

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0055.1
(241 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
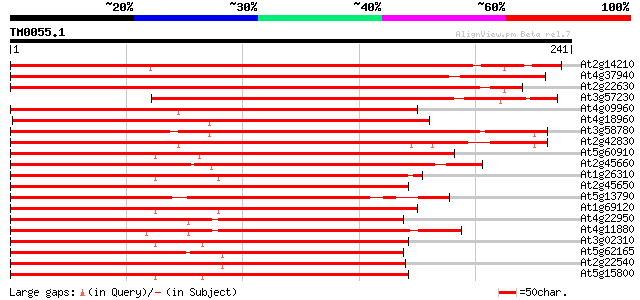
Score E
Sequences producing significant alignments: (bits) Value
At2g14210 pseudogene 280 4e-76
At4g37940 MADS-box protein AGL17 -like protein 273 6e-74
At2g22630 MADS-box protein AGL17 (AGL17) 255 2e-68
At3g57230 MADS-box transcription factor (AGL16) 169 2e-42
At4g09960 MADS-box protein AGL11 167 6e-42
At4g18960 floral homeotic protein agamous 166 1e-41
At3g58780 shatterproof 1 (SHP1)/ agamous -like 1 (AGL1) 166 1e-41
At2g42830 MADS box transcription factor (AGL5) 162 1e-40
At5g60910 floral homeotic protein AGL8 158 3e-39
At2g45660 MADS-box protein (AGL20) 158 3e-39
At1g26310 cauliflower (CAL) 158 3e-39
At2g45650 MADS-box protein (AGL6) 157 4e-39
At5g13790 floral homeotic protein AGL15 (sp|Q38847) 156 8e-39
At1g69120 unknown protein 156 8e-39
At4g22950 putative MADS Box / AGL protein 150 6e-37
At4g11880 MADS-box protein AGL14 150 6e-37
At3g02310 floral homeotic protein AGL4 149 1e-36
At5g62165 unknown protein 148 3e-36
At2g22540 putative MADS-box protein 147 6e-36
At5g15800 transcription factor (AGL2) 146 8e-36
>At2g14210 pseudogene
Length = 234
Score = 280 bits (717), Expect = 4e-76
Identities = 150/239 (62%), Positives = 191/239 (79%), Gaps = 8/239 (3%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAS- 59
MGRGKI IRRIDNSTSRQVTFSKRR+GLLKKAKELSILCDAEVG+I+FSSTGKLY+YAS
Sbjct: 1 MGRGKIVIRRIDNSTSRQVTFSKRRSGLLKKAKELSILCDAEVGVIIFSSTGKLYDYASN 60
Query: 60 TSMKSVIERYNKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLG 119
+SMK++IERYN++KEE +QL+N ASE+KFWQ E ASL+QQLQYLQECHR+L+GE LSG+
Sbjct: 61 SSMKTIIERYNRVKEEQHQLLNHASEIKFWQREVASLQQQLQYLQECHRKLVGEELSGMN 120
Query: 120 VKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQKENAE 179
+LQNLE+QL SLKGVR+KKDQ++T+EI+EL++KG +I +EN EL +++++KEN +
Sbjct: 121 ANDLQNLEDQLVTSLKGVRLKKDQLMTNEIRELNRKGQIIQKENHELQNIVDIMRKENIK 180
Query: 180 LQKKVYEARSTNEKDAASSPSCTIRNGYDLQA-ISLQLSQPQPQFSEPPAKAVKLGYSL 237
LQKKV+ + E +++ P I NG A LQL Q QP P K+++LG L
Sbjct: 181 LQKKVHGRTNAIEGNSSVDP---ISNGTTTYAPPQLQLIQLQP---APREKSIRLGLQL 233
>At4g37940 MADS-box protein AGL17 -like protein
Length = 228
Score = 273 bits (698), Expect = 6e-74
Identities = 142/230 (61%), Positives = 178/230 (76%), Gaps = 4/230 (1%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
MGRGKI I+RID+STSRQVTFSKRR GL+KKAKEL+ILCDAEVGLI+FSSTGKLY++AS+
Sbjct: 1 MGRGKIVIQRIDDSTSRQVTFSKRRKGLIKKAKELAILCDAEVGLIIFSSTGKLYDFASS 60
Query: 61 SMKSVIERYNKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLGV 120
SMKSVI+RYNK K E QL+NPASE+KFWQ EAA LRQ+L LQE HRQ+MGE L+GL V
Sbjct: 61 SMKSVIDRYNKSKIEQQQLLNPASEVKFWQREAAVLRQELHALQENHRQMMGEQLNGLSV 120
Query: 121 KELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQKENAEL 180
EL +LENQ+E+SL+G+RM+K+Q+LT EI+EL QK NLIHQEN +L +K++ I +EN EL
Sbjct: 121 NELNSLENQIEISLRGIRMRKEQLLTQEIQELSQKRNLIHQENLDLSRKVQRIHQENVEL 180
Query: 181 QKKVYEARSTNEKDAASSPSCTIRNGYDLQAISLQLSQPQPQFSEPPAKA 230
KK Y A + + + + + I LQLSQP+ + P +A
Sbjct: 181 YKKAYMANT----NGFTHREVAVADDESHTQIRLQLSQPEHSDYDTPPRA 226
>At2g22630 MADS-box protein AGL17 (AGL17)
Length = 227
Score = 255 bits (651), Expect = 2e-68
Identities = 135/221 (61%), Positives = 170/221 (76%), Gaps = 5/221 (2%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
MGRGKI I++ID+STSRQVTFSKRR GL+KKAKEL+ILCDAEV LI+FS+T KLY++AS+
Sbjct: 1 MGRGKIVIQKIDDSTSRQVTFSKRRKGLIKKAKELAILCDAEVCLIIFSNTDKLYDFASS 60
Query: 61 SMKSVIERYNKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLGV 120
S+KS IER+N K E +LMNPASE+KFWQ EA +LRQ+L LQE +RQL G L+GL V
Sbjct: 61 SVKSTIERFNTAKMEEQELMNPASEVKFWQREAETLRQELHSLQENYRQLTGVELNGLSV 120
Query: 121 KELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQKENAEL 180
KELQN+E+QLEMSL+G+RMK++QILT+EIKEL +K NL+H EN EL +K++ I +EN EL
Sbjct: 121 KELQNIESQLEMSLRGIRMKREQILTNEIKELTRKRNLVHHENLELSRKVQRIHQENVEL 180
Query: 181 QKKVYEARSTNEKDAASSPSCTIRNGYDLQA-ISLQLSQPQ 220
KK Y +TN Y+ A + LQLSQP+
Sbjct: 181 YKKAYGTSNTNGLGHHELVDAV----YESHAQVRLQLSQPE 217
>At3g57230 MADS-box transcription factor (AGL16)
Length = 179
Score = 169 bits (427), Expect = 2e-42
Identities = 92/179 (51%), Positives = 127/179 (70%), Gaps = 10/179 (5%)
Query: 62 MKSVIERYNKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLGVK 121
MKSVIERY+ K E + +PASE++FWQ EAA L++QL LQE HRQ+MGE LSGL V+
Sbjct: 1 MKSVIERYSDAKGETSSENDPASEIQFWQKEAAILKRQLHNLQENHRQMMGEELSGLSVE 60
Query: 122 ELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQKENAELQ 181
LQNLENQLE+SL+GVRMKKDQ+L +EI+ L+++GNL+HQEN +L+KK+ L+ ++N EL
Sbjct: 61 ALQNLENQLELSLRGVRMKKDQMLIEEIQVLNREGNLVHQENLDLHKKVNLMHQQNMELH 120
Query: 182 KKVYEARSTNEKDAASSPSCTIRNGYDL-----QAISLQLSQPQPQFSEPPAKAVKLGY 235
+KV E ++ + + NG D+ + + LQLSQPQ E +KA++L Y
Sbjct: 121 EKVSEVEGVK----IANKNSLLTNGLDMRDTSNEHVHLQLSQPQHD-HETHSKAIQLNY 174
>At4g09960 MADS-box protein AGL11
Length = 230
Score = 167 bits (422), Expect = 6e-42
Identities = 88/176 (50%), Positives = 122/176 (69%), Gaps = 1/176 (0%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
MGRGKI I+RI+NST+RQVTF KRRNGLLKKA ELS+LCDAEV LIVFS+ G+LYEYA+
Sbjct: 1 MGRGKIEIKRIENSTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSTRGRLYEYANN 60
Query: 61 SMKSVIERYNK-LKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLG 119
+++S IERY K + N ++Q E+A LRQQ+Q +Q +R LMG+ LS L
Sbjct: 61 NIRSTIERYKKACSDSTNTSTVQEINAAYYQQESAKLRQQIQTIQNSNRNLMGDSLSSLS 120
Query: 120 VKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQK 175
VKEL+ +EN+LE ++ +R KK ++L EI+ ++ + EN L K+ +++
Sbjct: 121 VKELKQVENRLEKAISRIRSKKHELLLVEIENAQKREIELDNENIYLRTKVAEVER 176
>At4g18960 floral homeotic protein agamous
Length = 284
Score = 166 bits (420), Expect = 1e-41
Identities = 87/180 (48%), Positives = 125/180 (69%), Gaps = 1/180 (0%)
Query: 2 GRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYASTS 61
GRGKI I+RI+N+T+RQVTF KRRNGLLKKA ELS+LCDAEV LIVFSS G+LYEY++ S
Sbjct: 50 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYSNNS 109
Query: 62 MKSVIERYNKLKEEHNQLMNPAS-ELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLGV 120
+K IERY K +++ + A +++Q E+A LRQQ+ +Q +RQLMGE + +
Sbjct: 110 VKGTIERYKKAISDNSNTGSVAEINAQYYQQESAKLRQQIISIQNSNRQLMGETIGSMSP 169
Query: 121 KELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQKENAEL 180
KEL+NLE +LE S+ +R KK+++L EI + ++ +H +N L K+ ++ N +
Sbjct: 170 KELRNLEGRLERSITRIRSKKNELLFSEIDYMQKREVDLHNDNQILRAKIAENERNNPSI 229
>At3g58780 shatterproof 1 (SHP1)/ agamous -like 1 (AGL1)
Length = 248
Score = 166 bits (420), Expect = 1e-41
Identities = 97/238 (40%), Positives = 147/238 (61%), Gaps = 12/238 (5%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
+GRGKI I+RI+N+T+RQVTF KRRNGLLKKA ELS+LCDAEV L++FS+ G+LYEYA+
Sbjct: 16 LGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVIFSTRGRLYEYANN 75
Query: 61 SMKSVIERYNKLKEEHNQLMNPAS----ELKFWQTEAASLRQQLQYLQECHRQLMGEGLS 116
S++ IERY K+ + +NP S +++Q EA+ LR+Q++ +Q +R ++GE L
Sbjct: 76 SVRGTIERY---KKACSDAVNPPSVTEANTQYYQQEASKLRRQIRDIQNSNRHIVGESLG 132
Query: 117 GLGVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQKE 176
L KEL+NLE +LE + VR KK+++L EI+ + ++ + N L K+ +
Sbjct: 133 SLNFKELKNLEGRLEKGISRVRSKKNELLVAEIEYMQKREMELQHNNMYLRAKIAEGARL 192
Query: 177 NAELQKKVYEARSTNEKDAASSPSCTIRNGYDLQAISLQLSQPQPQFS---EPPAKAV 231
N + Q+ +T + SS + Y+ I + L +P QFS +PP + V
Sbjct: 193 NPDQQESSVIQGTTVYESGVSSHDQS--QHYNRNYIPVNLLEPNQQFSGQDQPPLQLV 248
>At2g42830 MADS box transcription factor (AGL5)
Length = 246
Score = 162 bits (410), Expect = 1e-40
Identities = 96/240 (40%), Positives = 148/240 (61%), Gaps = 18/240 (7%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
+GRGKI I+RI+N+T+RQVTF KRRNGLLKKA ELS+LCDAEV L++FS+ G+LYEYA+
Sbjct: 16 IGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVIFSTRGRLYEYANN 75
Query: 61 SMKSVIERYNK-LKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLG 119
S++ IERY K + N + +++Q EA+ LR+Q++ +Q +R ++GE L L
Sbjct: 76 SVRGTIERYKKACSDAVNPPTITEANTQYYQQEASKLRRQIRDIQNLNRHILGESLGSLN 135
Query: 120 VKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKME----LIQK 175
KEL+NLE++LE + VR KK ++L EI+ + ++ + +N L K+ L Q+
Sbjct: 136 FKELKNLESRLEKGISRVRSKKHEMLVAEIEYMQKREIELQNDNMYLRSKITERTGLQQQ 195
Query: 176 ENAEL-QKKVYEARSTNEKDAASSPSCTIRNGYDLQAISLQLSQPQPQFS---EPPAKAV 231
E++ + Q VYE+ T+ + Y+ I++ L +P S +PP + V
Sbjct: 196 ESSVIHQGTVYESGVTSSHQSGQ---------YNRNYIAVNLLEPNQNSSNQDQPPLQLV 246
>At5g60910 floral homeotic protein AGL8
Length = 242
Score = 158 bits (399), Expect = 3e-39
Identities = 80/193 (41%), Positives = 131/193 (67%), Gaps = 2/193 (1%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
MGRG++ ++RI+N +RQVTFSKRR+GLLKKA E+S+LCDAEV LIVFSS GKL+EY++
Sbjct: 1 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAHEISVLCDAEVALIVFSSKGKLFEYSTD 60
Query: 61 S-MKSVIERYNKLKEEHNQLM-NPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGL 118
S M+ ++ERY++ QL+ S+ + W E A L+ +++ L++ R MGE L L
Sbjct: 61 SCMERILERYDRYLYSDKQLVGRDVSQSENWVLEHAKLKARVEVLEKNKRNFMGEDLDSL 120
Query: 119 GVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQKENA 178
+KELQ+LE+QL+ ++K +R +K+Q + + I L +K + N L KK++ +K+
Sbjct: 121 SLKELQSLEHQLDAAIKSIRSRKNQAMFESISALQKKDKALQDHNNSLLKKIKEREKKTG 180
Query: 179 ELQKKVYEARSTN 191
+ + ++ + +++
Sbjct: 181 QQEGQLVQCSNSS 193
>At2g45660 MADS-box protein (AGL20)
Length = 214
Score = 158 bits (399), Expect = 3e-39
Identities = 89/205 (43%), Positives = 134/205 (64%), Gaps = 7/205 (3%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
M RGK ++RI+N+TSRQVTFSKRRNGLLKKA ELS+LCDAEV LI+FS GKLYE+AS+
Sbjct: 1 MVRGKTQMKRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVSLIIFSPKGKLYEFASS 60
Query: 61 SMKSVIERYNKLKEEHNQLMNPASE--LKFWQTEAASLRQQLQYLQECHRQLMGEGLSGL 118
+M+ I+RY + ++ P SE ++ + EAA++ ++++ L+ R+L+GEG+
Sbjct: 61 NMQDTIDRYLRHTKDRVS-TKPVSEENMQHLKYEAANMMKKIEQLEASKRKLLGEGIGTC 119
Query: 119 GVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQKENA 178
++ELQ +E QLE S+K +R +K Q+ ++I++L QK + EN +L +K + E
Sbjct: 120 SIEELQQIEQQLEKSVKCIRARKTQVFKEQIEQLKQKEKALAAENEKLSEKWGSHESEVW 179
Query: 179 ELQKKVYEARSTNEKDAASSPSCTI 203
+ + ST D SSPS +
Sbjct: 180 SNKNQ----ESTGRGDEESSPSSEV 200
>At1g26310 cauliflower (CAL)
Length = 255
Score = 158 bits (399), Expect = 3e-39
Identities = 82/181 (45%), Positives = 124/181 (68%), Gaps = 6/181 (3%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
MGRG++ ++RI+N +RQVTFSKRR GLLKKA+E+S+LCDAEV LIVFS GKL+EY+S
Sbjct: 1 MGRGRVELKRIENKINRQVTFSKRRTGLLKKAQEISVLCDAEVSLIVFSHKGKLFEYSSE 60
Query: 61 S-MKSVIERYNKLKEEHNQLMNPASELKF---WQTEAASLRQQLQYLQECHRQLMGEGLS 116
S M+ V+ERY + QL+ P S + W E + L+ +++ L+ R +GE L
Sbjct: 61 SCMEKVLERYERYSYAERQLIAPDSHVNAQTNWSMEYSRLKAKIELLERNQRHYLGEELE 120
Query: 117 GLGVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQKE 176
+ +K+LQNLE QLE +LK +R +K+Q++ + + L +K I +EN+ L K+++ ++E
Sbjct: 121 PMSLKDLQNLEQQLETALKHIRSRKNQLMNESLNHLQRKEKEIQEENSMLTKQIK--ERE 178
Query: 177 N 177
N
Sbjct: 179 N 179
>At2g45650 MADS-box protein (AGL6)
Length = 252
Score = 157 bits (398), Expect = 4e-39
Identities = 81/171 (47%), Positives = 111/171 (64%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
MGRG++ ++RI+N +RQVTFSKRRNGLLKKA ELS+LCDAEV LI+FSS GKLYE+ S
Sbjct: 1 MGRGRVEMKRIENKINRQVTFSKRRNGLLKKAYELSVLCDAEVALIIFSSRGKLYEFGSV 60
Query: 61 SMKSVIERYNKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLGV 120
++S IERYN+ P + W E L+ + + L +R L+GE L +GV
Sbjct: 61 GIESTIERYNRCYNCSLSNNKPEETTQSWCQEVTKLKSKYESLVRTNRNLLGEDLGEMGV 120
Query: 121 KELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKME 171
KELQ LE QLE +L R +K Q++ +E+++L +K + N +L K E
Sbjct: 121 KELQALERQLEAALTATRQRKTQVMMEEMEDLRKKERQLGDINKQLKIKFE 171
>At5g13790 floral homeotic protein AGL15 (sp|Q38847)
Length = 268
Score = 156 bits (395), Expect = 8e-39
Identities = 93/189 (49%), Positives = 125/189 (65%), Gaps = 20/189 (10%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
MGRGKI I+RI+N+ SRQVTFSKRR+GLLKKA+ELS+LCDAEV +IVFS +GKL+EY+ST
Sbjct: 1 MGRGKIEIKRIENANSRQVTFSKRRSGLLKKARELSVLCDAEVAVIVFSKSGKLFEYSST 60
Query: 61 SMKSVIERYNKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLGV 120
MK + RY N + AS+ + E L+ QL LQE H QL G+GL+ L
Sbjct: 61 GMKQTLSRYG------NHQSSSASKAEEDCAEVDILKDQLSKLQEKHLQLQGKGLNPLTF 114
Query: 121 KELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQKENAEL 180
KELQ+LE QL +L VR +K+++LT++++E K ++ AEL EN L
Sbjct: 115 KELQSLEQQLYHALITVRERKERLLTNQLEESRLK-----EQRAEL---------ENETL 160
Query: 181 QKKVYEARS 189
+++V E RS
Sbjct: 161 RRQVQELRS 169
>At1g69120 unknown protein
Length = 256
Score = 156 bits (395), Expect = 8e-39
Identities = 79/177 (44%), Positives = 121/177 (67%), Gaps = 2/177 (1%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
MGRG++ ++RI+N +RQVTFSKRR GLLKKA E+S+LCDAEV L+VFS GKL+EY++
Sbjct: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALVVFSHKGKLFEYSTD 60
Query: 61 S-MKSVIERYNKLKEEHNQLMNPASELKF-WQTEAASLRQQLQYLQECHRQLMGEGLSGL 118
S M+ ++ERY + QL+ P S++ W E L+ +++ L+ R +GE L +
Sbjct: 61 SCMEKILERYERYSYAERQLIAPESDVNTNWSMEYNRLKAKIELLERNQRHYLGEDLQAM 120
Query: 119 GVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQK 175
KELQNLE QL+ +LK +R +K+Q++ + I EL +K I ++N+ L K+++ +K
Sbjct: 121 SPKELQNLEQQLDTALKHIRTRKNQLMYESINELQKKEKAIQEQNSMLSKQIKEREK 177
>At4g22950 putative MADS Box / AGL protein
Length = 219
Score = 150 bits (379), Expect = 6e-37
Identities = 81/171 (47%), Positives = 120/171 (69%), Gaps = 4/171 (2%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
M RGK ++RI+N+TSRQVTFSKRRNGLLKKA ELS+LCDAEV L++FS KLYE++S+
Sbjct: 1 MVRGKTEMKRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALVIFSPRSKLYEFSSS 60
Query: 61 SMKSVIERYNKLKEE--HNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGL 118
S+ + IERY + +E +N N S+ + E + L ++++ L+ R+L+GEG+
Sbjct: 61 SIAATIERYQRRIKEIGNNHKRNDNSQQA--RDETSGLTKKIEQLEISKRKLLGEGIDAC 118
Query: 119 GVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKK 169
++ELQ LENQL+ SL +R KK Q+L +EI++L + + +EN +L +K
Sbjct: 119 SIEELQQLENQLDRSLSRIRAKKYQLLREEIEKLKAEERNLVKENKDLKEK 169
>At4g11880 MADS-box protein AGL14
Length = 221
Score = 150 bits (379), Expect = 6e-37
Identities = 88/197 (44%), Positives = 129/197 (64%), Gaps = 8/197 (4%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEY-AS 59
M RGK ++RI+N+TSRQVTFSKRRNGLLKKA ELS+LCDAEV LI+FS GKLYE+ +S
Sbjct: 1 MVRGKTEMKRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALIIFSPRGKLYEFSSS 60
Query: 60 TSMKSVIERYNKLKEE--HNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSG 117
+S+ +ERY K ++ N N S+ + E L +++++L+ R++MGEGL
Sbjct: 61 SSIPKTVERYQKRIQDLGSNHKRNDNSQQS--KDETYGLARKIEHLEISTRKMMGEGLDA 118
Query: 118 LGVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQKEN 177
++ELQ LENQL+ SL +R KK Q+L +E ++L +K + EN L +K E+ +
Sbjct: 119 SSIEELQQLENQLDRSLMKIRAKKYQLLREETEKLKEKERNLIAENKMLMEKCEM---QG 175
Query: 178 AELQKKVYEARSTNEKD 194
+ ++ + ST+E D
Sbjct: 176 RGIIGRISSSSSTSELD 192
>At3g02310 floral homeotic protein AGL4
Length = 250
Score = 149 bits (377), Expect = 1e-36
Identities = 85/173 (49%), Positives = 113/173 (65%), Gaps = 2/173 (1%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
MGRG++ ++RI+N +RQVTF+KRRNGLLKKA ELS+LCDAEV LIVFS+ GKLYE+ ST
Sbjct: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVSLIVFSNRGKLYEFCST 60
Query: 61 S-MKSVIERYNKLKEEHNQLMN-PASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGL 118
S M +ERY K ++ N PA EL+ E L+ + + LQ R L+GE L L
Sbjct: 61 SNMLKTLERYQKCSYGSIEVNNKPAKELENSYREYLKLKGRYENLQRQQRNLLGEDLGPL 120
Query: 119 GVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKME 171
KEL+ LE QL+ SLK VR K Q + D++ +L K +++ N L K+E
Sbjct: 121 NSKELEQLERQLDGSLKQVRCIKTQYMLDQLSDLQGKEHILLDANRALSMKLE 173
>At5g62165 unknown protein
Length = 210
Score = 148 bits (373), Expect = 3e-36
Identities = 75/171 (43%), Positives = 123/171 (71%), Gaps = 3/171 (1%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
M RGKI +++I+N+TSRQVTFSKRRNGLLKKA ELS+LCDA++ LI+FS G+LYE++S+
Sbjct: 1 MVRGKIEMKKIENATSRQVTFSKRRNGLLKKAYELSVLCDAQLSLIIFSQRGRLYEFSSS 60
Query: 61 SMKSVIERYNKLKEEHNQLMNPASELKFWQ--TEAASLRQQLQYLQECHRQLMGEGLSGL 118
M+ IERY K ++H + N S++ Q EA+ + +++ L+ R+L+G+G++
Sbjct: 61 DMQKTIERYRKYTKDH-ETSNHDSQIHLQQLKQEASHMITKIELLEFHKRKLLGQGIASC 119
Query: 119 GVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKK 169
++ELQ +++QL+ SL VR +K Q+ +++++L K + +EN +L++K
Sbjct: 120 SLEELQEIDSQLQRSLGKVRERKAQLFKEQLEKLKAKEKQLLEENVKLHQK 170
>At2g22540 putative MADS-box protein
Length = 210
Score = 147 bits (370), Expect = 6e-36
Identities = 81/171 (47%), Positives = 113/171 (65%), Gaps = 1/171 (0%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
M R KI IR+IDN+T+RQVTFSKRR GL KKA+ELS+LCDA+V LI+FSSTGKL+E+ S+
Sbjct: 1 MAREKIQIRKIDNATARQVTFSKRRRGLFKKAEELSVLCDADVALIIFSSTGKLFEFCSS 60
Query: 61 SMKSVIERYNKLKEEHNQLMNPASELKFWQ-TEAASLRQQLQYLQECHRQLMGEGLSGLG 119
SMK V+ER+N + +L P+ EL+ + ++ A + +++ RQ+ GE L GL
Sbjct: 61 SMKEVLERHNLQSKNLEKLDQPSLELQLVENSDHARMSKEIADKSHRLRQMRGEELQGLD 120
Query: 120 VKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKM 170
++ELQ LE LE L V K + EI EL +KG + EN L +++
Sbjct: 121 IEELQQLEKALETGLTRVIETKSDKIMSEISELQKKGMQLMDENKRLRQQV 171
>At5g15800 transcription factor (AGL2)
Length = 251
Score = 146 bits (369), Expect = 8e-36
Identities = 81/173 (46%), Positives = 113/173 (64%), Gaps = 2/173 (1%)
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
MGRG++ ++RI+N +RQVTF+KRRNGLLKKA ELS+LCDAEV LI+FS+ GKLYE+ S+
Sbjct: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
Query: 61 S-MKSVIERYNKLKEEHNQLMN-PASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGL 118
S M ++RY K ++ N PA EL+ E L+ + + LQ R L+GE L L
Sbjct: 61 SNMLKTLDRYQKCSYGSIEVNNKPAKELENSYREYLKLKGRYENLQRQQRNLLGEDLGPL 120
Query: 119 GVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKME 171
KEL+ LE QL+ SLK VR K Q + D++ +L K ++ + N L K++
Sbjct: 121 NSKELEQLERQLDGSLKQVRSIKTQYMLDQLSDLQNKEQMLLETNRALAMKLD 173
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.129 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,903,172
Number of Sequences: 26719
Number of extensions: 197087
Number of successful extensions: 1055
Number of sequences better than 10.0: 206
Number of HSP's better than 10.0 without gapping: 125
Number of HSP's successfully gapped in prelim test: 81
Number of HSP's that attempted gapping in prelim test: 835
Number of HSP's gapped (non-prelim): 258
length of query: 241
length of database: 11,318,596
effective HSP length: 96
effective length of query: 145
effective length of database: 8,753,572
effective search space: 1269267940
effective search space used: 1269267940
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0055.1


