

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0055.3
(355 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
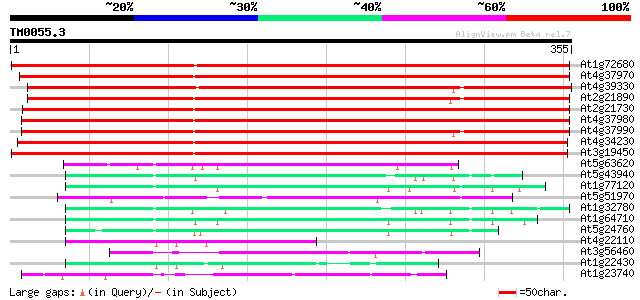
Score E
Sequences producing significant alignments: (bits) Value
At1g72680 putative cinnamyl-alcohol dehydrogenase 527 e-150
At4g37970 cinnamyl alcohol dehydrogenase -like protein, LCADa 356 1e-98
At4g39330 cinnamyl-alcohol dehydrogenase CAD1 350 9e-97
At2g21890 putative cinnamyl-alcohol dehydrogenase 347 7e-96
At2g21730 putative cinnamyl-alcohol dehydrogenase 340 7e-94
At4g37980 cinnamyl-alcohol dehydrogenase ELI3-1 333 7e-92
At4g37990 cinnamyl-alcohol dehydrogenase ELI3-2 327 6e-90
At4g34230 cinnamyl alcohol dehydrogenase like protein 325 3e-89
At3g19450 putative cinnamyl alcohol dehydrogenase 2 318 4e-87
At5g63620 alcohol dehydrogenase-like protein 94 1e-19
At5g43940 alcohol dehydrogenase (EC 1.1.1.1) class III (pir||S71... 92 3e-19
At1g77120 alcohol dehydrogenase 79 4e-15
At5g51970 sorbitol dehydrogenase-like protein 74 1e-13
At1g32780 alcohol dehydrogenase like protein 71 1e-12
At1g64710 alcohol dehydrogenase (EC 1.1.1.1) like protein 69 4e-12
At5g24760 alcohol dehydrogenase - like protein 65 6e-11
At4g22110 alcohol dehydrogenase like protein 60 2e-09
At3g56460 quinone reductase-like protein 60 2e-09
At1g22430 putative alcohol dehydrogenase 60 2e-09
At1g23740 auxin-induced protein like 59 4e-09
>At1g72680 putative cinnamyl-alcohol dehydrogenase
Length = 355
Score = 527 bits (1358), Expect = e-150
Identities = 253/353 (71%), Positives = 296/353 (83%), Gaps = 1/353 (0%)
Query: 2 SSEGASEECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNS 61
SSE EC+ WAA D SG+LSP+ +RR+V DDV + I+HCGVCYADV W+RN G+S
Sbjct: 3 SSESVENECMCWAARDPSGLLSPHTITRRSVTTDDVSLTITHCGVCYADVIWSRNQHGDS 62
Query: 62 VYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFT 121
YP VPGHEIAGIVTKVG NV F VGDHVGVGTYVNSCR+CE+C++G E +C KG VFT
Sbjct: 63 KYPLVPGHEIAGIVTKVGPNVQRFKVGDHVGVGTYVNSCRECEYCNEGQEVNCAKG-VFT 121
Query: 122 FNGVDHDGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQP 181
FNG+DHDG++TKGGYS IVVHERYC+ IP YPL SAAPLLCAGITVY+PMMRH MNQP
Sbjct: 122 FNGIDHDGSVTKGGYSSHIVVHERYCYKIPVDYPLESAAPLLCAGITVYAPMMRHNMNQP 181
Query: 182 GKSLGVIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRA 241
GKSLGVIGLGGLGHMAVKFGKAFGL+VTVFSTSISKKEEAL+LLGA+ FV+SSD ++M+A
Sbjct: 182 GKSLGVIGLGGLGHMAVKFGKAFGLSVTVFSTSISKKEEALNLLGAENFVISSDHDQMKA 241
Query: 242 LAKSLDFIVDTASGPHSFDPYMALLKSFGVLALVGFPGEIKIHPGLLIMGSRTVSGSGVG 301
L KSLDF+VDTASG H+FDPYM+LLK G LVGFP EIKI P L +G R ++GS G
Sbjct: 242 LEKSLDFLVDTASGDHAFDPYMSLLKIAGTYVLVGFPSEIKISPANLNLGMRMLAGSVTG 301
Query: 302 GTKEIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSL 354
GTK + M++FC A++I+PNIEV+PIQ NEAL+RV+KKD+KYRFVIDI+NSL
Sbjct: 302 GTKITQQMLDFCAAHKIYPNIEVIPIQKINEALERVVKKDIKYRFVIDIKNSL 354
>At4g37970 cinnamyl alcohol dehydrogenase -like protein, LCADa
Length = 363
Score = 356 bits (913), Expect = 1e-98
Identities = 174/349 (49%), Positives = 244/349 (69%), Gaps = 2/349 (0%)
Query: 7 SEECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCV 66
S E GWAA D+SG LSP+ FSRR G+++V VK+ +CG+C++D+ +N +S+YP V
Sbjct: 11 SVEAFGWAARDSSGHLSPFVFSRRKTGEEEVRVKVLYCGICHSDLHCLKNEWHSSIYPLV 70
Query: 67 PGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVD 126
PGHEI G V+++G+ V F++GD VGVG V+SCR CE C + E++C K ++ T+NGV
Sbjct: 71 PGHEIIGEVSEIGNKVSKFNLGDKVGVGCIVDSCRTCESCREDQENYCTK-AIATYNGVH 129
Query: 127 HDGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLG 186
HDGTI GGYS IVV ERY IP + PL SAAPLLCAGI++YSPM + P K +G
Sbjct: 130 HDGTINYGGYSDHIVVDERYAVKIPHTLPLVSAAPLLCAGISMYSPMKYFGLTGPDKHVG 189
Query: 187 VIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSL 246
++GLGGLGH+ V+F KAFG VTV S++ K ++AL LGAD F+VS+D+++M+A ++
Sbjct: 190 IVGLGGLGHIGVRFAKAFGTKVTVVSSTTGKSKDALDTLGADGFLVSTDEDQMKAAMGTM 249
Query: 247 DFIVDTASGPHSFDPYMALLKSFGVLALVGFPGE-IKIHPGLLIMGSRTVSGSGVGGTKE 305
D I+DT S HS P + LLKS G L L+G + I LI+G ++++GSG+GG +E
Sbjct: 250 DGIIDTVSASHSISPLIGLLKSNGKLVLLGATEKPFDISAFSLILGRKSIAGSGIGGMQE 309
Query: 306 IRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSL 354
++MI+F + I IE++ + Y N A+DR+ K DV+YRFVIDI N+L
Sbjct: 310 TQEMIDFAAEHGIKAEIEIISMDYVNTAMDRLAKGDVRYRFVIDISNTL 358
>At4g39330 cinnamyl-alcohol dehydrogenase CAD1
Length = 360
Score = 350 bits (897), Expect = 9e-97
Identities = 179/347 (51%), Positives = 236/347 (67%), Gaps = 6/347 (1%)
Query: 12 GWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEI 71
GW A D SGVLSP+ FSRR G++DV VKI CGVC+ D+ +N G S YP VPGHEI
Sbjct: 15 GWGARDKSGVLSPFHFSRRDNGENDVTVKILFCGVCHTDLHTIKNDWGYSYYPVVPGHEI 74
Query: 72 AGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHDGTI 131
GI TKVG NV F GD VGVG SC+ CE CD LE++C + S FT+N + DGT
Sbjct: 75 VGIATKVGKNVTKFKEGDRVGVGVISGSCQSCESCDQDLENYCPQMS-FTYNAIGSDGTK 133
Query: 132 TKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLG 191
GGYS+ IVV +R+ P++ P S APLLCAGITVYSPM + M + GK LGV GLG
Sbjct: 134 NYGGYSENIVVDQRFVLRFPENLPSDSGAPLLCAGITVYSPMKYYGMTEAGKHLGVAGLG 193
Query: 192 GLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLDFIVD 251
GLGH+AVK GKAFGL VTV S+S +K EEA++ LGAD F+V++D ++M+A ++D+I+D
Sbjct: 194 GLGHVAVKIGKAFGLKVTVISSSSTKAEEAINHLGADSFLVTTDPQKMKAAIGTMDYIID 253
Query: 252 TASGPHSFDPYMALLKSFGVLALVGFPG---EIKIHPGLLIMGSRTVSGSGVGGTKEIRD 308
T S H+ P + LLK G L +G P E+ + P L++G + V GS VGG KE ++
Sbjct: 254 TISAVHALYPLLGLLKVNGKLIALGLPEKPLELPMFP--LVLGRKMVGGSDVGGMKETQE 311
Query: 309 MIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSLN 355
M++FC + I +IE++ + N A++R+ K DV+YRFVID+ NSL+
Sbjct: 312 MLDFCAKHNITADIELIKMDEINTAMERLAKSDVRYRFVIDVANSLS 358
>At2g21890 putative cinnamyl-alcohol dehydrogenase
Length = 375
Score = 347 bits (889), Expect = 7e-96
Identities = 180/347 (51%), Positives = 233/347 (66%), Gaps = 7/347 (2%)
Query: 12 GWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEI 71
GWAA D SGVLSP+ FSRR G++DV VKI CGVC++D+ +N G S YP +PGHEI
Sbjct: 9 GWAANDESGVLSPFHFSRRENGENDVTVKILFCGVCHSDLHTIKNHWGFSRYPIIPGHEI 68
Query: 72 AGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHDGTI 131
GI TKVG NV F GD VGVG + SC+ CE C+ LE++C K VFT+N DGT
Sbjct: 69 VGIATKVGKNVTKFKEGDRVGVGVIIGSCQSCESCNQDLENYCPK-VVFTYNSRSSDGTR 127
Query: 132 TKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKM-NQPGKSLGVIGL 190
+GGYS IVV R+ IP P S APLLCAGITVYSPM + M + GK LGV GL
Sbjct: 128 NQGGYSDVIVVDHRFVLSIPDGLPSDSGAPLLCAGITVYSPMKYYGMTKESGKRLGVNGL 187
Query: 191 GGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLDFIV 250
GGLGH+AVK GKAFGL VTV S S K+ EA+ LGAD F+V++D ++M+ ++DFI+
Sbjct: 188 GGLGHIAVKIGKAFGLRVTVISRSSEKEREAIDRLGADSFLVTTDSQKMKEAVGTMDFII 247
Query: 251 DTASGPHSFDPYMALLKSFGVLALVGF---PGEIKIHPGLLIMGSRTVSGSGVGGTKEIR 307
DT S H+ P +LLK G L +G P ++ I P L++G + V GS +GG KE +
Sbjct: 248 DTVSAEHALLPLFSLLKVSGKLVALGLLEKPLDLPIFP--LVLGRKMVGGSQIGGMKETQ 305
Query: 308 DMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSL 354
+M+EFC ++I +IE++ + N A+DR++K DV+YRFVID+ NSL
Sbjct: 306 EMLEFCAKHKIVSDIELIKMSDINSAMDRLVKSDVRYRFVIDVANSL 352
>At2g21730 putative cinnamyl-alcohol dehydrogenase
Length = 376
Score = 340 bits (872), Expect = 7e-94
Identities = 178/349 (51%), Positives = 232/349 (66%), Gaps = 4/349 (1%)
Query: 9 ECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVPG 68
+ GWAA D SGVLSP+ FSRR G++DV VKI CGVC++D+ +N G S YP +PG
Sbjct: 6 KAFGWAANDESGVLSPFHFSRRENGENDVTVKILFCGVCHSDLHTIKNHWGFSRYPIIPG 65
Query: 69 HEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHD 128
HEI GI TKVG NV F GD VGVG + SC+ CE C+ LE++C K VFT+N D
Sbjct: 66 HEIVGIATKVGKNVTKFKEGDRVGVGVIIGSCQSCESCNQDLENYCPK-VVFTYNSRSSD 124
Query: 129 GTI-TKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKM-NQPGKSLG 186
GT +GGYS IVV R+ IP P S APLLCAGITVYSPM + M + GK LG
Sbjct: 125 GTSRNQGGYSDVIVVDHRFVLSIPDGLPSDSGAPLLCAGITVYSPMKYYGMTKESGKRLG 184
Query: 187 VIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSL 246
V GLGGLGH+AVK GKAFGL VTV S S K+ EA+ LGAD F+V++D ++M+ ++
Sbjct: 185 VNGLGGLGHIAVKIGKAFGLRVTVISRSSEKEREAIDRLGADSFLVTTDSQKMKEAVGTM 244
Query: 247 DFIVDTASGPHSFDPYMALLKSFGVLALVGFPGE-IKIHPGLLIMGSRTVSGSGVGGTKE 305
DFI+DT S H+ P +LLK G L +G P + + + L++G + V GS +GG KE
Sbjct: 245 DFIIDTVSAEHALLPLFSLLKVNGKLVALGLPEKPLDLPIFSLVLGRKMVGGSQIGGMKE 304
Query: 306 IRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSL 354
++M+EFC ++I +IE++ + N A+DR+ K DV+YRFVID+ NSL
Sbjct: 305 TQEMLEFCAKHKIVSDIELIKMSDINSAMDRLAKSDVRYRFVIDVANSL 353
>At4g37980 cinnamyl-alcohol dehydrogenase ELI3-1
Length = 357
Score = 333 bits (855), Expect = 7e-92
Identities = 167/348 (47%), Positives = 231/348 (65%), Gaps = 2/348 (0%)
Query: 8 EECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVP 67
+E G AA D SG+LSP+ FSRRA G+ DV K+ CG+C+ D++ +N G + YP VP
Sbjct: 7 KEAFGLAAKDESGILSPFSFSRRATGEKDVRFKVLFCGICHTDLSMAKNEWGLTTYPLVP 66
Query: 68 GHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDH 127
GHEI G+VT+VG+ V F+ GD VGVG SCR C+ C+DG E++C K + T +
Sbjct: 67 GHEIVGVVTEVGAKVKKFNAGDKVGVGYMAGSCRSCDSCNDGDENYCPK-MILTSGAKNF 125
Query: 128 DGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGV 187
D T+T GGYS +V E + IP + PL AAPLLCAG+TVYSPM H +++PG +GV
Sbjct: 126 DDTMTHGGYSDHMVCAEDFIIRIPDNLPLDGAAPLLCAGVTVYSPMKYHGLDKPGMHIGV 185
Query: 188 IGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLD 247
+GLGGLGH+AVKF KA G VTV STS K++EA++ LGAD F+VS D ++M+ ++D
Sbjct: 186 VGLGGLGHVAVKFAKAMGTKVTVISTSERKRDEAVTRLGADAFLVSRDPKQMKDAMGTMD 245
Query: 248 FIVDTASGPHSFDPYMALLKSFGVLALVGFPGE-IKIHPGLLIMGSRTVSGSGVGGTKEI 306
I+DT S H P + LLK+ G L +VG P E +++ LI G + V GS VGG KE
Sbjct: 246 GIIDTVSATHPLLPLLGLLKNKGKLVMVGAPAEPLELPVFPLIFGRKMVVGSMVGGIKET 305
Query: 307 RDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSL 354
++M++ + I +IE++ Y N A++R+ K DVKYRFVID+ N++
Sbjct: 306 QEMVDLAGKHNITADIELISADYVNTAMERLAKADVKYRFVIDVANTM 353
>At4g37990 cinnamyl-alcohol dehydrogenase ELI3-2
Length = 359
Score = 327 bits (838), Expect = 6e-90
Identities = 167/350 (47%), Positives = 231/350 (65%), Gaps = 6/350 (1%)
Query: 8 EECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVP 67
+E G AA D SGVLSP+ F+RR G+ DV K+ CG+C++D+ +N G S YP VP
Sbjct: 7 KEAFGLAAKDNSGVLSPFSFTRRETGEKDVRFKVLFCGICHSDLHMVKNEWGMSTYPLVP 66
Query: 68 GHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDH 127
GHEI G+VT+VG+ V F G+ VGVG V+SC C+ C +G+E++C K S+ T+ +
Sbjct: 67 GHEIVGVVTEVGAKVTKFKTGEKVGVGCLVSSCGSCDSCTEGMENYCPK-SIQTYGFPYY 125
Query: 128 DGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGV 187
D TIT GGYS +V E + IP + PL +AAPLLCAGITVYSPM H +++PG +GV
Sbjct: 126 DNTITYGGYSDHMVCEEGFVIRIPDNLPLDAAAPLLCAGITVYSPMKYHGLDKPGMHIGV 185
Query: 188 IGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLD 247
+GLGGLGH+ VKF KA G VTV STS K++EA++ LGAD F+VS D ++++ ++D
Sbjct: 186 VGLGGLGHVGVKFAKAMGTKVTVISTSEKKRDEAINRLGADAFLVSRDPKQIKDAMGTMD 245
Query: 248 FIVDTASGPHSFDPYMALLKSFGVLALVGFPG---EIKIHPGLLIMGSRTVSGSGVGGTK 304
I+DT S HS P + LLK G L +VG P E+ + P LI + V GS +GG K
Sbjct: 246 GIIDTVSATHSLLPLLGLLKHKGKLVMVGAPEKPLELPVMP--LIFERKMVMGSMIGGIK 303
Query: 305 EIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSL 354
E ++MI+ + I +IE++ Y N A++R+ K DV+YRFVID+ N+L
Sbjct: 304 ETQEMIDMAGKHNITADIELISADYVNTAMERLEKADVRYRFVIDVANTL 353
>At4g34230 cinnamyl alcohol dehydrogenase like protein
Length = 357
Score = 325 bits (832), Expect = 3e-89
Identities = 162/349 (46%), Positives = 229/349 (65%), Gaps = 2/349 (0%)
Query: 6 ASEECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPC 65
A + GWAA D SG+LSPY ++ R G +DV ++I CG+C+ D+ T+N LG S YP
Sbjct: 6 AERKTTGWAARDPSGILSPYTYTLRETGPEDVNIRIICCGICHTDLHQTKNDLGMSNYPM 65
Query: 66 VPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGV 125
VPGHE+ G V +VGS+V F+VGD VGVG V C C C+ LE +C K ++++N V
Sbjct: 66 VPGHEVVGEVVEVGSDVSKFTVGDIVGVGCLVGCCGGCSPCERDLEQYCPK-KIWSYNDV 124
Query: 126 DHDGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSL 185
+G T+GG++K VVH+++ IP+ + AAPLLCAG+TVYSP+ + QPG
Sbjct: 125 YINGQPTQGGFAKATVVHQKFVVKIPEGMAVEQAAPLLCAGVTVYSPLSHFGLKQPGLRG 184
Query: 186 GVIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKS 245
G++GLGG+GHM VK KA G +VTV S+S K+EEAL LGAD +V+ SDQ +M LA S
Sbjct: 185 GILGLGGVGHMGVKIAKAMGHHVTVISSSNKKREEALQDLGADDYVIGSDQAKMSELADS 244
Query: 246 LDFIVDTASGPHSFDPYMALLKSFGVLALVG-FPGEIKIHPGLLIMGSRTVSGSGVGGTK 304
LD+++DT H+ +PY++LLK G L L+G ++ LL++G + ++GS +G K
Sbjct: 245 LDYVIDTVPVHHALEPYLSLLKLDGKLILMGVINNPLQFLTPLLMLGRKVITGSFIGSMK 304
Query: 305 EIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENS 353
E +M+EFC + IEVV + Y N A +R+ K DV+YRFV+D+E S
Sbjct: 305 ETEEMLEFCKEKGLSSIIEVVKMDYVNTAFERLEKNDVRYRFVVDVEGS 353
>At3g19450 putative cinnamyl alcohol dehydrogenase 2
Length = 365
Score = 318 bits (814), Expect = 4e-87
Identities = 163/353 (46%), Positives = 226/353 (63%), Gaps = 2/353 (0%)
Query: 2 SSEGASEECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNS 61
S E ++ LGWAA D SGVLSPY ++ R+ G DDVY+K+ CG+C+ D+ +N LG S
Sbjct: 3 SVEAGEKKALGWAARDPSGVLSPYSYTLRSTGADDVYIKVICCGICHTDIHQIKNDLGMS 62
Query: 62 VYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFT 121
YP VPGHE+ G V +VGS+V F+VGD VGVG V C C+ C LE +C K +++
Sbjct: 63 NYPMVPGHEVVGEVLEVGSDVSKFTVGDVVGVGVVVGCCGSCKPCSSELEQYCNK-RIWS 121
Query: 122 FNGVDHDGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQP 181
+N V DG T+GG++ ++V++++ IP+ + AAPLLCAG+TVYSP+ +
Sbjct: 122 YNDVYTDGKPTQGGFADTMIVNQKFVVKIPEGMAVEQAAPLLCAGVTVYSPLSHFGLMAS 181
Query: 182 GKSLGVIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRA 241
G G++GLGG+GHM VK KA G +VTV S+S KKEEA+ LGAD +VVSSD EM+
Sbjct: 182 GLKGGILGLGGVGHMGVKIAKAMGHHVTVISSSDKKKEEAIEHLGADDYVVSSDPAEMQR 241
Query: 242 LAKSLDFIVDTASGPHSFDPYMALLKSFGVLALVG-FPGEIKIHPGLLIMGSRTVSGSGV 300
LA SLD+I+DT H DPY+A LK G L L+G ++ L+I+G + +SGS +
Sbjct: 242 LADSLDYIIDTVPVFHPLDPYLACLKLDGKLILMGVINTPLQFVTPLVILGRKVISGSFI 301
Query: 301 GGTKEIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENS 353
G KE +++ FC + IE V I N A +R+ K DV+YRFV+D+ S
Sbjct: 302 GSIKETEEVLAFCKEKGLTSTIETVKIDELNIAFERLRKNDVRYRFVVDVAGS 354
>At5g63620 alcohol dehydrogenase-like protein
Length = 427
Score = 94.0 bits (232), Expect = 1e-19
Identities = 72/280 (25%), Positives = 127/280 (44%), Gaps = 32/280 (11%)
Query: 35 DDVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVG-----SNVHGFSVGD 89
+++ +K CGVC++D+ + + + PCV GHEI G V + G ++ F +G
Sbjct: 81 NEILIKTKACGVCHSDLHVMKGEIPFAS-PCVIGHEITGEVVEHGPLTDHKIINRFPIGS 139
Query: 90 HVGVGTYVNSCRDCEFCDDGLEHHC--------VKGSVF---TFNGVDHDGT----ITKG 134
V VG ++ C C +C G + C KG+++ T + HD + + G
Sbjct: 140 RV-VGAFIMPCGTCSYCAKGHDDLCEDFFAYNRAKGTLYDGETRLFLRHDDSPVYMYSMG 198
Query: 135 GYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLG 194
G +++ V +P+S P + +A L CA T Y M +PG S+ VIG+GG+G
Sbjct: 199 GMAEYCVTPAHGLAPLPESLPYSESAILGCAVFTAYGAMAHAAEIRPGDSIAVIGIGGVG 258
Query: 195 HMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAK-----SLDFI 249
++ +AFG + + K + LGA V ++ ++ + + + +D
Sbjct: 259 SSCLQIARAFGASDIIAVDVQDDKLQKAKTLGATHIVNAAKEDAVERIREITGGMGVDVA 318
Query: 250 VDTASGPHSFDPYMALLKSFGVLALVGFP-----GEIKIH 284
V+ P +F +K G ++G GEI I+
Sbjct: 319 VEALGKPQTFMQCTLSVKDGGKAVMIGLSQAGSVGEIDIN 358
>At5g43940 alcohol dehydrogenase (EC 1.1.1.1) class III
(pir||S71244)
Length = 379
Score = 92.4 bits (228), Expect = 3e-19
Identities = 84/318 (26%), Positives = 127/318 (39%), Gaps = 37/318 (11%)
Query: 36 DVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGT 95
+V +KI + +C+ D ++PC+ GHE AGIV VG V GDHV +
Sbjct: 36 EVRIKILYTALCHTDAYTWSGKDPEGLFPCILGHEAAGIVESVGEGVTEVQAGDHV-IPC 94
Query: 96 YVNSCRDCEFCDDGLEHHCVK--------------GSVFTFNGVDHDGTITKGGYSKFIV 141
Y CR+C+FC G + C K S F+ NG + +S++ V
Sbjct: 95 YQAECRECKFCKSGKTNLCGKVRSATGVGIMMNDRKSRFSVNGKPIYHFMGTSTFSQYTV 154
Query: 142 VHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLGHMAVKFG 201
VH+ I + PL L C T + +PG ++ + GLG +G +
Sbjct: 155 VHDVSVAKIDPTAPLDKVCLLGCGVPTGLGAVWNTAKVEPGSNVAIFGLGTVGLAVAEGA 214
Query: 202 KAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLDFIVDTASG--PHSF 259
K G + + SKK E G + FV D ++ + IVD G +SF
Sbjct: 215 KTAGASRIIGIDIDSKKYETAKKFGVNEFVNPKDHDK-----PIQEVIVDLTDGGVDYSF 269
Query: 260 D----------PYMALLKSFGVLALVGFPG---EIKIHPGLLIMGSRTVSGSGVGGTKEI 306
+ K +G +VG EI P L+ G R G+ GG K
Sbjct: 270 ECIGNVSVMRAALECCHKGWGTSVIVGVAASGQEISTRPFQLVTG-RVWKGTAFGGFKS- 327
Query: 307 RDMIEFCVANEIHPNIEV 324
R + + V ++ I+V
Sbjct: 328 RTQVPWLVEKYMNKEIKV 345
>At1g77120 alcohol dehydrogenase
Length = 379
Score = 79.0 bits (193), Expect = 4e-15
Identities = 80/334 (23%), Positives = 132/334 (38%), Gaps = 32/334 (9%)
Query: 36 DVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGT 95
+V +KI +C+ DV + ++P + GHE GIV VG V GDHV +
Sbjct: 36 EVRIKILFTSLCHTDVYFWEAKGQTPLFPRIFGHEAGGIVESVGEGVTDLQPGDHV-LPI 94
Query: 96 YVNSCRDCEFCDDGLEHHCVKGSVFT-FNGVDHDGT-------------ITKGGYSKFIV 141
+ C +C C + C + T G+ HDG + +S++ V
Sbjct: 95 FTGECGECRHCHSEESNMCDLLRINTERGGMIHDGESRFSINGKPIYHFLGTSTFSEYTV 154
Query: 142 VHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLGHMAVKFG 201
VH I PL + C T + + G+S+ + GLG +G A +
Sbjct: 155 VHSGQVAKINPDAPLDKVCIVSCGLSTGLGATLNVAKPKKGQSVAIFGLGAVGLGAAEGA 214
Query: 202 KAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEE--MRALAKSLDFIVD-----TAS 254
+ G + + SK+ + G V D ++ + +A+ D VD T S
Sbjct: 215 RIAGASRIIGVDFNSKRFDQAKEFGVTECVNPKDHDKPIQQVIAEMTDGGVDRSVECTGS 274
Query: 255 GPHSFDPYMALLKSFGVLALVGFPGE---IKIHPGLLIMGSRTVSGSGVGGTK---EIRD 308
+ + +GV LVG P + K HP + + RT+ G+ G K +I
Sbjct: 275 VQAMIQAFECVHDGWGVAVLVGVPSKDDAFKTHP-MNFLNERTLKGTFFGNYKPKTDIPG 333
Query: 309 MIEFCVANEIHPN---IEVVPIQYANEALDRVIK 339
++E + E+ VP N+A D ++K
Sbjct: 334 VVEKYMNKELELEKFITHTVPFSEINKAFDYMLK 367
>At5g51970 sorbitol dehydrogenase-like protein
Length = 364
Score = 73.9 bits (180), Expect = 1e-13
Identities = 78/301 (25%), Positives = 125/301 (40%), Gaps = 24/301 (7%)
Query: 31 AVGDDDVYVKISHCGVCYADVAWTRNTLGNSVY---PCVPGHEIAGIVTKVGSNVHGFSV 87
+VG DV V++ G+C +DV + + P V GHE AGI+ +VG V V
Sbjct: 38 SVGPHDVRVRMKAVGICGSDVHYLKTMRCADFVVKEPMVIGHECAGIIEEVGEEVKHLVV 97
Query: 88 GDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHDGTITKGGYSKFIVVHERYC 147
GD V + + SC C C +G + C + F V G + +V C
Sbjct: 98 GDRVALEPGI-SCWRCNLCREGRYNLCPEMKFFATPPV-------HGSLANQVVHPADLC 149
Query: 148 FLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLGHMAVKFGKAFGLN 207
F +P++ L A +C ++V R P ++ V+G G +G + + +AF +
Sbjct: 150 FKLPENVSLEEGA--MCEPLSVGVHACRRAEVGPETNVLVMGAGPIGLVTMLAARAFSVP 207
Query: 208 VTVFSTSISKKEEALSLLGADRFV--------VSSDQEEM-RALAKSLDFIVDTASGPHS 258
V + LGAD V V S+ E++ +A+ ++D D A +
Sbjct: 208 RIVIVDVDENRLAVAKQLGADEIVQVTTNLEDVGSEVEQIQKAMGSNIDVTFDCAGFNKT 267
Query: 259 FDPYMALLKSFGVLALVGFPGEIKIHPGLLIMGSRTVSGSGVGGTKEIRDM-IEFCVANE 317
+A + G + LVG I P L +R V GV K + +EF + +
Sbjct: 268 MSTALAATRCGGKVCLVGMGHGIMTVP-LTPAAAREVDVVGVFRYKNTWPLCLEFLTSGK 326
Query: 318 I 318
I
Sbjct: 327 I 327
>At1g32780 alcohol dehydrogenase like protein
Length = 394
Score = 70.9 bits (172), Expect = 1e-12
Identities = 83/365 (22%), Positives = 145/365 (38%), Gaps = 55/365 (15%)
Query: 36 DVYVKISHCGVCYADVA-WTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVG 94
+V VKI + +C+ D+ W +P + GHE GIV VG V GD+V +
Sbjct: 37 EVRVKILYSSICHTDLGCWNGTNEAERAFPRILGHEAVGIVESVGEGVKDVKEGDYV-IP 95
Query: 95 TYVNSCRDCEFCDDGLEHHC-------VKGSVFTFNGVDHDGTITKGG------------ 135
T+ C +C+ C + C +K + G TI K G
Sbjct: 96 TFNGECGECKVCKREESNLCERYHVDPMKRVMVNDGGTRFSTTINKDGGSSQSQPIYHFL 155
Query: 136 ----YSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLG 191
++++ V+ I + PL + L C T + GKS V GLG
Sbjct: 156 NTSTFTEYTVLDSACVVKIDPNSPLKQMSLLSCGVSTGVGAAWNIANVKEGKSTAVFGLG 215
Query: 192 GLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLDFIVD 251
+G + +A G + + + + K E L+G F+ D L K + ++
Sbjct: 216 SVGLAVAEGARARGASRIIGVDANASKFEKGKLMGVTDFINPKD------LTKPVHQMIR 269
Query: 252 TASG---PHSF----------DPYMALLKSFGVLALVGF---PGEIKIHPGLLIMGSRTV 295
+G +SF + +++ +G LVG P + +HP L G R +
Sbjct: 270 EITGGGVDYSFECTGNVDVLREAFLSTHVGWGSTVLVGIYPTPRTLPLHPMELFDG-RRI 328
Query: 296 SGSGVGGTK---EIRDMIEFCVAN--EIHPNI-EVVPIQYANEALDRVIKKDVKYRFVID 349
+GS GG K ++ + + C+ ++ P I +P + N+A ++++ R ++
Sbjct: 329 TGSVFGGFKPKSQLPNFAQQCMKGVVKLEPFITNELPFEKINDAF-QLLRDGKSLRCILQ 387
Query: 350 IENSL 354
I L
Sbjct: 388 ISKLL 392
>At1g64710 alcohol dehydrogenase (EC 1.1.1.1) like protein
Length = 380
Score = 68.9 bits (167), Expect = 4e-12
Identities = 70/332 (21%), Positives = 128/332 (38%), Gaps = 35/332 (10%)
Query: 36 DVYVKISHCGVCYADV-AWTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVG 94
+V ++I +C+ D+ AW YP + GHE AGIV VG V GDHV +
Sbjct: 36 EVRIRILFTSICHTDLSAWKGENEAQRAYPRILGHEAAGIVESVGEGVEEMMAGDHV-LP 94
Query: 95 TYVNSCRDCEFCDDGLEHHCVK-------------GSVFTFNGVDHDGT---ITKGGYSK 138
+ C DC C + C + G F D+ + +S+
Sbjct: 95 IFTGECGDCRVCKRDGANLCERFRVDPMKKVMVTDGKTRFFTSKDNKPIYHFLNTSTFSE 154
Query: 139 FIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLGHMAV 198
+ V+ + +PL + L C T QP ++ + GLG +G
Sbjct: 155 YTVIDSACVLKVDPLFPLEKISLLSCGVSTGVGAAWNVADIQPASTVAIFGLGAVGLAVA 214
Query: 199 KFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEE------MRALAKSLDFIVDT 252
+ +A G + + K + G F+ + ++ M +++ +
Sbjct: 215 EGARARGASKIIGIDINPDKFQLGREAGISEFINPKESDKAVHERVMEITEGGVEYSFEC 274
Query: 253 ASGPHSF-DPYMALLKSFGVLALVGF---PGEIKIHPGLLIMGSRTVSGSGVGGTK---E 305
A + + +++ GV ++G P + IHP L G R+++ S GG K +
Sbjct: 275 AGSIEALREAFLSTNSGVGVTVMLGVHASPQLLPIHPMELFQG-RSITASVFGGFKPKTQ 333
Query: 306 IRDMIEFCVANEIHPNIEV---VPIQYANEAL 334
+ I C+ ++ ++ + +P NEA+
Sbjct: 334 LPFFITQCLQGLLNLDLFISHQLPFHDINEAM 365
>At5g24760 alcohol dehydrogenase - like protein
Length = 381
Score = 65.1 bits (157), Expect = 6e-11
Identities = 70/298 (23%), Positives = 109/298 (36%), Gaps = 30/298 (10%)
Query: 36 DVYVKISHCGVCYADV-AWTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVG 94
++ +K+ +C +D+ AW S+ P + GHE AGIV +G V F GDHV +
Sbjct: 42 EIRIKVVCTSLCRSDLSAWE----SQSLLPRIFGHEAAGIVESIGEGVTEFEKGDHV-LA 96
Query: 95 TYVNSCRDCEFCDDGLEHHCV------KGSV-------FTFNGVDHDGTITKGGYSKFIV 141
+ C C C G + C KG + F+ G +S++ V
Sbjct: 97 VFTGECGSCRHCISGKSNMCQVLGMERKGLMHSDQKTRFSIKGKPVYHYCAVSSFSEYTV 156
Query: 142 VHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLGHMAVKFG 201
VH + PL L C Q G S+ + GLG +G +
Sbjct: 157 VHSGCAVKVDPLAPLHKICLLSCGVAAGLGAAWNVADVQKGSSVVIFGLGTVGLSVAQGA 216
Query: 202 KAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEE------MRALAKSLDFIVDTASG 255
K G + K E G F+ S+D E R DF +
Sbjct: 217 KLRGAAQILGVDINPAKAEQAKTFGVTDFINSNDLSEPIPQVIKRMTGGGADFSFECVGD 276
Query: 256 PH-SFDPYMALLKSFGVLALVGFP---GEIKIHPGLLIMGSRTVSGSGVGGTKEIRDM 309
+ + +G+ +G P E+ H GL + G +++ G+ GG K D+
Sbjct: 277 TGIATTALQSCSDGWGMTVTLGVPKAKPEVSAHYGLFLSG-KSLKGTLFGGWKPKSDL 333
>At4g22110 alcohol dehydrogenase like protein
Length = 389
Score = 60.1 bits (144), Expect = 2e-09
Identities = 47/174 (27%), Positives = 76/174 (43%), Gaps = 15/174 (8%)
Query: 36 DVYVKISHCGVCYADVAWTRNTLGNSV-YPCVPGHEIAGIVTKVGSNVHGFSVGDHV--G 92
+V +KI +C+ DV++++ G +P + GHE G++ +G +V+GF GD V
Sbjct: 43 EVRIKIICTSLCHTDVSFSKIDSGPLARFPRILGHEAVGVIESIGEHVNGFQQGDVVLPV 102
Query: 93 VGTYVNSCRDCE---------FCDDGLEHHCVKGSVFTFN---GVDHDGTITKGGYSKFI 140
+ CRDC+ F DD L + G F G D + +S++
Sbjct: 103 FHPHCEECRDCKSSKSNWCARFADDFLSNTRRYGMTSRFKDSFGEDIYHFLFVSSFSEYT 162
Query: 141 VVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLG 194
VV + I P+ AA L C T + + G ++ V GLG +G
Sbjct: 163 VVDIAHLVKISPDIPVDKAALLSCGVSTGIGAAWKVANVEKGSTVAVFGLGAVG 216
>At3g56460 quinone reductase-like protein
Length = 348
Score = 59.7 bits (143), Expect = 2e-09
Identities = 65/245 (26%), Positives = 106/245 (42%), Gaps = 49/245 (20%)
Query: 64 PCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFN 123
P +PG + +GIV +G V F VGD V C F D
Sbjct: 69 PFIPGSDYSGIVDAIGPAVTKFRVGDRV-----------CSFAD---------------- 101
Query: 124 GVDHDGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGK 183
G +++FIV + FL+P+ + +AA L A T + ++ G+
Sbjct: 102 ---------LGSFAQFIVADQSRLFLVPERCDMVAAAALPVAFGTSHVALVHRARLTSGQ 152
Query: 184 SLGVIG-LGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRF-------VVSSD 235
L V+G GG+G AV+ GK G V + ++K + L +G D V+SS
Sbjct: 153 VLLVLGAAGGVGLAAVQIGKVCGAIVIAVARG-TEKIQLLKSMGVDHVVDLGTENVISSV 211
Query: 236 QEEMRA-LAKSLDFIVDTASGPHSFDPYMALLKSFGVLALVGF-PGEIKIHP-GLLIMGS 292
+E ++ K +D + D G + + M +LK + ++GF GEI + P + ++ +
Sbjct: 212 KEFIKTRKLKGVDVLYDPVGGKLTKES-MKVLKWGAQILVIGFASGEIPVIPANIALVKN 270
Query: 293 RTVSG 297
TV G
Sbjct: 271 WTVHG 275
>At1g22430 putative alcohol dehydrogenase
Length = 388
Score = 59.7 bits (143), Expect = 2e-09
Identities = 61/253 (24%), Positives = 101/253 (39%), Gaps = 37/253 (14%)
Query: 36 DVYVKISHCGVCYADVAWTRNTLGN-SVYPCVPGHEIAGIVTKVGSNVHGFSVGDHV--G 92
+V +KI +C+ D+ + + + G S +P + GHE G+V +G NV GF GD V
Sbjct: 42 EVRIKILCTSLCHTDLTFWKLSFGPISRFPRILGHEAVGVVESIGENVDGFKQGDVVLPV 101
Query: 93 VGTYVNSCRDCE---------FCDDGLEHHCVKGSVFTFNGVDHDGTITK-----GGYSK 138
Y C+DC+ + +D + + G F D G + +S+
Sbjct: 102 FHPYCEECKDCKSSKTNWCDRYAEDFISNTRRYGMASRFK--DSSGEVIHHFLFVSSFSE 159
Query: 139 FIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLGHMAV 198
+ VV + I P+ AA L C T + + G ++ + GLG +G +AV
Sbjct: 160 YTVVDIAHLVKISPEIPVDKAALLSCGVSTGIGAAWKVANVEEGSTIAIFGLGAVG-LAV 218
Query: 199 KFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLDFIVDTASGPHS 258
G ++ A ++G D +SD+ E+ DFI T G
Sbjct: 219 AEG--------------ARLRGAAKIIGID---TNSDKFELGKKFGFTDFINPTLCGEKK 261
Query: 259 FDPYMALLKSFGV 271
+ + GV
Sbjct: 262 ISEVIKEMTEGGV 274
>At1g23740 auxin-induced protein like
Length = 386
Score = 58.9 bits (141), Expect = 4e-09
Identities = 73/277 (26%), Positives = 114/277 (40%), Gaps = 41/277 (14%)
Query: 8 EECLGWAATDTSGVLSPYKFSRRAV----GDDDVYVKISHCGVCYADVAWTRNTLG--NS 61
+E W +D GV K V +D V +K+ + D + +S
Sbjct: 76 KEMKAWVYSDYGGV-DVLKLESNIVVPEIKEDQVLIKVVAAALNPVDAKRRQGKFKATDS 134
Query: 62 VYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFT 121
P VPG+++AG+V KVGS V GD V Y N + LE
Sbjct: 135 PLPTVPGYDVAGVVVKVGSAVKDLKEGDEV----YAN------VSEKALE---------- 174
Query: 122 FNGVDHDGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQP 181
G G +++ V E+ L PK+ A AA L A T ++R + +
Sbjct: 175 -------GPKQFGSLAEYTAVEEKLLALKPKNIDFAQAAGLPLAIETADEGLVRTEFS-A 226
Query: 182 GKSLGVI-GLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMR 240
GKS+ V+ G GG+G + ++ K V +T+ ++K E + LGAD + +E +
Sbjct: 227 GKSILVLNGAGGVGSLVIQLAKHVYGASKVAATASTEKLELVRSLGAD-LAIDYTKENIE 285
Query: 241 ALAKSLDFIVDTASGPHSFDPYMALLKSFG-VLALVG 276
L D + D D + ++K G V+AL G
Sbjct: 286 DLPDKYDVVFDAIG---MCDKAVKVIKEGGKVVALTG 319
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.139 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,134,628
Number of Sequences: 26719
Number of extensions: 360341
Number of successful extensions: 790
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 718
Number of HSP's gapped (non-prelim): 47
length of query: 355
length of database: 11,318,596
effective HSP length: 100
effective length of query: 255
effective length of database: 8,646,696
effective search space: 2204907480
effective search space used: 2204907480
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0055.3


