEDRKVVAWG 381
Score = 35.4 bits (80), Expect = 0.075
Identities = 22/75 (29%), Positives = 36/75 (47%), Gaps = 3/75 (4%)
Query: 377 GTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCT-SPSGTLFA 435
G+ S+AFS DG L + G DG + +D T+ + G + + C+ S G
Sbjct: 302 GSINSIAFSNDGAHLATVGRDGYLRIFDFLTQKLVCGG--KSYYGALLCCSWSMDGKYIL 359
Query: 436 AGSDSGIVNVYNRED 450
G + +V V++ ED
Sbjct: 360 TGGEDDLVQVWSMED 374
>At3g15610 unknown protein
Length = 341
Score = 38.5 bits (88), Expect = 0.009
Identities = 52/247 (21%), Positives = 101/247 (40%), Gaps = 34/247 (13%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
V + F ++ + L+T G ++ LR F ++ + + I IR ++L G Q ILS
Sbjct: 105 VRACAFSQDTKYLITGGFEKILRVFDLN-RLDAPPTEIDKSPGSIRTLTWL-HGDQTILS 162
Query: 309 ------GRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLV 362
G R + + TLE P+ E + ++D + F + L+
Sbjct: 163 SCTDIGGVRLWDVRSGKIVQTLETKSPVTSAEVSQDGRYITTADGSTVKFWDANHFGLVK 222
Query: 363 STKTKQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCI-- 420
S + S + + G K ++ G D V +D H G + GC
Sbjct: 223 SYDMPCNIESASLE--------PKSGNKFVAGGEDMWVRLFDF------HTGKEIGCNKG 268
Query: 421 -NSTALCT--SPSGTLFAAGSDSGIVNVY-----NREDFLGGKRKPIKTIENLITKVDFM 472
+ C +P+G +A+GS+ G + ++ N E+ + KP ++++ + K++
Sbjct: 269 HHGPVHCVRFAPTGESYASGSEDGTIRIWQTGPVNPEEI--SESKPKQSVDEVARKIEGF 326
Query: 473 RFNHDSQ 479
N + +
Sbjct: 327 HINKEGK 333
>At5g14530 unknown protein
Length = 330
Score = 38.1 bits (87), Expect = 0.012
Identities = 52/245 (21%), Positives = 92/245 (37%), Gaps = 54/245 (22%)
Query: 247 GPVNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVI 306
G ++SV FHR LL+T+ D LR F I + KI + + + F S +I
Sbjct: 25 GKIHSVGFHRTDDLLVTSSEDDSLRLFDIANAKQLKI--TYHKKHGTDRVCFTHHPSSLI 82
Query: 307 LSGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKT 366
S R LE G +SL + + + F G++ ++
Sbjct: 83 CSSRY-----------NLESTG-------ESLRYLSMYDNRILRYFKGHKDRVV------ 118
Query: 367 KQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCIN---ST 423
SL S +S D V WDLR C +G ++
Sbjct: 119 --------------SLCMSPINDSFMSGSLDRSVRLWDLRVNAC------QGILHLRGRP 158
Query: 424 ALCTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKT--IENLITKVDFMRFNHDSQIL 481
A+ G +FA + G V +++ + G P T + +V+ ++F++D + +
Sbjct: 159 AVAYDQQGLVFAIAMEGGAVKLFDSRCYDKG---PFDTFLVGGDTAEVNDIKFSNDGKSM 215
Query: 482 AICSS 486
+ ++
Sbjct: 216 LLTTT 220
Score = 37.4 bits (85), Expect = 0.020
Identities = 30/100 (30%), Positives = 47/100 (47%), Gaps = 5/100 (5%)
Query: 309 GRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSD--SQMIAFVGNEGYILLVSTKT 366
G K F S KG + LVG + + + S+D S ++ N Y+L
Sbjct: 176 GAVKLFDSRCYDKGPFDTF--LVGGDTAEVNDIKFSNDGKSMLLTTTNNNIYVLDAYRGE 233
Query: 367 KQLVGSLKMS-GTTRSLAFSEDGQKLLSAGGDGQVYHWDL 405
K+ SL+ S GT F+ DG+ +LS GDG ++ W++
Sbjct: 234 KKCGFSLEPSQGTPIEATFTPDGKYVLSGSGDGTLHAWNI 273
>At4g02730 putative WD-repeat protein
Length = 333
Score = 38.1 bits (87), Expect = 0.012
Identities = 38/169 (22%), Positives = 76/169 (44%), Gaps = 14/169 (8%)
Query: 252 VQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILSGRR 311
V F+ + L+++ D+ +R +++ K ++ I PI F DGS ++ +
Sbjct: 134 VNFNPPSNLIVSGSFDETIRIWEV--KTGKCVRMIKAHSMPISSVHFNRDGSLIVSASHD 191
Query: 312 KFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLVG 371
+D +GT K L+ + ++ + S + + I + + L + T +
Sbjct: 192 GSCKIWDAKEGTCLKT--LIDDKSPAVSFAKFSPNGKFILVATLDSTLKLSNYATGKF-- 247
Query: 372 SLKM-SGTTRSL-----AFS-EDGQKLLSAGGDGQVYHWDLRTRTCIHK 413
LK+ +G T + AFS +G+ ++S D VY WDL+ R + +
Sbjct: 248 -LKVYTGHTNKVFCITSAFSVTNGKYIVSGSEDNCVYLWDLQARNILQR 295
Score = 33.1 bits (74), Expect = 0.37
Identities = 22/74 (29%), Positives = 35/74 (46%), Gaps = 1/74 (1%)
Query: 359 ILLVSTKTKQLVGSLKM-SGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDE 417
I + KT + V +K S S+ F+ DG ++SA DG WD + TC+ +D+
Sbjct: 152 IRIWEVKTGKCVRMIKAHSMPISSVHFNRDGSLIVSASHDGSCKIWDAKEGTCLKTLIDD 211
Query: 418 GCINSTALCTSPSG 431
+ SP+G
Sbjct: 212 KSPAVSFAKFSPNG 225
>At1g73720 unknown protein
Length = 511
Score = 38.1 bits (87), Expect = 0.012
Identities = 50/219 (22%), Positives = 87/219 (38%), Gaps = 9/219 (4%)
Query: 296 ASFLPDGSQVILSGRRKFFYSFDLVKGTLEKIGPLVGREEKSLE-----VFELSSDSQMI 350
A F PDG + S F +D + G L+K E + + S DS+M+
Sbjct: 219 ARFSPDGQFLASSSVDGFIEVWDYISGKLKKDLQYQADESFMMHDDPVLCIDFSRDSEML 278
Query: 351 AFVGNEGYILLVSTKTKQLVGSLKM-SGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRT 409
A +G I + +T + S SL+FS DG +LLS D L++
Sbjct: 279 ASGSQDGKIKIWRIRTGVCIRRFDAHSQGVTSLSFSRDGSQLLSTSFDQTARIHGLKSGK 338
Query: 410 CIHKGVDEGCINSTALCTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKV 469
+ + + A+ TS + A SD + ++ KP + V
Sbjct: 339 LLKEFRGHTSYVNHAIFTSDGSRIITASSDCTVKVWDSKTTDCLQTFKPPPPLRGTDASV 398
Query: 470 DFMR-FNHDSQILAICSSMKKSSLKLIHIPSYTVFSNSS 507
+ + F +++ + +C+ K SS+ ++ + V S SS
Sbjct: 399 NSIHLFPKNTEHIVVCN--KTSSIYIMTLQGQVVKSFSS 435
Score = 31.2 bits (69), Expect = 1.4
Identities = 25/84 (29%), Positives = 36/84 (42%), Gaps = 9/84 (10%)
Query: 339 EVFELSSDSQMIAFVGNEGYILL---VSTKTK-----QLVGSLKM-SGTTRSLAFSEDGQ 389
E S D Q +A +G+I + +S K K Q S M + FS D +
Sbjct: 217 ECARFSPDGQFLASSSVDGFIEVWDYISGKLKKDLQYQADESFMMHDDPVLCIDFSRDSE 276
Query: 390 KLLSAGGDGQVYHWDLRTRTCIHK 413
L S DG++ W +RT CI +
Sbjct: 277 MLASGSQDGKIKIWRIRTGVCIRR 300
>At5g64730 unknown protein
Length = 299
Score = 37.4 bits (85), Expect = 0.020
Identities = 38/169 (22%), Positives = 75/169 (43%), Gaps = 22/169 (13%)
Query: 246 NGPVNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQV 305
+G VN+V+F+ ++ ++++AG D+ LR + R+ ++ + + D +FL V
Sbjct: 102 DGEVNAVKFNDSSSVVVSAGFDRSLRVWDC---RSHSVEPVQIID------TFLDTVMSV 152
Query: 306 ILSGRRKFFYSFDLVKGTLEKIGPLVGRE-----EKSLEVFELSSDSQMIAFVGNEGYIL 360
+L+ S D GT+ +GRE + + +S+D + + +
Sbjct: 153 VLTKTEIIGGSVD---GTVRTFDMRIGREMSDNLGQPVNCISISNDGNCVLAGCLDSTLR 209
Query: 361 LVSTKTKQLV----GSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDL 405
L+ T +L+ G + S T + D ++ DG V+ WDL
Sbjct: 210 LLDRTTGELLQVYKGHISKSFKTDCCLTNSDAH-VIGGSEDGLVFFWDL 257
Score = 36.6 bits (83), Expect = 0.034
Identities = 29/101 (28%), Positives = 44/101 (42%), Gaps = 3/101 (2%)
Query: 344 SSDSQMIAFVGNEGYILLVSTKTKQLVGSLKMSGT-TRSLAFSEDGQKLLSAGGDGQVYH 402
+ D G + I L + L+ + K G R + + D K S GGD QVY+
Sbjct: 27 NGDGNYALTCGKDRTIRLWNPHRGILIKTYKSHGREVRDVHVTSDNAKFCSCGGDRQVYY 86
Query: 403 WDLRTRTCIHK-GVDEGCINSTALCTSPSGTLFAAGSDSGI 442
WD+ T I K +G +N+ S S + +AG D +
Sbjct: 87 WDVSTGRVIRKFRGHDGEVNAVKFNDS-SSVVVSAGFDRSL 126
Score = 34.3 bits (77), Expect = 0.17
Identities = 25/115 (21%), Positives = 51/115 (43%), Gaps = 4/115 (3%)
Query: 296 ASFLPDGSQVILSGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGN 355
A F DG+ + G+ + ++ +G L K GRE + + V +SD+ G
Sbjct: 24 ARFNGDGNYALTCGKDRTIRLWNPHRGILIKTYKSHGREVRDVHV---TSDNAKFCSCGG 80
Query: 356 EGYILLVSTKTKQLVGSLK-MSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRT 409
+ + T +++ + G ++ F++ ++SAG D + WD R+ +
Sbjct: 81 DRQVYYWDVSTGRVIRKFRGHDGEVNAVKFNDSSSVVVSAGFDRSLRVWDCRSHS 135
>At5g55660 putative protein
Length = 759
Score = 37.4 bits (85), Expect = 0.020
Identities = 29/114 (25%), Positives = 56/114 (48%), Gaps = 12/114 (10%)
Query: 9 ARPKIEEKEVVDREENSDVDTVKSKKRKRDRKREEHVVDMVEQVREMRKLESFLFGSLYS 68
A P++E+K+ ++EN D + + K+ +++ ++EE D ++ +++K G
Sbjct: 231 AEPEVEDKKTESKDENEDKE--EEKEDEKEDEKEESNDDKEDKKEDIKKSNKRGKGKTEK 288
Query: 69 PLESGK--EVDGEVEPSDLFFTDR------SADSVLSVCDEDG--EFSDGSGDG 112
K E ++EP FF+DR S + +++V D+D EF G G
Sbjct: 289 TRGKTKSDEEKKDIEPKTPFFSDRPVRERKSVERLVAVVDKDSSREFHVEKGKG 342
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.134 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,674,644
Number of Sequences: 26719
Number of extensions: 549380
Number of successful extensions: 2336
Number of sequences better than 10.0: 165
Number of HSP's better than 10.0 without gapping: 54
Number of HSP's successfully gapped in prelim test: 113
Number of HSP's that attempted gapping in prelim test: 2009
Number of HSP's gapped (non-prelim): 404
length of query: 509
length of database: 11,318,596
effective HSP length: 104
effective length of query: 405
effective length of database: 8,539,820
effective search space: 3458627100
effective search space used: 3458627100
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0055.8


