

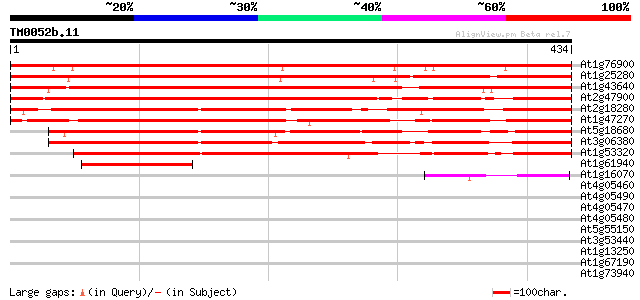
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0052b.11
(434 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g76900 unknown protein 613 e-176
At1g25280 unknown protein 609 e-175
At1g43640 unknown protein (At1g43640) 582 e-166
At2g47900 Tub family like protein 458 e-129
At2g18280 tubby-like protein 2 419 e-117
At1g47270 unknown protein 411 e-115
At5g18680 tub family-like protein 375 e-104
At3g06380 unknown protein 371 e-103
At1g53320 tubby like protein (Tub family protein) 338 3e-93
At1g61940 hypothetical protein, 5' partial 111 7e-25
At1g16070 unknown protein 58 9e-09
At4g05460 N7 like-protein 38 0.012
At4g05490 N7 -like protein 37 0.016
At4g05470 N7 -like protein 36 0.047
At4g05480 N7 -like protein 35 0.081
At5g55150 putative protein 34 0.18
At3g53440 unknown protein 33 0.24
At1g13250 unknown protein 33 0.40
At1g67190 unknown protein 32 0.52
At1g73940 unknown protein 32 0.89
>At1g76900 unknown protein
Length = 455
Score = 613 bits (1582), Expect = e-176
Identities = 327/455 (71%), Positives = 367/455 (79%), Gaps = 21/455 (4%)
Query: 1 MSFRSIVRDVRDGFGSLSRRSFDVILPGHSRS--KSRSSVHELHDQPLV--IQSSRWASL 56
MSFRSIVRDVRD GSLSRRSFD L ++ KSR SV + H++ LV IQ + WA+L
Sbjct: 1 MSFRSIVRDVRDSIGSLSRRSFDFKLSSLNKEGGKSRGSVQDSHEEQLVVTIQETPWANL 60
Query: 57 PPELLRDVIKRLETNESAWPGRKHVVACAAVCKSWREMCKEIVGSPEFCGKITFPVSLKQ 116
PPELLRDVIKRLE +ES WP R+HVVACA+VC+SWR+MCKEIV SPE GKITFPVSLKQ
Sbjct: 61 PPELLRDVIKRLEESESVWPARRHVVACASVCRSWRDMCKEIVQSPELSGKITFPVSLKQ 120
Query: 117 PGYRDGPIQCFIKRDKSKLTYHLFLCLSPALLVENGKFLLSAKRTRRTTCTEYIISMDAD 176
PG RD +QCFIKRDKS LTYHL+LCLSPALLVENGKFLLSAKR RRTT TEY+ISM AD
Sbjct: 121 PGPRDATMQCFIKRDKSNLTYHLYLCLSPALLVENGKFLLSAKRIRRTTYTEYVISMHAD 180
Query: 177 NISRSSSTYIGKLRSNFLGTKFIIYDTQPPYNN---AQLSPPGRSRRFNSKKVSPKVPSG 233
ISRSS+TYIGK+RSNFLGTKFIIYDTQP YN+ + P G SRRF SK+VSPKVPSG
Sbjct: 181 TISRSSNTYIGKIRSNFLGTKFIIYDTQPAYNSNIARAVQPVGLSRRFYSKRVSPKVPSG 240
Query: 234 SYNIAQVTYELNVLGTRGPRRMNCTMHSIPASSLDLGGSVPGKPELL-PRA-LEDSFRSI 291
SY IAQV+YELNVLGTRGPRRM+C M+SIPASSL GG+VPG+P+++ PR+ L++SFRSI
Sbjct: 241 SYKIAQVSYELNVLGTRGPRRMHCAMNSIPASSLAEGGTVPGQPDIIVPRSILDESFRSI 300
Query: 292 SFARS----IDNSTEFSSARFSDIIGAGNEEEE---EEGKER---PLVLKNKSPRWHEQL 341
+ + S D S +FSSARFSDI+G +E++E EEGKER PLVLKNK PRWHEQL
Sbjct: 301 TSSSSRKITYDYSNDFSSARFSDILGPLSEDQEVVLEEGKERNSPPLVLKNKPPRWHEQL 360
Query: 342 QCWCLNFRGRVTVASVKNFQLIAATQPASGGGAPPTGTGAA--PPSQPPSDPDKIILQFG 399
QCWCLNFRGRVTVASVKNFQLIAA QP P PS PDKIILQFG
Sbjct: 361 QCWCLNFRGRVTVASVKNFQLIAANQPQPQPQPQPQPQPLTQPQPSGQTDGPDKIILQFG 420
Query: 400 KVGKDMFTMDYRYPLSAFQAFAICLTSFDTKLACE 434
KVGKDMFTMD+RYPLSAFQAFAICL+SFDTKLACE
Sbjct: 421 KVGKDMFTMDFRYPLSAFQAFAICLSSFDTKLACE 455
>At1g25280 unknown protein
Length = 445
Score = 609 bits (1571), Expect = e-175
Identities = 317/450 (70%), Positives = 365/450 (80%), Gaps = 21/450 (4%)
Query: 1 MSFRSIVRDVRDGFGSLSRRSFDVILPGHSRSKSRSSVHELHDQ------PLVIQSSRWA 54
MSFR IV+D+RDGFGSLSRRSFD L + K++ S + P+++Q+SRWA
Sbjct: 1 MSFRGIVQDLRDGFGSLSRRSFDFRLSSLHKGKAQGSSFREYSSSRDLLSPVIVQTSRWA 60
Query: 55 SLPPELLRDVIKRLETNESAWPGRKHVVACAAVCKSWREMCKEIVGSPEFCGKITFPVSL 114
+LPPELL DVIKRLE +ES WP RKHVVACA+VC+SWR MC+EIV PE CGK+TFPVSL
Sbjct: 61 NLPPELLFDVIKRLEESESNWPARKHVVACASVCRSWRAMCQEIVLGPEICGKLTFPVSL 120
Query: 115 KQPGYRDGPIQCFIKRDKSKLTYHLFLCLSPALLVENGKFLLSAKRTRRTTCTEYIISMD 174
KQPG RD IQCFIKRDKSKLT+HLFLCLSPALLVENGKFLLSAKRTRRTT TEYIISMD
Sbjct: 121 KQPGPRDAMIQCFIKRDKSKLTFHLFLCLSPALLVENGKFLLSAKRTRRTTRTEYIISMD 180
Query: 175 ADNISRSSSTYIGKLRSNFLGTKFIIYDTQPPYN---NAQLSPPGRSRRFNSKKVSPKVP 231
ADNISRSS++Y+GKLRSNFLGTKF++YDTQPP N +A ++ RF+S++VSPKVP
Sbjct: 181 ADNISRSSNSYLGKLRSNFLGTKFLVYDTQPPPNTSSSALITDRTSRSRFHSRRVSPKVP 240
Query: 232 SGSYNIAQVTYELNVLGTRGPRRMNCTMHSIPASSLDLGGSVPGKPELL---PRALEDSF 288
SGSYNIAQ+TYELNVLGTRGPRRM+C M+SIP SSL+ GGSVP +PE L P +L+DSF
Sbjct: 241 SGSYNIAQITYELNVLGTRGPRRMHCIMNSIPISSLEPGGSVPNQPEKLVPAPYSLDDSF 300
Query: 289 RS-ISFARSI--DNSTEFSSARFSDIIGAGNEEEEEEGKERPLVLKNKSPRWHEQLQCWC 345
RS ISF++S S +FSS+RFS+ +G ++ EEE RPL+LKNK PRWHEQLQCWC
Sbjct: 301 RSNISFSKSSFDHRSLDFSSSRFSE-MGISCDDNEEEASFRPLILKNKQPRWHEQLQCWC 359
Query: 346 LNFRGRVTVASVKNFQLIAATQPASGGGAPPTGTGAAPPSQPP-SDPDKIILQFGKVGKD 404
LNFRGRVTVASVKNFQL+AA QP G TG AAP S P + DK+ILQFGKVGKD
Sbjct: 360 LNFRGRVTVASVKNFQLVAARQPQPQG----TGAAAAPTSAPAHPEQDKVILQFGKVGKD 415
Query: 405 MFTMDYRYPLSAFQAFAICLTSFDTKLACE 434
MFTMDYRYPLSAFQAFAICL+SFDTKLACE
Sbjct: 416 MFTMDYRYPLSAFQAFAICLSSFDTKLACE 445
>At1g43640 unknown protein (At1g43640)
Length = 429
Score = 582 bits (1499), Expect = e-166
Identities = 301/442 (68%), Positives = 352/442 (79%), Gaps = 21/442 (4%)
Query: 1 MSFRSIVRDVRDGFGSLSRRSFDVILPG---HSRSKSRSSVHELHDQPLVIQSSRWASLP 57
MSF SIVRDVRD GS SRRSFDV + H RSKS + D +VI+++RWA+LP
Sbjct: 1 MSFLSIVRDVRDTVGSFSRRSFDVRVSNGTTHQRSKSHGVEAHIEDL-IVIKNTRWANLP 59
Query: 58 PELLRDVIKRLETNESAWPGRKHVVACAAVCKSWREMCKEIVGSPEFCGKITFPVSLKQP 117
LLRDV+K+L+ +ES WP RK VVACA VCK+WR MCK+IV SPEF GK+TFPVSLKQP
Sbjct: 60 AALLRDVMKKLDESESTWPARKQVVACAGVCKTWRLMCKDIVKSPEFSGKLTFPVSLKQP 119
Query: 118 GYRDGPIQCFIKRDKSKLTYHLFLCLSPALLVENGKFLLSAKRTRRTTCTEYIISMDADN 177
G RDG IQC+IKRDKS +TYHL+L LSPA+LVE+GKFLLSAKR+RR T TEY+ISMDADN
Sbjct: 120 GPRDGIIQCYIKRDKSNMTYHLYLSLSPAILVESGKFLLSAKRSRRATYTEYVISMDADN 179
Query: 178 ISRSSSTYIGKLRSNFLGTKFIIYDTQPPYNNAQ-LSPPGRSRRFNSKKVSPKVPSGSYN 236
ISRSSSTYIGKL+SNFLGTKFI+YDT P YN++Q LSPP RSR FNSKKVSPKVPSGSYN
Sbjct: 180 ISRSSSTYIGKLKSNFLGTKFIVYDTAPAYNSSQILSPPNRSRSFNSKKVSPKVPSGSYN 239
Query: 237 IAQVTYELNVLGTRGPRRMNCTMHSIPASSLDLGGSVPGKPELLPRALEDSFRSISFARS 296
IAQVTYELN+LGTRGPRRMNC MHSIP+ +L+ GG+VP +PE L R+L++SFRSI ++
Sbjct: 240 IAQVTYELNLLGTRGPRRMNCIMHSIPSLALEPGGTVPSQPEFLQRSLDESFRSIGSSKI 299
Query: 297 IDNSTEFSSARFSDIIGAGNEEEEEEGKERPLVLKNKSPRWHEQLQCWCLNFRGRVTVAS 356
+++S +F+ +EEEGK RPLVLK K PRW + L+CWCLNF+GRVTVAS
Sbjct: 300 VNHSGDFT------------RPKEEEGKVRPLVLKTKPPRWLQPLRCWCLNFKGRVTVAS 347
Query: 357 VKNFQLIAA--TQPASG--GGAPPTGTGAAPPSQPPSDPDKIILQFGKVGKDMFTMDYRY 412
VKNFQL++A QP SG GGA T +P S+ DKIIL FGKVGKDMFTMDYRY
Sbjct: 348 VKNFQLMSAATVQPGSGSDGGALATRPSLSPQQPEQSNHDKIILHFGKVGKDMFTMDYRY 407
Query: 413 PLSAFQAFAICLTSFDTKLACE 434
PLSAFQAFAI L++FDTKLACE
Sbjct: 408 PLSAFQAFAISLSTFDTKLACE 429
>At2g47900 Tub family like protein
Length = 406
Score = 458 bits (1178), Expect = e-129
Identities = 235/434 (54%), Positives = 308/434 (70%), Gaps = 28/434 (6%)
Query: 1 MSFRSIVRDVRDGFGSLSRRSFDVILPGHSRSKSRSSVHELHDQPLVIQSSRWASLPPEL 60
MSF+S+++D+R GS+SR+ FDV G+ RS+S+ V + + S WAS+PPEL
Sbjct: 1 MSFKSLIQDMRGELGSISRKGFDVRF-GYGRSRSQRVVQDTSVPVDAFKQSCWASMPPEL 59
Query: 61 LRDVIKRLETNESAWPGRKHVVACAAVCKSWREMCKEIVGSPEFCGKITFPVSLKQPGYR 120
LRDV+ R+E +E WP RK+VV+CA VC++WRE+ KEIV PE K+TFP+SLKQPG R
Sbjct: 60 LRDVLMRIEQSEDTWPSRKNVVSCAGVCRNWREIVKEIVRVPELSSKLTFPISLKQPGPR 119
Query: 121 DGPIQCFIKRDKSKLTYHLFLCLSPALLVENGKFLLSAKRTRRTTCTEYIISMDADNISR 180
+QC+I R++S TY+L+L L+ A ++GKFLL+AKR RR TCT+YIIS++ D++SR
Sbjct: 120 GSLVQCYIMRNRSNQTYYLYLGLNQAASNDDGKFLLAAKRFRRPTCTDYIISLNCDDVSR 179
Query: 181 SSSTYIGKLRSNFLGTKFIIYDTQPPYNNAQLSPPGRSRRFNSKKVSPKVPSGSYNIAQV 240
S+TYIGKLRSNFLGTKF +YD QP Q++ SR + K+VSP++PSG+Y +A +
Sbjct: 180 GSNTYIGKLRSNFLGTKFTVYDAQPTNPGTQVTRTRSSRLLSLKQVSPRIPSGNYPVAHI 239
Query: 241 TYELNVLGTRGPRRMNCTMHSIPASSLDLGGSVPGKPELLPRALEDSFRSISFARSIDNS 300
+YELNVLG+RGPRRM C M +IPAS+++ GG+ P + EL+ L DSF S SF RS
Sbjct: 240 SYELNVLGSRGPRRMQCVMDAIPASAVEPGGTAPTQTELVHSNL-DSFPSFSFFRS---- 294
Query: 301 TEFSSARFSDIIGAGNEEEEEEGKERPLVLKNKSPRWHEQLQCWCLNFRGRVTVASVKNF 360
S R + + ++EG LVLKNK+PRWHEQLQCWCLNF GRVTVASVKNF
Sbjct: 295 ---KSIRAESLPSGPSSAAQKEGL---LVLKNKAPRWHEQLQCWCLNFNGRVTVASVKNF 348
Query: 361 QLIAATQPASGGGAPPTGTGAAPPSQPPSDPDKIILQFGKVGKDMFTMDYRYPLSAFQAF 420
QL+AA P +G P + + +ILQFGKVGKD+FTMDY+YP+SAFQAF
Sbjct: 349 QLVAA--PENGPAGP--------------EHENVILQFGKVGKDVFTMDYQYPISAFQAF 392
Query: 421 AICLTSFDTKLACE 434
ICL+SFDTK+ACE
Sbjct: 393 TICLSSFDTKIACE 406
>At2g18280 tubby-like protein 2
Length = 394
Score = 419 bits (1077), Expect = e-117
Identities = 232/440 (52%), Positives = 287/440 (64%), Gaps = 53/440 (12%)
Query: 1 MSFRSIVRD---VRDGFGSLSRRSFDVILPGHSRSKSRSSVHELHDQPLV-IQSSRWASL 56
MS +SI+RD VRDG G +S+RS+ SKS + PL I S WASL
Sbjct: 1 MSLKSILRDLKEVRDGLGGISKRSW---------SKSSHIAPDQTTPPLDNIPQSPWASL 51
Query: 57 PPELLRDVIKRLETNESAWPGRKHVVACAAVCKSWREMCKEIVGSPEFCGKITFPVSLKQ 116
PPELL D+I R+E +E+AWP R VV+CA+VCKSWR + EIV PE CGK+TFP+SLKQ
Sbjct: 52 PPELLHDIIWRVEESETAWPARAAVVSCASVCKSWRGITMEIVRIPEQCGKLTFPISLKQ 111
Query: 117 PGYRDGPIQCFIKRDKSKLTYHLFLCLSPALLVENGKFLLSAKRTRRTTCTEYIISMDAD 176
PG RD PIQCFIKR+++ TY L+ L P+ EN K LL+A+R RR TCT++IIS+ A
Sbjct: 112 PGPRDSPIQCFIKRNRATATYILYYGLMPS-ETENDKLLLAARRIRRATCTDFIISLSAK 170
Query: 177 NISRSSSTYIGKLRSNFLGTKFIIYDTQPPYNNAQLSPPGRSRRFNSKKVSPKVPSGSYN 236
N SRSSSTY+GKLRS FLGTKF IYD Q + AQ P +RR + K+ +PK+P+ S
Sbjct: 171 NFSRSSSTYVGKLRSGFLGTKFTIYDNQTASSTAQAQP---NRRLHPKQAAPKLPTNSST 227
Query: 237 IAQVTYELNVLGTRGPRRMNCTMHSIPASSLDLGGSVPGKPELLPRALEDSFRSISFARS 296
+ +TYELNVL TRGPRRM+C M SIP S SV +P S +
Sbjct: 228 VGNITYELNVLRTRGPRRMHCAMDSIPLS------SVIAEP--------------SVVQG 267
Query: 297 IDNSTEFSSARFSDIIGAGNE--EEEEEGKERPLVLKNKSPRWHEQLQCWCLNFRGRVTV 354
I+ S + + I E + +++PLVLKNKSPRWHEQLQCWCLNF+GRVTV
Sbjct: 268 IEEEVSSSPSPKGETITTDKEIPDNSPSLRDQPLVLKNKSPRWHEQLQCWCLNFKGRVTV 327
Query: 355 ASVKNFQLIAATQPASGGGAPPTGTGAAPPSQPPSDPDKIILQFGKVGKDMFTMDYRYPL 414
ASVKNFQL+A A PP + +++ILQFGK+GKD+FTMDYRYPL
Sbjct: 328 ASVKNFQLVAEID--------------ASLDAPPEEHERVILQFGKIGKDIFTMDYRYPL 373
Query: 415 SAFQAFAICLTSFDTKLACE 434
SAFQAFAIC++SFDTK ACE
Sbjct: 374 SAFQAFAICISSFDTKPACE 393
>At1g47270 unknown protein
Length = 388
Score = 411 bits (1056), Expect = e-115
Identities = 226/436 (51%), Positives = 280/436 (63%), Gaps = 50/436 (11%)
Query: 1 MSFRSIVRDVRDGFGSLSRRSFDVILPGHSRSKSRSSVHELHDQPLVIQSSRWASLPPEL 60
MS ++IV++ + ++ RR I P S S S +E +Q + W LPPEL
Sbjct: 1 MSLKNIVKNK---YKAIGRRGRSHIAPEGSSVSSSLSTNEGLNQSI------WVDLPPEL 51
Query: 61 LRDVIKRLETNESAWPGRKHVVACAAVCKSWREMCKEIVGSPEFCGKITFPVSLKQPGYR 120
L D+I+R+E+ +S WPGR+ VVACA+VCKSWREM KE+V PE G ITFP+SL+QPG R
Sbjct: 52 LLDIIQRIESEQSLWPGRRDVVACASVCKSWREMTKEVVKVPELSGLITFPISLRQPGPR 111
Query: 121 DGPIQCFIKRDKSKLTYHLFLCLSPALLVENGKFLLSAKRTRRTTCTEYIISMDADNISR 180
D PIQCFIKR+++ Y L+L LSPAL + K LLSAKR RR T E+++S+ ++ SR
Sbjct: 112 DAPIQCFIKRERATGIYRLYLGLSPALSGDKSKLLLSAKRVRRATGAEFVVSLSGNDFSR 171
Query: 181 SSSTYIGKLRSNFLGTKFIIYDTQPPYNNAQLSPPGRSRRFNSKKVSPKV--PSGSYNIA 238
SSS YIGKLRSNFLGTKF +Y+ QPP N +L P S +VSP V S SYNIA
Sbjct: 172 SSSNYIGKLRSNFLGTKFTVYENQPPPFNRKLPP--------SMQVSPWVSSSSSSYNIA 223
Query: 239 QVTYELNVLGTRGPRRMNCTMHSIPASSLDLGGSVPGKPELLPRALEDSFRSISFARSID 298
+ YELNVL TRGPRRM C MHSIP S++ GG + E + + + F
Sbjct: 224 SILYELNVLRTRGPRRMQCIMHSIPISAIQEGGKIQSPTEFTNQGKKKKKPLMDFC---- 279
Query: 299 NSTEFSSARFSDIIGAGNEEEEEEGKERPLVLKNKSPRWHEQLQCWCLNFRGRVTVASVK 358
+GN E KE PL+LKNKSPRWHEQLQCWCLNF+GRVTVASVK
Sbjct: 280 ---------------SGNLGGESVIKE-PLILKNKSPRWHEQLQCWCLNFKGRVTVASVK 323
Query: 359 NFQLIAATQPASGGGAPPTGTGAAPPSQPPSDPDKIILQFGKVGKDMFTMDYRYPLSAFQ 418
NFQL+AA A + P + D++ILQFGK+GKD+FTMDYRYP+SAFQ
Sbjct: 324 NFQLVAAAAEAGKN-----------MNIPEEEQDRVILQFGKIGKDIFTMDYRYPISAFQ 372
Query: 419 AFAICLTSFDTKLACE 434
AFAICL+SFDTK CE
Sbjct: 373 AFAICLSSFDTKPVCE 388
>At5g18680 tub family-like protein
Length = 389
Score = 375 bits (964), Expect = e-104
Identities = 205/413 (49%), Positives = 271/413 (64%), Gaps = 43/413 (10%)
Query: 31 RSKSRSSVHEL------HDQPLVIQSSRWASLPPELLRDVIKRLETNESA-WPGRKHVVA 83
RS+ VH+L + Q RW+ +P ELLR+++ R+E + WP R+ VVA
Sbjct: 11 RSRPHRVVHDLAAAAAADSTSVSSQDYRWSEIPEELLREILIRVEAADGGGWPSRRSVVA 70
Query: 84 CAAVCKSWREMCKEIVGSPEFCGKITFPVSLKQPGYRDGPIQCFIKRDKSKLTYHLFLCL 143
CA VC+ WR + E V PE K+TFP+SLKQPG RD +QCFIKR++ +YHL+L L
Sbjct: 71 CAGVCRGWRLLMNETVVVPEISSKLTFPISLKQPGPRDSLVQCFIKRNRITQSYHLYLGL 130
Query: 144 SPALLVENGKFLLSAKRTRRTTCTEYIISMDADNISRSSSTYIGKLRSNFLGTKFIIYDT 203
+ + L ++GKFLL+A + + TTCT+YIIS+ +D++SR S Y+GK+RSNFLGTKF ++D
Sbjct: 131 TNS-LTDDGKFLLAACKLKHTTCTDYIISLRSDDMSRRSQAYVGKVRSNFLGTKFTVFDG 189
Query: 204 Q--PPYNNAQLSPPGRSRRFNSKKVSPKVPSGSYNIAQVTYELNVLGTRGPRRMNCTMHS 261
P A+L +SR +N KVS KVP GSY +A +TYELNVLG+RGPR+M C M +
Sbjct: 190 NLLPSTGAAKLR---KSRSYNPAKVSAKVPLGSYPVAHITYELNVLGSRGPRKMQCLMDT 246
Query: 262 IPASSLDLGGSVPGKPELLPRALEDSFRSISFARSIDNSTEFSSARFSDIIGAGNEEEEE 321
IP S+++ G V +P P S S S ++ + +S+
Sbjct: 247 IPTSTMEPQG-VASEPSEFPLLGTRSTLSRSQSKPLRSSSSHL----------------- 288
Query: 322 EGKERPLVLKNKSPRWHEQLQCWCLNFRGRVTVASVKNFQLIAATQPASGGGAPPTGTGA 381
KE PLVL NK+PRWHEQL+CWCLNF GRVTVASVKNFQL+AA G + +GTG
Sbjct: 289 --KETPLVLSNKTPRWHEQLRCWCLNFHGRVTVASVKNFQLVAA------GASCGSGTGM 340
Query: 382 APPSQPPSDPDKIILQFGKVGKDMFTMDYRYPLSAFQAFAICLTSFDTKLACE 434
+P Q ++IILQFGKVGKDMFTMDY YP+SAFQAFAICL+SF+T++ACE
Sbjct: 341 SPERQ----SERIILQFGKVGKDMFTMDYGYPISAFQAFAICLSSFETRIACE 389
>At3g06380 unknown protein
Length = 380
Score = 371 bits (953), Expect = e-103
Identities = 202/405 (49%), Positives = 263/405 (64%), Gaps = 36/405 (8%)
Query: 31 RSKSRSSVHELHDQPLVIQSSRWASLPPELLRDVIKRLETNESA-WPGRKHVVACAAVCK 89
RS+ VH W+ LP ELLR+++ R+ET + WP R++VVACA VC+
Sbjct: 11 RSRPHRVVHAAASTANSSDPFSWSELPEELLREILIRVETVDGGDWPSRRNVVACAGVCR 70
Query: 90 SWREMCKEIVGSPEFCGKITFPVSLKQPGYRDGPIQCFIKRDKSKLTYHLFLCLSPALLV 149
SWR + KEIV PEF K+TFP+SLKQ G RD +QCFIKR+++ +YHL+L L+ + L
Sbjct: 71 SWRILTKEIVAVPEFSSKLTFPISLKQSGPRDSLVQCFIKRNRNTQSYHLYLGLTTS-LT 129
Query: 150 ENGKFLLSAKRTRRTTCTEYIISMDADNISRSSSTYIGKLRSNFLGTKFIIYDTQPPYNN 209
+NGKFLL+A + +R TCT+YIIS+ +D+IS+ S+ Y+G++RSNFLGTKF ++D +
Sbjct: 130 DNGKFLLAASKLKRATCTDYIISLRSDDISKRSNAYLGRMRSNFLGTKFTVFDG----SQ 185
Query: 210 AQLSPPGRSRRFNSKKVSPKVPSGSYNIAQVTYELNVLGTRGPRRMNCTMHSIPASSLDL 269
+ +SR N KVSP+VP GSY IA ++YELNVLG+RGPRRM C M +IP S ++
Sbjct: 186 TGAAKMQKSRSSNFIKVSPRVPQGSYPIAHISYELNVLGSRGPRRMRCIMDTIPMSIVES 245
Query: 270 GGSVPGKPELLPRALEDSFRSISFARSIDNSTEFSSARFSDIIGAGNEEEEEEGKERPLV 329
G V S RS RS +SA SD +GN + PLV
Sbjct: 246 RGVVAS-----TSISSFSSRSSPVFRSHSKPLRSNSASCSD---SGNNLGDP-----PLV 292
Query: 330 LKNKSPRWHEQLQCWCLNFRGRVTVASVKNFQLIAATQPASGGGAPPTGTGAAPPSQPPS 389
L NK+PRWHEQL+CWCLNF GRVTVASVKNFQL+A + +G
Sbjct: 293 LSNKAPRWHEQLRCWCLNFHGRVTVASVKNFQLVAVSDCEAG-----------------Q 335
Query: 390 DPDKIILQFGKVGKDMFTMDYRYPLSAFQAFAICLTSFDTKLACE 434
++IILQFGKVGKDMFTMDY YP+SAFQAFAICL+SF+T++ACE
Sbjct: 336 TSERIILQFGKVGKDMFTMDYGYPISAFQAFAICLSSFETRIACE 380
>At1g53320 tubby like protein (Tub family protein)
Length = 379
Score = 338 bits (867), Expect = 3e-93
Identities = 185/387 (47%), Positives = 241/387 (61%), Gaps = 50/387 (12%)
Query: 50 SSRWASLPPELLRDVIKRLETNESAWPGRKHVVACAAVCKSWREMCKEIVGSPEFCGKIT 109
SS W+++ PELL ++I+R+E E WP R+ VV CA V K WRE+ + S GKIT
Sbjct: 41 SSSWSAMLPELLGEIIRRVEETEDRWPQRRDVVTCACVSKKWREITHDFARSSLNSGKIT 100
Query: 110 FPVSLKQPGYRDGPIQCFIKRDKSKLTYHLFLCLSPALLVENGKFLLSAKRTRRTTCTEY 169
FP LK PG RD QC IKR+K T++L+L L+P+ + GKFLL+A+R R TEY
Sbjct: 101 FPSCLKLPGPRDFSNQCLIKRNKKTSTFYLYLALTPSF-TDKGKFLLAARRFRTGAYTEY 159
Query: 170 IISMDADNISRSSSTYIGKLRSNFLGTKFIIYDTQPPYNNAQLSPPGRSRRFNSKKVSPK 229
IIS+DAD+ S+ S+ Y+GKLRS+FLGT F +YD+QPP+N A+ S SRRF SK++SP+
Sbjct: 160 IISLDADDFSQGSNAYVGKLRSDFLGTNFTVYDSQPPHNGAKPSNGKASRRFASKQISPQ 219
Query: 230 VPSGSYNIAQVTYELNVLGTRGPRRMNCTMH--SIPASSLDLGGSVPGKPELLPRALEDS 287
VP+G++ + V+Y+ N+L +RGPRRM T+ S SS G S KP + + ++
Sbjct: 220 VPAGNFEVGHVSYKFNLLKSRGPRRMVSTLRCPSPSPSSSSAGLSSDQKPCDVTKIMK-- 277
Query: 288 FRSISFARSIDNSTEFSSARFSDIIGAGNEEEEEEGKERPLVLKNKSPRWHEQLQCWCLN 347
+ ++G +LKNK+PRWHE LQCWCLN
Sbjct: 278 ------------------------------KPNKDGSSLT-ILKNKAPRWHEHLQCWCLN 306
Query: 348 FRGRVTVASVKNFQLIAATQPASGGGAPPTGTGAAPPSQPPSDPDKIILQFGKVGKDMFT 407
F GRVTVASVKNFQL+A + P+G G D + ++LQFGKVG D FT
Sbjct: 307 FHGRVTVASVKNFQLVATVDQSQ-----PSGKG---------DEETVLLQFGKVGDDTFT 352
Query: 408 MDYRYPLSAFQAFAICLTSFDTKLACE 434
MDYR PLSAFQAFAICLTSF TKLACE
Sbjct: 353 MDYRQPLSAFQAFAICLTSFGTKLACE 379
>At1g61940 hypothetical protein, 5' partial
Length = 265
Score = 111 bits (278), Expect = 7e-25
Identities = 47/86 (54%), Positives = 65/86 (74%)
Query: 56 LPPELLRDVIKRLETNESAWPGRKHVVACAAVCKSWREMCKEIVGSPEFCGKITFPVSLK 115
+PPELLRDV+ R+E +E WP RK+VV+C VCK+WR++ KEIV PE K TFP+SLK
Sbjct: 1 MPPELLRDVLMRIERSEDTWPSRKNVVSCVGVCKNWRQIFKEIVNVPEVSSKFTFPISLK 60
Query: 116 QPGYRDGPIQCFIKRDKSKLTYHLFL 141
QPG +QC++KR++S T++L+L
Sbjct: 61 QPGPGGSLVQCYVKRNRSNQTFYLYL 86
Score = 34.3 bits (77), Expect = 0.14
Identities = 30/98 (30%), Positives = 38/98 (38%), Gaps = 34/98 (34%)
Query: 253 RRMNCTMHSIPASSLDLGGSVPGKPELLPRALEDSFRSISFARSIDNSTEFSSARFSDII 312
R +C M +I AS++ GG+ + EL DN F S
Sbjct: 109 RETHCVMDAISASAVKPGGTATTQTEL------------------DNFVSFRSPSGQ--- 147
Query: 313 GAGNEEEEEEGKERPLVLKNKSPRWHEQLQCWCLNFRG 350
KE LVLK+K PR E Q WCL+F G
Sbjct: 148 -----------KEGVLVLKSKVPRLEE--QSWCLDFNG 172
>At1g16070 unknown protein
Length = 397
Score = 58.2 bits (139), Expect = 9e-09
Identities = 39/117 (33%), Positives = 55/117 (46%), Gaps = 28/117 (23%)
Query: 322 EGKERPLVLKNKSPRWHEQLQCWCLNFRGRVTV-----ASVKNFQLIAATQPASGGGAPP 376
E KE+ L ++ P +++ + L+FR R +SVKNFQL P
Sbjct: 302 ENKEKIQKLCSRIPHYNKISKQHELDFRDRGRTGLRIQSSVKNFQLTLTETPR------- 354
Query: 377 TGTGAAPPSQPPSDPDKIILQFGKVGKDMFTMDYRYPLSAFQAFAICLTSFDTKLAC 433
+ ILQ G+V K + +D+RYP S +QAF ICL S D+KL C
Sbjct: 355 ----------------QTILQMGRVDKARYVIDFRYPFSGYQAFCICLASIDSKLCC 395
>At4g05460 N7 like-protein
Length = 302
Score = 37.7 bits (86), Expect = 0.012
Identities = 18/49 (36%), Positives = 26/49 (52%), Gaps = 7/49 (14%)
Query: 49 QSSRWASLPPELLRDVIKRLETNESAWPGRKHVVACAAVCKSWREMCKE 97
+S+ W LPPEL ++ RL E +K VC+SWR +CK+
Sbjct: 15 ESTNWTELPPELTSAILHRLGAIEILENAQK-------VCRSWRRVCKD 56
>At4g05490 N7 -like protein
Length = 307
Score = 37.4 bits (85), Expect = 0.016
Identities = 19/47 (40%), Positives = 25/47 (52%), Gaps = 7/47 (14%)
Query: 51 SRWASLPPELLRDVIKRLETNESAWPGRKHVVACAAVCKSWREMCKE 97
S WA LPP+LL ++ RL E RK VC+SWR + K+
Sbjct: 25 SNWAELPPDLLSSILLRLSPLEILENARK-------VCRSWRRVSKD 64
>At4g05470 N7 -like protein
Length = 304
Score = 35.8 bits (81), Expect = 0.047
Identities = 17/52 (32%), Positives = 25/52 (47%), Gaps = 7/52 (13%)
Query: 46 LVIQSSRWASLPPELLRDVIKRLETNESAWPGRKHVVACAAVCKSWREMCKE 97
L+ + W LPPEL ++ RL + +K VCK WR +CK+
Sbjct: 39 LMKERRNWVDLPPELTTSILLRLSLTDILDNAQK-------VCKEWRRICKD 83
>At4g05480 N7 -like protein
Length = 322
Score = 35.0 bits (79), Expect = 0.081
Identities = 15/45 (33%), Positives = 23/45 (50%), Gaps = 7/45 (15%)
Query: 53 WASLPPELLRDVIKRLETNESAWPGRKHVVACAAVCKSWREMCKE 97
W LPPEL ++ RL + RK +C++WR +CK+
Sbjct: 40 WVDLPPELTTSILLRLSVTDILDNARK-------LCRAWRRICKD 77
>At5g55150 putative protein
Length = 340
Score = 33.9 bits (76), Expect = 0.18
Identities = 17/59 (28%), Positives = 26/59 (43%), Gaps = 7/59 (11%)
Query: 46 LVIQSSRWASLPPELLRDVIKRLETNESAWPGRKHVVACAAVCKSWREMCKEIVGSPEF 104
+ + SS W+ PELL V L + ++ CA VC SW++ + S F
Sbjct: 1 MALPSSSWSEFLPELLNTVFHNLND-------ARDILNCATVCSSWKDSSSAVYYSRTF 52
>At3g53440 unknown protein
Length = 512
Score = 33.5 bits (75), Expect = 0.24
Identities = 18/46 (39%), Positives = 26/46 (56%), Gaps = 2/46 (4%)
Query: 299 NSTEFSSARFSDIIGAGNEEEEEEGKERPLVLKNKSPR--WHEQLQ 342
N E + A+ ++ G EEEEEE +E L KNK + W E+L+
Sbjct: 221 NEEENAEAKADEVKRKGKEEEEEEDEEEGLKAKNKLEKGGWTEELE 266
>At1g13250 unknown protein
Length = 345
Score = 32.7 bits (73), Expect = 0.40
Identities = 26/80 (32%), Positives = 35/80 (43%), Gaps = 11/80 (13%)
Query: 27 PGHSRSKSRSSVHELHDQPLVIQSSRWASLPPELLRDVIKRLETNESAWPGRKHVVACAA 86
P RSK SS+ DQPL A L P +R VI +V
Sbjct: 125 PNLVRSKISSSIRRALDQPLNYARIYLADLLPIAVRRVIY----------FDSDLVVVDD 174
Query: 87 VCKSWR-EMCKEIVGSPEFC 105
V K WR ++ + +VG+PE+C
Sbjct: 175 VAKLWRIDLRRHVVGAPEYC 194
>At1g67190 unknown protein
Length = 419
Score = 32.3 bits (72), Expect = 0.52
Identities = 20/66 (30%), Positives = 32/66 (48%), Gaps = 14/66 (21%)
Query: 56 LPPELLRDVIKRLETNESAWPGRKHVVACAAVCKSWREMCKEIVGSPEFCGKITFPVSLK 115
LP E++ +++ RL G + VV +A C+ WRE C++ + + F S
Sbjct: 4 LPVEVIGNILSRLG-------GARDVVIASATCRKWREACRKHLQTLSF-------NSAD 49
Query: 116 QPGYRD 121
P YRD
Sbjct: 50 WPFYRD 55
>At1g73940 unknown protein
Length = 151
Score = 31.6 bits (70), Expect = 0.89
Identities = 16/46 (34%), Positives = 26/46 (55%), Gaps = 1/46 (2%)
Query: 200 IYDTQPPYNNAQLSPPGRSRRFNSKKVSPKVP-SGSYNIAQVTYEL 244
I+D + YN+ + R R N +K++PKVP +Y I+ Y+L
Sbjct: 19 IFDNEISYNSLLVRYMSRERAVNVRKINPKVPIQEAYAISNSLYDL 64
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,569,961
Number of Sequences: 26719
Number of extensions: 487030
Number of successful extensions: 1998
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 1897
Number of HSP's gapped (non-prelim): 64
length of query: 434
length of database: 11,318,596
effective HSP length: 102
effective length of query: 332
effective length of database: 8,593,258
effective search space: 2852961656
effective search space used: 2852961656
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0052b.11


