

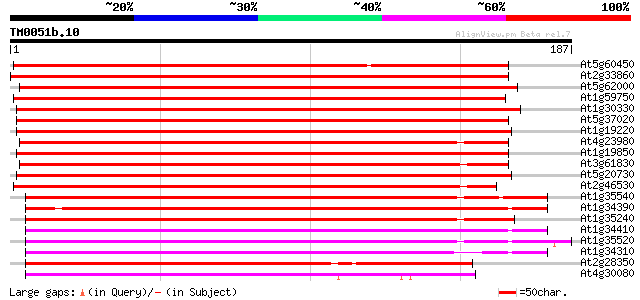
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0051b.10
(187 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g60450 auxin response factor 4 259 4e-70
At2g33860 auxin response transcription factor 3 (ETTIN/ARF3) 207 3e-54
At5g62000 auxin response factor - like protein 196 5e-51
At1g59750 auxin response factor 1 190 4e-49
At1g30330 putative protein 188 1e-48
At5g37020 auxin response factor 8 (ARF8) 186 5e-48
At1g19220 auxin response factor (ARF) like protein 174 2e-44
At4g23980 auxin response factor 9 (ARF9) 173 4e-44
At1g19850 transcription factor monopteros (MP) 172 7e-44
At3g61830 auxin response factor-like protein 171 3e-43
At5g20730 auxin response factor 7 (ARF7) 168 2e-42
At2g46530 ARF1 family auxin responsive transcription factor like... 165 2e-41
At1g35540 auxin response factor, putative 149 7e-37
At1g34390 auxin response factor, putative 146 6e-36
At1g35240 hypothetical protein 144 2e-35
At1g34410 auxin response factor, putative 144 3e-35
At1g35520 auxin response factor, putative 143 6e-35
At1g34310 hypothetical protein 137 4e-33
At2g28350 unknown protein 134 4e-32
At4g30080 transcription factor like protein 126 6e-30
>At5g60450 auxin response factor 4
Length = 788
Score = 259 bits (663), Expect = 4e-70
Identities = 127/165 (76%), Positives = 141/165 (84%), Gaps = 1/165 (0%)
Query: 2 TSHMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHI 61
T HMFCKTLTA DT TH GFSVPRRAAEDC PLDYKQQRPSQEL+AKDLHGVEWKF HI
Sbjct: 173 TPHMFCKTLTASDTSTHGGFSVPRRAAEDCFAPLDYKQQRPSQELIAKDLHGVEWKFRHI 232
Query: 62 YRG*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGN 121
YRG PR HLLTTGWSIFVSQ NLVSGDAV FLR E GELRLGIRR+ PRNGLP+S +
Sbjct: 233 YRGQPRRHLLTTGWSIFVSQKNLVSGDAVLFLRDEGGELRLGIRRAARPRNGLPDSII-E 291
Query: 122 QNSYPHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSI 166
+NS +ILS+VAN +ST+SMFHVFYSPRA++++FV PY+KY SI
Sbjct: 292 KNSCSNILSLVANAVSTKSMFHVFYSPRATHAEFVIPYEKYITSI 336
>At2g33860 auxin response transcription factor 3 (ETTIN/ARF3)
Length = 608
Score = 207 bits (526), Expect = 3e-54
Identities = 102/166 (61%), Positives = 124/166 (74%)
Query: 1 STSHMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCH 60
+T HMFCKTLTA DT TH GFSVPRRAAEDC PPLDY Q RPSQEL+A+DLHG+EW+F H
Sbjct: 154 NTPHMFCKTLTASDTSTHGGFSVPRRAAEDCFPPLDYSQPRPSQELLARDLHGLEWRFRH 213
Query: 61 IYRG*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVG 120
IYRG PR HLLTTGWS FV++ LVSGDAV FLRG++G+LRLG+RR+ S
Sbjct: 214 IYRGQPRRHLLTTGWSAFVNKKKLVSGDAVLFLRGDDGKLRLGVRRASQIEGTAALSAQY 273
Query: 121 NQNSYPHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSI 166
NQN + S VA+ IST S+F + Y+P+AS S+F+ P K+ K +
Sbjct: 274 NQNMNHNNFSEVAHAISTHSVFSISYNPKASWSNFIIPAPKFLKVV 319
>At5g62000 auxin response factor - like protein
Length = 371
Score = 196 bits (499), Expect = 5e-51
Identities = 91/166 (54%), Positives = 122/166 (72%)
Query: 4 HMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIYR 63
H FCKTLTA DT TH GFSV RR A++CLPPLD +Q P+QELVAKDLH EW+F HI+R
Sbjct: 160 HSFCKTLTASDTSTHGGFSVLRRHADECLPPLDMSRQPPTQELVAKDLHANEWRFRHIFR 219
Query: 64 G*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQN 123
G PR HLL +GWS+FVS LV+GDA FLRGENGELR+G+RR++ + +P S + + +
Sbjct: 220 GQPRRHLLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHS 279
Query: 124 SYPHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSIHDN 169
+ +L+ + IST +MF V+Y PR S S+F+ P+ +Y +S+ +N
Sbjct: 280 MHLGVLATAWHAISTGTMFTVYYKPRTSPSEFIVPFDQYMESVKNN 325
>At1g59750 auxin response factor 1
Length = 665
Score = 190 bits (482), Expect = 4e-49
Identities = 91/164 (55%), Positives = 115/164 (69%)
Query: 2 TSHMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHI 61
T H FCKTLTA DT TH GFSV RR A+DCLPPLD QQ P QELVA DLH EW F HI
Sbjct: 120 TVHSFCKTLTASDTSTHGGFSVLRRHADDCLPPLDMSQQPPWQELVATDLHNSEWHFRHI 179
Query: 62 YRG*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGN 121
+RG PR HLLTTGWS+FVS LV+GDA FLRGEN ELR+G+RR + + +P S + +
Sbjct: 180 FRGQPRRHLLTTGWSVFVSSKKLVAGDAFIFLRGENEELRVGVRRHMRQQTNIPSSVISS 239
Query: 122 QNSYPHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKS 165
+ + +L+ A+ I+T ++F VFY PR S S+F+ +Y ++
Sbjct: 240 HSMHIGVLATAAHAITTGTIFSVFYKPRTSRSEFIVSVNRYLEA 283
>At1g30330 putative protein
Length = 933
Score = 188 bits (478), Expect = 1e-48
Identities = 89/168 (52%), Positives = 120/168 (70%)
Query: 3 SHMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIY 62
++ FCKTLTA DT TH GFSVPRRAAE PPLDY QQ P+QEL+A+DLH EWKF HI+
Sbjct: 124 TNYFCKTLTASDTSTHGGFSVPRRAAEKVFPPLDYSQQPPAQELMARDLHDNEWKFRHIF 183
Query: 63 RG*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQ 122
RG P+ HLLTTGWS+FVS LV+GD+V F+ + +L LGIRR+ P+ +P S + +
Sbjct: 184 RGQPKRHLLTTGWSVFVSAKRLVAGDSVLFIWNDKNQLLLGIRRANRPQTVMPSSVLSSD 243
Query: 123 NSYPHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSIHDNK 170
+ + +L+ A+ +T S F +FY+PRAS S+FV P KY K+++ +
Sbjct: 244 SMHLGLLAAAAHAAATNSRFTIFYNPRASPSEFVIPLAKYVKAVYHTR 291
>At5g37020 auxin response factor 8 (ARF8)
Length = 811
Score = 186 bits (473), Expect = 5e-48
Identities = 90/164 (54%), Positives = 116/164 (69%)
Query: 3 SHMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIY 62
S+ FCKTLTA DT TH GFSVPRRAAE PPLDY Q P+QEL+A+DLH VEWKF HI+
Sbjct: 123 SNYFCKTLTASDTSTHGGFSVPRRAAEKVFPPLDYTLQPPAQELIARDLHDVEWKFRHIF 182
Query: 63 RG*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQ 122
RG P+ HLLTTGWS+FVS LV+GD+V F+R E +L LGIR + P+ +P S + +
Sbjct: 183 RGQPKRHLLTTGWSVFVSAKRLVAGDSVIFIRNEKNQLFLGIRHATRPQTIVPSSVLSSD 242
Query: 123 NSYPHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSI 166
+ + +L+ A+ +T S F VF+ PRAS S+FV KY K++
Sbjct: 243 SMHIGLLAAAAHASATNSCFTVFFHPRASQSEFVIQLSKYIKAV 286
>At1g19220 auxin response factor (ARF) like protein
Length = 1086
Score = 174 bits (442), Expect = 2e-44
Identities = 86/165 (52%), Positives = 111/165 (67%)
Query: 3 SHMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIY 62
+ FCKTLTA DT TH GFSVPRRAAE PPLD+ Q P+QE+VAKDLH W F HIY
Sbjct: 123 TEFFCKTLTASDTSTHGGFSVPRRAAEKIFPPLDFSMQPPAQEIVAKDLHDTTWTFRHIY 182
Query: 63 RG*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQ 122
RG P+ HLLTTGWS+FVS L +GD+V F+R E +L LGIRR+ L S + +
Sbjct: 183 RGQPKRHLLTTGWSVFVSTKRLFAGDSVLFVRDEKSQLMLGIRRANRQTPTLSSSVISSD 242
Query: 123 NSYPHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSIH 167
+ + IL+ A+ + S F +F++PRAS S+FV P KY K+++
Sbjct: 243 SMHIGILAAAAHANANSSPFTIFFNPRASPSEFVVPLAKYNKALY 287
>At4g23980 auxin response factor 9 (ARF9)
Length = 638
Score = 173 bits (439), Expect = 4e-44
Identities = 83/163 (50%), Positives = 112/163 (67%), Gaps = 2/163 (1%)
Query: 4 HMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIYR 63
H F K LTA DT TH GFSV R+ A +CLPPLD QQ P+QELVA+D+HG +WKF HI+R
Sbjct: 114 HSFSKVLTASDTSTHGGFSVLRKHATECLPPLDMTQQTPTQELVAEDVHGYQWKFKHIFR 173
Query: 64 G*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQN 123
G PR HLLTTGWS FV+ LV+GD FLRGENGELR+G+RR+ ++ +P S + + +
Sbjct: 174 GQPRRHLLTTGWSTFVTSKRLVAGDTFVFLRGENGELRVGVRRANLQQSSMPSSVISSHS 233
Query: 124 SYPHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSI 166
+ +L+ + T++MF V+Y PR S F+ KY +++
Sbjct: 234 MHLGVLATARHATQTKTMFIVYYKPR--TSQFIISLNKYLEAM 274
>At1g19850 transcription factor monopteros (MP)
Length = 902
Score = 172 bits (437), Expect = 7e-44
Identities = 83/164 (50%), Positives = 109/164 (65%)
Query: 3 SHMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIY 62
+ FCKTLTA DT TH GFSVPRRAAE PPLDY Q P+QELV +DLH W F HIY
Sbjct: 155 TEFFCKTLTASDTSTHGGFSVPRRAAEKLFPPLDYSAQPPTQELVVRDLHENTWTFRHIY 214
Query: 63 RG*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQ 122
RG P+ HLLTTGWS+FV L +GD+V F+R E +L +G+RR+ + LP S +
Sbjct: 215 RGQPKRHLLTTGWSLFVGSKRLRAGDSVLFIRDEKSQLMVGVRRANRQQTALPSSVLSAD 274
Query: 123 NSYPHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSI 166
+ + +L+ A+ + R+ F +FY+PRA ++FV P KY K+I
Sbjct: 275 SMHIGVLAAAAHATANRTPFLIFYNPRACPAEFVIPLAKYRKAI 318
>At3g61830 auxin response factor-like protein
Length = 602
Score = 171 bits (432), Expect = 3e-43
Identities = 84/163 (51%), Positives = 111/163 (67%), Gaps = 2/163 (1%)
Query: 4 HMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIYR 63
H F K LTA DT TH GFSV R+ A +CLP LD Q P+QELV +DLHG EW+F HI+R
Sbjct: 126 HSFVKILTASDTSTHGGFSVLRKHATECLPSLDMTQATPTQELVTRDLHGFEWRFKHIFR 185
Query: 64 G*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQN 123
G PR HLLTTGWS FVS LV+GDA FLRGENG+LR+G+RR ++ +P S + +Q+
Sbjct: 186 GQPRRHLLTTGWSTFVSSKRLVAGDAFVFLRGENGDLRVGVRRLARHQSTMPTSVISSQS 245
Query: 124 SYPHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSI 166
+ +L+ ++ + T ++F VFY PR S F+ KY ++I
Sbjct: 246 MHLGVLATASHAVRTTTIFVVFYKPRI--SQFIVGVNKYMEAI 286
>At5g20730 auxin response factor 7 (ARF7)
Length = 1164
Score = 168 bits (425), Expect = 2e-42
Identities = 82/165 (49%), Positives = 109/165 (65%)
Query: 3 SHMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIY 62
+ FCKTLTA DT TH GFSVPRRAAE P LD+ Q P QELVAKD+H W F HIY
Sbjct: 124 NEFFCKTLTASDTSTHGGFSVPRRAAEKIFPALDFSMQPPCQELVAKDIHDNTWTFRHIY 183
Query: 63 RG*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQ 122
RG P+ HLLTTGWS+FVS L +GD+V F+R +L LGIRR+ + L S + +
Sbjct: 184 RGQPKRHLLTTGWSVFVSTKRLFAGDSVLFIRDGKAQLLLGIRRANRQQPALSSSVISSD 243
Query: 123 NSYPHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSIH 167
+ + +L+ A+ + S F +FY+PRA+ ++FV P KY K+++
Sbjct: 244 SMHIGVLAAAAHANANNSPFTIFYNPRAAPAEFVVPLAKYTKAMY 288
>At2g46530 ARF1 family auxin responsive transcription factor like
protein
Length = 622
Score = 165 bits (417), Expect = 2e-41
Identities = 82/161 (50%), Positives = 108/161 (66%), Gaps = 2/161 (1%)
Query: 2 TSHMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHI 61
T F K LTA DT TH GFSV R+ A +CLP LD Q P+QELVA+DLHG EW+F HI
Sbjct: 141 TVDSFVKILTASDTSTHGGFSVLRKHATECLPSLDMTQPTPTQELVARDLHGYEWRFKHI 200
Query: 62 YRG*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGN 121
+RG PR HLLTTGWS FV+ LV+GDA FLRGE G+LR+G+RR ++ +P S + +
Sbjct: 201 FRGQPRRHLLTTGWSTFVTSKRLVAGDAFVFLRGETGDLRVGVRRLAKQQSTMPASVISS 260
Query: 122 QNSYPHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKY 162
Q+ +L+ ++ ++T ++F VFY PR S F+ KY
Sbjct: 261 QSMRLGVLATASHAVTTTTIFVVFYKPRI--SQFIISVNKY 299
>At1g35540 auxin response factor, putative
Length = 661
Score = 149 bits (377), Expect = 7e-37
Identities = 78/174 (44%), Positives = 106/174 (60%), Gaps = 3/174 (1%)
Query: 6 FCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIYRG* 65
F K LTA DT H GFSVP++ A +CLPPLD Q P+QE++A DLHG +W+F HIYRG
Sbjct: 182 FTKVLTASDTSVHGGFSVPKKHAIECLPPLDMSQPLPTQEILAIDLHGNQWRFRHIYRGT 241
Query: 66 PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQNSY 125
+ HLLT GW+ F + LV GD + F+RGE GELR+GIRR+ + +P S V ++
Sbjct: 242 AQRHLLTIGWNAFTTSKKLVEGDVIVFVRGETGELRVGIRRAGHQQGNIPSSIVSIESMR 301
Query: 126 PHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSIHDNKRNEPSHRVM 179
I++ + + MF V Y PR +S F+ Y K+ + +NK N S M
Sbjct: 302 HGIIASAKHAFDNQCMFIVVYKPR--SSQFIVSYDKFL-DVVNNKFNVGSRFTM 352
>At1g34390 auxin response factor, putative
Length = 600
Score = 146 bits (369), Expect = 6e-36
Identities = 75/174 (43%), Positives = 106/174 (60%), Gaps = 3/174 (1%)
Query: 6 FCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIYRG* 65
F K LTA DT GF VP++ A +CLPPLD Q P+QEL+A DLHG +W+F H YRG
Sbjct: 126 FTKVLTASDT--SGGFFVPKKHAIECLPPLDMSQPLPTQELLATDLHGNQWRFNHNYRGT 183
Query: 66 PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQNSY 125
P+ HLLTTGW+ F + LV+GD + F+RGE GELR+GIRR+ + +P S + ++
Sbjct: 184 PQRHLLTTGWNAFTTSKKLVAGDVIVFVRGETGELRVGIRRAGHQQGNIPSSIISIESMR 243
Query: 126 PHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSIHDNKRNEPSHRVM 179
+++ + + MF V Y P +S F+ Y K+ ++ +NK N S M
Sbjct: 244 HGVIASAKHAFDNQCMFIVVYKPSIRSSQFIVSYDKFLDAV-NNKFNVGSRFTM 296
>At1g35240 hypothetical protein
Length = 615
Score = 144 bits (364), Expect = 2e-35
Identities = 71/163 (43%), Positives = 102/163 (62%), Gaps = 2/163 (1%)
Query: 6 FCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIYRG* 65
F K LTA DT + GF VP++ A +CLPPLD Q P+QEL+AKDLHG +W+F H YRG
Sbjct: 126 FTKVLTASDTSAYGGFFVPKKHAIECLPPLDMSQPLPAQELLAKDLHGNQWRFRHSYRGT 185
Query: 66 PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQNSY 125
P+ H LTTGW+ F + LV GD + F+RGE GELR+GIRR+ + +P S V
Sbjct: 186 PQRHSLTTGWNEFTTSKKLVKGDVIVFVRGETGELRVGIRRARHQQGNIPSSIVSIDCMR 245
Query: 126 PHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSIHD 168
+++ + + + +F V Y PR +S F+ Y K+ ++++
Sbjct: 246 HGVIASAKHALDNQCIFIVVYKPR--SSQFIVSYDKFLDAMNN 286
>At1g34410 auxin response factor, putative
Length = 620
Score = 144 bits (363), Expect = 3e-35
Identities = 73/174 (41%), Positives = 104/174 (58%), Gaps = 1/174 (0%)
Query: 6 FCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIYRG* 65
F K LTA DT + GFSVP++ A +CLPPLD Q P+QE++A DLH +W+F H YRG
Sbjct: 138 FTKVLTASDTSAYGGFSVPKKHAIECLPPLDMSQPLPAQEILAIDLHDNQWRFRHNYRGT 197
Query: 66 PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQNSY 125
P+ H LTTGW+ F++ LV GD + F+RGE GELR+GIRR+ + +P S V
Sbjct: 198 PQRHSLTTGWNEFITSKKLVKGDVIVFVRGETGELRVGIRRARHQQGNIPSSIVSIDCMR 257
Query: 126 PHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSIHDNKRNEPSHRVM 179
+++ + + +F V Y P +S F+ Y K+ ++ +NK N S M
Sbjct: 258 HGVIASAKHAFDNQCIFIVVYKPSIRSSQFIVSYDKFLDAV-NNKFNVGSRFTM 310
>At1g35520 auxin response factor, putative
Length = 576
Score = 143 bits (360), Expect = 6e-35
Identities = 80/183 (43%), Positives = 107/183 (57%), Gaps = 4/183 (2%)
Query: 6 FCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIYRG* 65
F K LTA D + FSVP++ A +CLPPLD Q P+QEL+A DLHG +W F H YRG
Sbjct: 126 FTKVLTASDISANGVFSVPKKHAIECLPPLDMSQPLPAQELLAIDLHGNQWSFRHSYRGT 185
Query: 66 PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQNSY 125
P+ HLLTTGW+ F + LV GD + F+RGE GELR+GIRR+ + +P S V
Sbjct: 186 PQRHLLTTGWNEFTTSKKLVKGDVIVFVRGETGELRVGIRRARHQQGNIPSSIVSIDCMR 245
Query: 126 PHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSIHDNKRNEPSHRVME-ERDH 184
+++ + + MF V Y PR +S F+ Y K+ ++ +NK N S M E D
Sbjct: 246 HGVIASAKHAFDNQCMFIVVYKPR--SSQFIVSYDKFLDAV-NNKFNVGSRFTMRFEGDD 302
Query: 185 LYE 187
L E
Sbjct: 303 LSE 305
>At1g34310 hypothetical protein
Length = 591
Score = 137 bits (344), Expect = 4e-33
Identities = 74/174 (42%), Positives = 100/174 (56%), Gaps = 10/174 (5%)
Query: 6 FCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIYRG* 65
F K LTA DT H GF VP++ A +CLP LD Q P+QEL+A DLHG +W+F H YRG
Sbjct: 131 FTKVLTASDTSAHGGFFVPKKHAIECLPSLDMSQPLPAQELLAIDLHGNQWRFNHNYRGT 190
Query: 66 PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQNSY 125
P+ HLLTTGW+ F + LV+GD + F+RGE GELR+GIRR+ + +P S V
Sbjct: 191 PQRHLLTTGWNAFTTSKKLVAGDVIVFVRGETGELRVGIRRARHQQGNIPSSIVSIDCMR 250
Query: 126 PHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSIHDNKRNEPSHRVM 179
+++ + + MF V Y P Y K+ ++ +NK N S M
Sbjct: 251 HGVVASAKHAFDNQCMFTVVYKP---------SYDKFLDAV-NNKFNVGSRFTM 294
>At2g28350 unknown protein
Length = 693
Score = 134 bits (336), Expect = 4e-32
Identities = 72/149 (48%), Positives = 91/149 (60%), Gaps = 3/149 (2%)
Query: 6 FCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIYRG* 65
F KTLT D GFSVPR AE P LDY + P Q ++AKD+HG WKF HIYRG
Sbjct: 115 FAKTLTQSDANNGGGFSVPRYCAETIFPRLDYSAEPPVQTVIAKDIHGETWKFRHIYRGT 174
Query: 66 PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQNSY 125
PR HLLTTGWS FV+Q L++GD++ FLR E+G+L +GIRR+ R GL S G+ N Y
Sbjct: 175 PRRHLLTTGWSTFVNQKKLIAGDSIVFLRSESGDLCVGIRRA--KRGGL-GSNAGSDNPY 231
Query: 126 PHILSVVANVISTRSMFHVFYSPRASNSD 154
P + + ST + + R N+D
Sbjct: 232 PGFSGFLRDDESTTTTSKLMMMKRNGNND 260
>At4g30080 transcription factor like protein
Length = 670
Score = 126 bits (317), Expect = 6e-30
Identities = 77/183 (42%), Positives = 96/183 (52%), Gaps = 33/183 (18%)
Query: 6 FCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIYRG* 65
F KTLT D GFSVPR AE P LDY + P Q ++AKD+HG WKF HIYRG
Sbjct: 120 FAKTLTQSDANNGGGFSVPRYCAETIFPRLDYNAEPPVQTILAKDVHGDVWKFRHIYRGT 179
Query: 66 PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSV------GPRNGLPESTV 119
PR HLLTTGWS FV+Q LV+GD++ F+R ENG+L +GIRR+ GP + +
Sbjct: 180 PRRHLLTTGWSNFVNQKKLVAGDSIVFMRAENGDLCVGIRRAKRGGIGNGPEYSAGWNPI 239
Query: 120 GNQNSYPHIL-------------------------SVV--ANVISTRSMFHVFYSPRASN 152
G Y +L SV+ A + + F V Y PRAS
Sbjct: 240 GGSCGYSSLLREDESNSLRRSNCSLADRKGKVTAESVIEAATLAISGRPFEVVYYPRAST 299
Query: 153 SDF 155
S+F
Sbjct: 300 SEF 302
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.137 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,326,067
Number of Sequences: 26719
Number of extensions: 169074
Number of successful extensions: 399
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 41
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 345
Number of HSP's gapped (non-prelim): 54
length of query: 187
length of database: 11,318,596
effective HSP length: 93
effective length of query: 94
effective length of database: 8,833,729
effective search space: 830370526
effective search space used: 830370526
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0051b.10


