

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0052a.3
(242 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
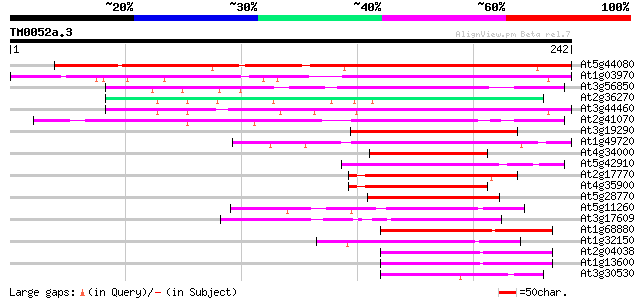
Score E
Sequences producing significant alignments: (bits) Value
At5g44080 bZIP protein AtbZIP13 196 7e-51
At1g03970 G-box binding bZip transcription factor GBF4 / AtbZip40 190 7e-49
At3g56850 ABA-responsive element binding bZip transcription fact... 86 2e-17
At2g36270 bZip transcription factor AtbZip39 75 4e-14
At3g44460 bZIP transcription factor DPBF2 / AtbZip67 73 1e-13
At2g41070 bZIP transcription factor AtbZIP12 / DPBF4 73 2e-13
At3g19290 abscisic acid responsive elements-binding factor (ABRE... 71 6e-13
At1g49720 abscisic acid responsive elements-binding factor (ABRE... 69 3e-12
At4g34000 abscisic acid responsive elements-binding factor (ABRE... 62 4e-10
At5g42910 bzip protein AtbZip15 55 3e-08
At2g17770 bZIP transcription factor atbzip27 54 1e-07
At4g35900 bZIP transcription factor AtbZIP14 53 2e-07
At5g28770 bZip transcription factor BZO2H3 / AtbZip63 50 1e-06
At5g11260 bZip transcription factor HY5 / AtbZip56 50 1e-06
At3g17609 bZip transcription factor AtbZip64 49 3e-06
At1g68880 bZip transcription factor AtbZip8 48 4e-06
At1g32150 putative G-Box binding protein 48 4e-06
At2g04038 bZip transcription factor AtbZip48 48 5e-06
At1g13600 bZip transcription factor AtbZip58 47 7e-06
At3g30530 bZip transcription factor AtbZip42 47 1e-05
>At5g44080 bZIP protein AtbZIP13
Length = 315
Score = 196 bits (499), Expect = 7e-51
Identities = 119/231 (51%), Positives = 157/231 (67%), Gaps = 14/231 (6%)
Query: 20 SSPPPSSMPQFTTTNFIDHADSTLTDPYVPRTVDDVWQEIIAGDHHQCKEEIPDEMMTLE 79
+SP P + T ++ +DH T T ++VD++W+E+++G+ KEE +E+MTLE
Sbjct: 91 ASPAPMEITTTTASDVVDHGGGTETTRG-GKSVDEIWREMVSGEGKGMKEETSEEIMTLE 149
Query: 80 DFLVKAG-----AVDGDDEDDEVKMSMPLTERLSGVFSLDGSSFQGMEGVEGGSASVGGF 134
DFL KA AV ED +VK+ P+T + + FQ ++ VEG S+ F
Sbjct: 150 DFLAKAAVEDETAVTASAEDLDVKI--PVTNYGFDHSAPPHNPFQMIDKVEG---SIVAF 204
Query: 135 GNGVEVVEG-TRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAV 193
GNG++V G RGKR R +VE LDKAA QRQRRMIKNRESAARSRERKQAYQVELE+LA
Sbjct: 205 GNGLDVYGGGARGKRARVMVEPLDKAAAQRQRRMIKNRESAARSRERKQAYQVELEALAA 264
Query: 194 KLEEENDKLLKEKAERTKERFKQLMEEVIPVVE--KRRPPRPLRRINSMQW 242
KLEEEN+ L KE ++ KER+++LME VIPVVE K++PPR LRRI S++W
Sbjct: 265 KLEEENELLSKEIEDKRKERYQKLMEFVIPVVEKPKQQPPRFLRRIRSLEW 315
>At1g03970 G-box binding bZip transcription factor GBF4 / AtbZip40
Length = 270
Score = 190 bits (482), Expect = 7e-49
Identities = 130/289 (44%), Positives = 169/289 (57%), Gaps = 66/289 (22%)
Query: 1 MASSKMMMSSSKSLDHQQPSSPPPSSMPQFTTTNFI------DHA-----------DSTL 43
MAS K+M SS+ L + SS SS P +++ + DH+ D T
Sbjct: 1 MASFKLMSSSNSDLSRRNSSSA--SSSPSIRSSHHLRPNPHADHSRISFAYGGGVNDYTF 58
Query: 44 TDPYVP--------------------RTVDDVWQEIIAGDHH--QCKEEIPDEMMTLEDF 81
P ++VDDVW+EI++G+ KEE P+++MTLEDF
Sbjct: 59 ASDSKPFEMAIDVDRSIGDRNSVNNGKSVDDVWKEIVSGEQKTIMMKEEEPEDIMTLEDF 118
Query: 82 LVKAGAVDGDDEDDEVKMSMPLTERLS--GVFSLD-----GSSFQGMEGVEGGSASVGGF 134
L KA +G ++ +VK+ TERL+ G ++ D SSFQ +EG GG
Sbjct: 119 LAKAEMDEGASDEIDVKIP---TERLNNDGSYTFDFPMQRHSSFQMVEGSMGGGV----- 170
Query: 135 GNGVEVVEGTRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVK 194
TRGKRGR ++E +DKAA QRQ+RMIKNRESAARSRERKQAYQVELE+LA K
Sbjct: 171 ---------TRGKRGRVMMEAMDKAAAQRQKRMIKNRESAARSRERKQAYQVELETLAAK 221
Query: 195 LEEENDKLLKEKAERTKERFKQLMEEVIPVVEKRRPP-RPLRRINSMQW 242
LEEEN++LLKE E TKER+K+LME +IPV EK RPP RPL R +S++W
Sbjct: 222 LEEENEQLLKEIEESTKERYKKLMEVLIPVDEKPRPPSRPLSRSHSLEW 270
>At3g56850 ABA-responsive element binding bZip transcription factor
AREB3 / AtbZip66
Length = 297
Score = 85.5 bits (210), Expect = 2e-17
Identities = 76/233 (32%), Positives = 108/233 (45%), Gaps = 56/233 (24%)
Query: 42 TLTDPYVPRTVDDVWQEII----AGDHHQCKEEIPD-EMMTLEDFLVKAGAVD----GDD 92
TL +TVD+VW++I G H+ +++ P MTLED L+KAG V G +
Sbjct: 83 TLPRDLSKKTVDEVWKDIQQNKNGGSAHERRDKQPTLGEMTLEDLLLKAGVVTETIPGSN 142
Query: 93 EDDEVK--------------------------MSMPLTERLSGVFSLDGSSFQGMEGVEG 126
D V SMP + D M+ +
Sbjct: 143 HDGPVGGGSAGSGAGLGQNITQVGPWIQYHQLPSMPQPQAFMPYPVSD------MQAMVS 196
Query: 127 GSASVGGFGNGVEVVEGTRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQV 186
S+ +GG + T G++ E ++K ++RQ+RMIKNRESAARSR RKQAY
Sbjct: 197 QSSLMGGLSD-----TQTPGRKRVASGEVVEKTVERRQKRMIKNRESAARSRARKQAYTH 251
Query: 187 ELESLAVKLEEENDKLLKEKAERTKERFKQLMEEVIPVVEKRRPPRPLRRINS 239
ELE +LEEEN++L K+K +E+++P V P R LRR +S
Sbjct: 252 ELEIKVSRLEEENERLRKQKE----------VEKILPSVPPPDPKRQLRRTSS 294
>At2g36270 bZip transcription factor AtbZip39
Length = 442
Score = 74.7 bits (182), Expect = 4e-14
Identities = 81/279 (29%), Positives = 110/279 (39%), Gaps = 90/279 (32%)
Query: 42 TLTDPYVPRTVDDVWQEIIAG---------------DHHQCKEEIPDEM---------MT 77
TL P +TVD+VW EI G + Q + E MT
Sbjct: 147 TLPAPLCRKTVDEVWSEIHRGGGSGNGGDSNGRSSSSNGQNNAQNGGETAARQPTFGEMT 206
Query: 78 LEDFLVKAGAV------------DGDDEDDEVKMSMPLTERLSGVFS------------- 112
LEDFLVKAG V +++ + ++L GVF
Sbjct: 207 LEDFLVKAGVVREHPTNPKPNPNPNQNQNPSSVIPAAAQQQLYGVFQGTGDPSFPGQAMG 266
Query: 113 ----------LDGSSFQGMEGVEGGSASVGGFGNG------------------------V 138
G +Q V+ G GG G G V
Sbjct: 267 VGDPSGYAKRTGGGGYQQAPPVQAGVCYGGGVGFGAGGQQMGMVGPLSPVSSDGLGHGQV 326
Query: 139 EVVEGTRGK-----RGRPVVEN--LDKAAQQRQRRMIKNRESAARSRERKQAYQVELESL 191
+ + G G RGR V + ++K ++RQRRMIKNRESAARSR RKQAY VELE+
Sbjct: 327 DNIGGQYGVDMGGLRGRKRVVDGPVEKVVERRQRRMIKNRESAARSRARKQAYTVELEAE 386
Query: 192 AVKLEEENDKLLKEKAERTKERFKQLMEEVIPVVEKRRP 230
+L+EEN +L AE ++R +Q E + + + P
Sbjct: 387 LNQLKEENAQLKHALAELERKRKQQYFESLKSRAQPKLP 425
>At3g44460 bZIP transcription factor DPBF2 / AtbZip67
Length = 331
Score = 73.2 bits (178), Expect = 1e-13
Identities = 76/237 (32%), Positives = 107/237 (45%), Gaps = 41/237 (17%)
Query: 42 TLTDPYVPRTVDDVWQEIIAG---------DHHQCKEEIPDEM----MTLEDFLVKAGAV 88
+L P +TVD+VW EI G E I + +TLEDFLVKAG V
Sbjct: 100 SLPVPLCKKTVDEVWLEIQNGVQQHPPSSNSGQNSAENIRRQQTLGEITLEDFLVKAGVV 159
Query: 89 DGDDEDDEVKMSMPLTERLSGVFSLDG------------------SSFQGMEGVEGGSAS 130
+ +K +M ++ G G S + V G S+S
Sbjct: 160 Q-----EPLKTTMRMSSSDFGYNPEFGVGLHCQNQNNYGDNRSVYSENRPFYSVLGESSS 214
Query: 131 -VGGFGNGVEVVEGTRGKR--GRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVE 187
+ G G + + G R R + + ++RQRRMIKNRESAARSR R+QAY VE
Sbjct: 215 CMTGNGRSNQYLTGLDAFRIKKRIIDGPPEILMERRQRRMIKNRESAARSRARRQAYTVE 274
Query: 188 LESLAVKLEEENDKLLKEKAERTKERFKQLMEEVIPVVEKRRPP--RPLRRINSMQW 242
LE L EEN KL + E K+R ++++ V +++ R +RR+ S W
Sbjct: 275 LELELNNLTEENTKLKEIVEENEKKRRQEIISRSKQVTKEKSGDKLRKIRRMASAGW 331
>At2g41070 bZIP transcription factor AtbZIP12 / DPBF4
Length = 262
Score = 72.8 bits (177), Expect = 2e-13
Identities = 73/245 (29%), Positives = 104/245 (41%), Gaps = 44/245 (17%)
Query: 11 SKSLDHQQPSSPPPSSMPQFTTTNFIDHADSTLTDPYVPRTVDDVWQEIIAGDHHQCKEE 70
S +LD + PP+ + TL +TVD+VW++I +
Sbjct: 43 SMNLDELLKTVLPPAE------EGLVRQGSLTLPRDLSKKTVDEVWRDIQQDKNGNGTST 96
Query: 71 IPDEM------MTLEDFLVKAGAVDGDDEDDEVKMSMPLT----------ERLSGVFSLD 114
+TLED L++AG V E +++ ++ G +
Sbjct: 97 TTTHKQPTLGEITLEDLLLRAGVVTETVVPQENVVNIASNGQWVEYHHQPQQQQGFMTYP 156
Query: 115 GSSFQGMEGVEGGSASVGGFGNGVEVVEGTRGKRGRPVVENLDKAAQQRQRRMIKNRESA 174
Q M + G S + G R R E ++K ++RQ+RMIKNRESA
Sbjct: 157 VCEMQDMVMMGGLSDTPQAPG------------RKRVAGEIVEKTVERRQKRMIKNRESA 204
Query: 175 ARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTKERFKQLMEEVIPVVEKRRPPRPL 234
ARSR RKQAY ELE +LEEEN+KL R KE +E+++P P L
Sbjct: 205 ARSRARKQAYTHELEIKVSRLEEENEKL-----RRLKE-----VEKILPSEPPPDPKWKL 254
Query: 235 RRINS 239
RR NS
Sbjct: 255 RRTNS 259
>At3g19290 abscisic acid responsive elements-binding factor
(ABRE/ABF4) / bZip transcription factor AtbZip38
Length = 431
Score = 70.9 bits (172), Expect = 6e-13
Identities = 38/72 (52%), Positives = 51/72 (70%)
Query: 148 RGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKA 207
RGR L+K ++RQRRMIKNRESAARSR RKQAY +ELE+ KL++ N +L K++A
Sbjct: 339 RGRRSNTGLEKVIERRQRRMIKNRESAARSRARKQAYTLELEAEIEKLKKTNQELQKKQA 398
Query: 208 ERTKERFKQLME 219
E + + +L E
Sbjct: 399 EMVEMQKNELKE 410
Score = 30.8 bits (68), Expect = 0.68
Identities = 38/151 (25%), Positives = 57/151 (37%), Gaps = 36/151 (23%)
Query: 2 ASSKMMMSSSKSLDHQQPSS--PPPSSMPQFTTTNFIDHADSTLTDPYVPRTVDDVWQEI 59
A + M S+ + QP + PPP N TL +TVD+VW+ +
Sbjct: 77 AQAMAMTSAPAATAVAQPGAGIPPPGG-------NLQRQGSLTLPRTISQKTVDEVWKCL 129
Query: 60 IAGDHHQ-------CKEEIPDEM------MTLEDFLVKAGAVDGDDEDDEVKMSMPLTER 106
I D + + +P MTLE+FL +AG V D+ ++
Sbjct: 130 ITKDGNMEGSSGGGGESNVPPGRQQTLGEMTLEEFLFRAGVVREDN----------CVQQ 179
Query: 107 LSGVFSLDGSSFQGMEGVEGGSASVGGFGNG 137
+ V + + F G GG GFG G
Sbjct: 180 MGQVNGNNNNGFYGNSTAAGGL----GFGFG 206
>At1g49720 abscisic acid responsive elements-binding factor
(ABRE/ABF1)/ bZip transcription factor AtbZip35
Length = 392
Score = 68.6 bits (166), Expect = 3e-12
Identities = 59/159 (37%), Positives = 80/159 (50%), Gaps = 20/159 (12%)
Query: 97 VKMSMPLTERLSGVF--SLDGSSFQGMEGVEG---------GSASVGGFGNGVEVVEGTR 145
V + P+ G+F S DG + M G G SA + + V V G
Sbjct: 241 VTFAAPVNMVNRGLFETSADGPANSNMGGAGGTVTATSPGTSSAENNTWSSPVPYVFG-- 298
Query: 146 GKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKE 205
RGR L+K ++RQ+RMIKNRESAARSR RKQAY +ELE+ L+ N L K+
Sbjct: 299 --RGRRSNTGLEKVVERRQKRMIKNRESAARSRARKQAYTLELEAEIESLKLVNQDLQKK 356
Query: 206 KAERTKERFKQLME--EVIPVVEKRRPPRPLRRINSMQW 242
+AE K +L E + P++ KR + LRR + W
Sbjct: 357 QAEIMKTHNSELKEFSKQPPLLAKR---QCLRRTLTGPW 392
Score = 30.4 bits (67), Expect = 0.89
Identities = 30/133 (22%), Positives = 55/133 (40%), Gaps = 14/133 (10%)
Query: 42 TLTDPYVPRTVDDVWQEIIA--GDHHQCKEEIPDEM-----MTLEDFLVKAGAVDGDDED 94
TL +TVD+VW+ + + G + + + MTLEDFL++AG V D+
Sbjct: 98 TLPRTLSQKTVDEVWKYLNSKEGSNGNTGTDALERQQTLGEMTLEDFLLRAGVVKEDNTQ 157
Query: 95 DEVKMSMPLTERLSGVFSLDGSSFQGMEGVEGGSASVGGFGNGVEVVEGTRGKRGRPVVE 154
S SG ++ +G++ + S+ GN ++ G V
Sbjct: 158 QNENSS-------SGFYANNGAAGLEFGFGQPNQNSISFNGNNSSMIMNQAPGLGLKVGG 210
Query: 155 NLDKAAQQRQRRM 167
+ + Q Q+++
Sbjct: 211 TMQQQQQPHQQQL 223
>At4g34000 abscisic acid responsive elements-binding factor
(ABRE/ABF3) / bZip transcription factor AtbZip37
Length = 449
Score = 61.6 bits (148), Expect = 4e-10
Identities = 30/51 (58%), Positives = 42/51 (81%)
Query: 156 LDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEK 206
L+K ++RQ+RMIKNRESAARSR RKQAY +ELE+ +L+E N++L K++
Sbjct: 368 LEKVIERRQKRMIKNRESAARSRARKQAYTMELEAEIAQLKELNEELQKKQ 418
Score = 34.3 bits (77), Expect = 0.062
Identities = 43/171 (25%), Positives = 67/171 (39%), Gaps = 35/171 (20%)
Query: 30 FTTTNFIDHADSTLTDPYVPRTVDDVWQEIIAGDH------HQCKEEIPDEM-----MTL 78
FT + TL + VDDVW+E++ D + IP MTL
Sbjct: 116 FTGGSLQRQGSLTLPRTISQKRVDDVWKELMKEDDIGNGVVNGGTSGIPQRQQTLGEMTL 175
Query: 79 EDFLVKAGAVDGDDEDDEVKMSMPLTERLSGVFSLDGSSFQGMEGVEGGSASVGGF---- 134
E+FLV+AG V + + E V + +G F G G + GF
Sbjct: 176 EEFLVRAGVVREEPQPVE------------SVTNFNG-GFYGFGSNGGLGTASNGFVANQ 222
Query: 135 -----GNGVEVVEGTRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRER 180
GNGV V + + +P+ + QQ Q M++ + +++ER
Sbjct: 223 PQDLSGNGVAVRQDLLTAQTQPLQMQQPQMVQQPQ--MVQQPQQLIQTQER 271
>At5g42910 bzip protein AtbZip15
Length = 370
Score = 55.1 bits (131), Expect = 3e-08
Identities = 38/96 (39%), Positives = 55/96 (56%), Gaps = 5/96 (5%)
Query: 144 TRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLL 203
TRG + + + ++ RR IKNRESAARSR RKQA +E+E L+++ ++LL
Sbjct: 277 TRGGKINSEITAEKQFVDKKLRRKIKNRESAARSRARKQAQTMEVEVELENLKKDYEELL 336
Query: 204 KEKAERTKERFKQLMEEVIPVVEKRRPPRPLRRINS 239
K+ E K +Q+ +I + E RP R LRR S
Sbjct: 337 KQHVELRK---RQMEPGMISLHE--RPERKLRRTKS 367
>At2g17770 bZIP transcription factor atbzip27
Length = 234
Score = 53.5 bits (127), Expect = 1e-07
Identities = 34/78 (43%), Positives = 48/78 (60%), Gaps = 8/78 (10%)
Query: 147 KRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEK 206
KRG+ ++ D +R +RMIKNRESAARSR RKQAY ELE L+ EN +L ++
Sbjct: 153 KRGQ---DSDDTRGDRRYKRMIKNRESAARSRARKQAYTNELELEIAHLQTENARLKIQQ 209
Query: 207 -----AERTKERFKQLME 219
AE T+ + K+ ++
Sbjct: 210 EQLKIAEATQNQVKKTLQ 227
>At4g35900 bZIP transcription factor AtbZIP14
Length = 285
Score = 52.8 bits (125), Expect = 2e-07
Identities = 29/60 (48%), Positives = 41/60 (68%), Gaps = 3/60 (5%)
Query: 147 KRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEK 206
KRG+ ++ + + +R +RMIKNRESAARSR RKQAY ELE L+ EN +L +++
Sbjct: 204 KRGQ---DSNEGSGNRRHKRMIKNRESAARSRARKQAYTNELELEVAHLQAENARLKRQQ 260
>At5g28770 bZip transcription factor BZO2H3 / AtbZip63
Length = 307
Score = 50.1 bits (118), Expect = 1e-06
Identities = 26/57 (45%), Positives = 37/57 (64%)
Query: 155 NLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTK 211
N++ +R +RM+ NRESA RSR RKQA+ ELE+ +L EN KL+K + T+
Sbjct: 139 NMNPTNVKRVKRMLSNRESARRSRRRKQAHLSELETQVSQLRVENSKLMKGLTDVTQ 195
>At5g11260 bZip transcription factor HY5 / AtbZip56
Length = 168
Score = 50.1 bits (118), Expect = 1e-06
Identities = 42/132 (31%), Positives = 66/132 (49%), Gaps = 17/132 (12%)
Query: 96 EVKMSMPLTERLSGVFSLDGSSF-QGMEGVEGGSASVGGFGNGVEVVEGTRG----KRGR 150
E+K + E + V G + + G E GSA+ G E + T G KRGR
Sbjct: 29 EIKEGIESDEEIRRVPEFGGEAVGKETSGRESGSAT------GQERTQATVGESQRKRGR 82
Query: 151 PVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERT 210
E +K R +R+++NR SA ++RERK+AY ELE+ LE +N +L E+ T
Sbjct: 83 TPAEKENK----RLKRLLRNRVSAQQARERKKAYLSELENRVKDLENKNSEL--EERLST 136
Query: 211 KERFKQLMEEVI 222
+ Q++ ++
Sbjct: 137 LQNENQMLRHIL 148
>At3g17609 bZip transcription factor AtbZip64
Length = 149
Score = 48.5 bits (114), Expect = 3e-06
Identities = 38/121 (31%), Positives = 60/121 (49%), Gaps = 12/121 (9%)
Query: 92 DEDDEVKMSMPLTERLSGVFSLDGSSFQGMEGVEGGSASVGGFGNGVEVVEGTRGKRGRP 151
+E DE + +P E L S+ G+ E NGV + RG+ P
Sbjct: 22 EESDEELLMVPDMEAAGSTCVLSSSADDGVNNPELDQTQ-----NGVSTAKRRRGRN--P 74
Query: 152 VVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTK 211
V DK + +R+++NR SA ++RERK+ Y +LES A +L+ ND+L ++ + T
Sbjct: 75 V----DKEYRSL-KRLLRNRVSAQQARERKKVYVSDLESRANELQNNNDQLEEKISTLTN 129
Query: 212 E 212
E
Sbjct: 130 E 130
>At1g68880 bZip transcription factor AtbZip8
Length = 138
Score = 48.1 bits (113), Expect = 4e-06
Identities = 26/74 (35%), Positives = 48/74 (64%), Gaps = 1/74 (1%)
Query: 161 QQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTKERFKQLMEE 220
++++RR + NRESA RSR RKQ + EL S+ V+L +N L+ E ++ +E +++++EE
Sbjct: 46 ERKRRRKVSNRESARRSRMRKQRHMEELWSMLVQLINKNKSLVDELSQ-ARECYEKVIEE 104
Query: 221 VIPVVEKRRPPRPL 234
+ + E+ R +
Sbjct: 105 NMKLREENSKSRKM 118
>At1g32150 putative G-Box binding protein
Length = 389
Score = 48.1 bits (113), Expect = 4e-06
Identities = 34/90 (37%), Positives = 48/90 (52%), Gaps = 3/90 (3%)
Query: 133 GFGNGVEVVEGT--RGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELES 190
G GN V G G + +P ++ D+ +RQRR NRESA RSR RKQA EL
Sbjct: 266 GHGNVSGAVPGVVVDGSQSQPWLQVSDEREIKRQRRKQSNRESARRSRLRKQAECDELAQ 325
Query: 191 LAVKLEEENDKLLKEKAERTKERFKQLMEE 220
A L EN L+ + + K ++++L+ E
Sbjct: 326 RAEVLNGENSS-LRAEINKLKSQYEELLAE 354
>At2g04038 bZip transcription factor AtbZip48
Length = 166
Score = 47.8 bits (112), Expect = 5e-06
Identities = 29/74 (39%), Positives = 44/74 (59%), Gaps = 1/74 (1%)
Query: 161 QQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTKERFKQLMEE 220
+++QRRM+ NRESA RSR RKQ + EL S ++L EN+ L+ +K R E +++E
Sbjct: 73 ERKQRRMLSNRESARRSRMRKQRHLDELWSQVIRLRNENNCLI-DKLNRVSETQNCVLKE 131
Query: 221 VIPVVEKRRPPRPL 234
+ E+ R L
Sbjct: 132 NSKLKEEASDLRQL 145
>At1g13600 bZip transcription factor AtbZip58
Length = 196
Score = 47.4 bits (111), Expect = 7e-06
Identities = 29/74 (39%), Positives = 43/74 (57%), Gaps = 1/74 (1%)
Query: 161 QQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTKERFKQLMEE 220
+++QRRMI NRESA RSR RKQ + EL S ++L +N L+ +K R E + ++E
Sbjct: 85 ERKQRRMISNRESARRSRMRKQRHLDELWSQVIRLRTDNHCLM-DKLNRVSESHELALKE 143
Query: 221 VIPVVEKRRPPRPL 234
+ E+ R L
Sbjct: 144 NAKLKEETSDLRQL 157
>At3g30530 bZip transcription factor AtbZip42
Length = 173
Score = 46.6 bits (109), Expect = 1e-05
Identities = 31/84 (36%), Positives = 48/84 (56%), Gaps = 16/84 (19%)
Query: 161 QQRQRRMIKNRESAARSRERKQAYQVELESLAV--------------KLEEENDKLLKEK 206
+++QRRMI NRESA RSR RKQ + EL S + L E +DK+L+E
Sbjct: 80 ERKQRRMISNRESARRSRMRKQRHLDELWSQVMWLRIENHQLLDKLNNLSESHDKVLQEN 139
Query: 207 AERTKERFKQLMEEVIPVVEKRRP 230
A+ +E F+ +++VI ++ + P
Sbjct: 140 AQLKEETFE--LKQVISDMQIQSP 161
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,581,546
Number of Sequences: 26719
Number of extensions: 255256
Number of successful extensions: 1957
Number of sequences better than 10.0: 237
Number of HSP's better than 10.0 without gapping: 130
Number of HSP's successfully gapped in prelim test: 107
Number of HSP's that attempted gapping in prelim test: 1642
Number of HSP's gapped (non-prelim): 362
length of query: 242
length of database: 11,318,596
effective HSP length: 96
effective length of query: 146
effective length of database: 8,753,572
effective search space: 1278021512
effective search space used: 1278021512
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0052a.3


