

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0051a.1
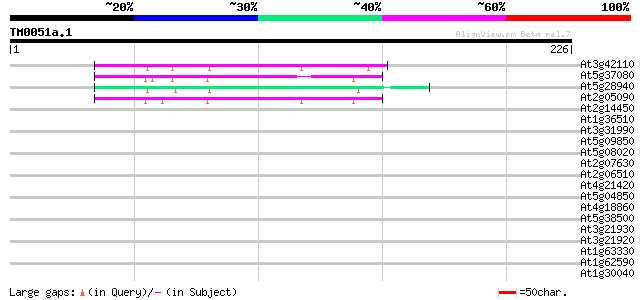
(226 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g42110 putative protein 49 2e-06
At5g37080 putative protein 46 1e-05
At5g28940 44 7e-05
At2g05090 putative replication protein A1 41 4e-04
At2g14450 putative replication protein A1 37 0.011
At1g36510 hypothetical protein 36 0.019
At3g31990 hypothetical protein 35 0.032
At5g09850 unknown protein 34 0.072
At5g08020 replication factor A - like protein 32 0.36
At2g07630 putative replication protein A1 31 0.47
At2g06510 replication protein A1 like 31 0.47
At4g21420 hypothetical protein 29 2.3
At5g04850 unknown protein 28 3.9
At4g18860 hypothetical protein 28 3.9
At5g38500 putative protein 28 5.1
At3g21930 hypothetical protein 28 5.1
At3g21920 hypothetical protein 28 5.1
At1g63330 unknown protein 28 5.1
At1g62590 putative membrane-associated salt-inducible protein (A... 28 5.1
At1g30040 unknown protein 28 5.1
>At3g42110 putative protein
Length = 448
Score = 49.3 bits (116), Expect = 2e-06
Identities = 38/141 (26%), Positives = 62/141 (43%), Gaps = 23/141 (16%)
Query: 35 IYATIKKIESTNDWYYHDCE-CNSAAYSDGA------------QYYCEKCNKHVTPTI-R 80
+ TI I++ WYY C+ CN A +++C+ C VT I R
Sbjct: 212 VLCTIYAIDTDWAWYYISCKTCNKKVNHIHAGVHGVNNKGKKPRFWCDTCKTVVTNVIPR 271
Query: 81 YKLLLNVADDTASATFTVFYREGCYLMNKKGNEILE-KMEKIGSMEHPPQEIVDLVHKEF 139
Y L NV D+T A F +F L + +L+ ++++ E+ P+ + +L+ K F
Sbjct: 272 YMLYANVMDNTGEAKFLLFDSICSELTGESATSVLDGSLDEVVDPENLPEPVKNLIGKTF 331
Query: 140 LFKV--------EGKNICGRW 152
LF V +GK + W
Sbjct: 332 LFLVWVEKEHISDGKEVYKVW 352
>At5g37080 putative protein
Length = 566
Score = 46.2 bits (108), Expect = 1e-05
Identities = 39/138 (28%), Positives = 58/138 (41%), Gaps = 27/138 (19%)
Query: 35 IYATIKKIESTNDWYYHDC-ECN------SAAYSDGA--------QYYCEKCNKHVTPT- 78
I +I I++ W+Y C +CN + S G +++C+KC + +T
Sbjct: 290 ILGSIFAIDTDWGWFYFGCPKCNRKTELVKESTSTGKMVKTPMKPKFWCDKCQESITNVE 349
Query: 79 IRYKLLLNVADDTASATFTVFYREGCYLMNKKGNEILEKMEKIGSMEHPPQEIVDLVHK- 137
RYKL + V D TA VF L+ K E+++ G E P I D++
Sbjct: 350 ARYKLHVRVMDQTAEIKLMVFENNATKLIGKSSEELVD-----GQYEEDPTIISDVITNL 404
Query: 138 -----EFLFKVEGKNICG 150
FL VE NI G
Sbjct: 405 CGKTFHFLVSVEKANIYG 422
>At5g28940
Length = 485
Score = 43.9 bits (102), Expect = 7e-05
Identities = 37/151 (24%), Positives = 61/151 (39%), Gaps = 18/151 (11%)
Query: 35 IYATIKKIESTNDWYYHDCE-CNSAAYSDGAQ------------YYCEKCNKHVTPTI-R 80
+ I IE W+Y C+ C+ + Y+C KCN ++T + R
Sbjct: 247 VMCKIHTIEGDFGWFYVACKKCSKKVEMNNKDKTAIADKPTKTTYWCPKCNLNITQVVPR 306
Query: 81 YKLLLNVADDTASATFTVFYREGCYLMNKKGNEILEKMEKIGSMEHPPQEIVDLVHKEF- 139
+KL + V D T +F ++ ++L+ + E P I +LV K F
Sbjct: 307 FKLHVGVIDQTGDIKCIMFDSAASKMIGHSAFDLLDGVYDDDDPERMPPSIKNLVGKTFQ 366
Query: 140 -LFKVEGKNICGRWFNPAFNVLKICKDQQII 169
L +E N+ GR N + V K+ I+
Sbjct: 367 LLVCIENDNLSGR--NDTYMVGKVWPGVDIV 395
>At2g05090 putative replication protein A1
Length = 521
Score = 41.2 bits (95), Expect = 4e-04
Identities = 37/135 (27%), Positives = 55/135 (40%), Gaps = 19/135 (14%)
Query: 35 IYATIKKIESTNDWYYHDC-ECNSAAY--------------SDGAQYYCEKCNKHVTPT- 78
I +I I++ W+Y C +CN +++C+K + +T
Sbjct: 239 ILGSIFAIDTDWGWFYFGCPKCNRKIELVKESTSTVKRIQAPTKPKFWCDKYQESITNVE 298
Query: 79 IRYKLLLNVADDTASATFTVFYREGCYLMNKKGNEILE-KMEKIGSMEHPPQEIVDLVHK 137
RYKL + V D T VF L+ K E+L+ + E+I P I +L K
Sbjct: 299 ARYKLHIRVMDQTGEIKLMVFENNATNLIRKSSEELLDGQYEEIEDPTIKPDVITNLCGK 358
Query: 138 --EFLFKVEGKNICG 150
FL VE NI G
Sbjct: 359 TFHFLVSVEKANIYG 373
>At2g14450 putative replication protein A1
Length = 527
Score = 36.6 bits (83), Expect = 0.011
Identities = 51/213 (23%), Positives = 81/213 (37%), Gaps = 38/213 (17%)
Query: 35 IYATIKKIESTNDWYYHDCE-CNSAAYSDGA------------QYYCEKCNKHVTPTIRY 81
+ TI I++ WYY +C+ CN A +++CE C VT
Sbjct: 176 VLCTIYAIDTDWAWYYINCKTCNKKVNHIHAGVNGVNNKGKKPRFWCETCKSFVT----- 230
Query: 82 KLLLNVADDTASATFTVFYREGCYLMNKKGNEILE-KMEKIGSMEHPPQEIVDLVHKEFL 140
NV T A +F ++ + +L +++I E P + +L+ K FL
Sbjct: 231 ----NVVSSTGEAKLLLFDSICSEIIGEYATSVLNGSVDEIEDPEDLPDSVKNLISKTFL 286
Query: 141 FK--VEGKNIC-GRWFNPAFNVL---KICKDQQIISKFKTKTITKSVKRNTPPPILE--- 191
F VE NI G+ VL + ++Q + + V + P +LE
Sbjct: 287 FLVWVEKDNISDGKEIYKVSKVLLKDGLLEEQLLEDSTEHVNHASIVSGDKVPLMLENGN 346
Query: 192 ---DSNTPIKRRSDEGSITEVVGSNDAEMFKKI 221
DS TP +R E GS + KK+
Sbjct: 347 GSPDSTTPSSKRV---YARETSGSEGSSSSKKV 376
>At1g36510 hypothetical protein
Length = 351
Score = 35.8 bits (81), Expect = 0.019
Identities = 48/196 (24%), Positives = 76/196 (38%), Gaps = 29/196 (14%)
Query: 34 IIYATIKKIESTNDWYYHDCECNSAAYSDGAQYYCEKCNKHVTPTIRYKLLLNVADDTAS 93
+ A + IE+ WYY C+ + A G C C+ T L+ A AS
Sbjct: 157 VTIARVCFIETMPKWYYIACKAHGDA---GPICSCGVCDADATD------LILRASYGAS 207
Query: 94 ATFTVFYREGCY--LMNKKGNEILEKMEKIGSMEHPPQEIVDLVHKEFLFKVEGKNICGR 151
+ + +G L+ K E ++ K P+ + DL+ K LFK+
Sbjct: 208 PEVKLLFFDGLAQCLIGKTAAEFFAEVPKESDPSILPEVLADLMGKTMLFKL-------- 259
Query: 152 WFNPAFNVLKICKDQQIISKF--KTKTITKSVKRNTPPPILEDSNTPI---KRRSDEGSI 206
+ + LK K ++ KF KT + K K T ++ DS I K E S+
Sbjct: 260 --SIGTDNLKSTKAAYVVEKFWEKTDMVEKFAKELT---VVSDSACDIINPKIEMIESSV 314
Query: 207 TEVVGSNDAEMFKKIK 222
+ S D +F +K
Sbjct: 315 VDDSPSTDKRIFDDVK 330
>At3g31990 hypothetical protein
Length = 450
Score = 35.0 bits (79), Expect = 0.032
Identities = 43/210 (20%), Positives = 77/210 (36%), Gaps = 54/210 (25%)
Query: 32 IFIIYATIKKIESTNDWYYHDCECNSAAYSDGAQYYCEKCNKHVTPTI------------ 79
+F + TI I+ WYY C+KCNK VT +
Sbjct: 221 MFDVLCTIYDIDRDWSWYY---------------IACKKCNKKVTKVVTTSLKAQFWCET 265
Query: 80 ----------RYKLLLNVADDTASATFTVFYREGCYLMNKKGNEILE-KMEKIGSMEHPP 128
RYKL + V D + +F ++ N +L+ ++I ++ P
Sbjct: 266 CRAPVNNVLARYKLHVKVMDSSGEMKLMLFDAMASEIVGCPANNLLDGSFDEIEDPDNIP 325
Query: 129 QEIVDLVHK--EFLFKVEGKNICGRWFNPAFNVLKICKDQQIISKFKTKTITKSVKRNTP 186
I +L+ K ++L VE +N+ +N L K ++++ + P
Sbjct: 326 GSIKNLIGKTYQYLVCVENENV--------WNGLDTYKVSRVLA------YGGLGEEEIP 371
Query: 187 PPILEDSNTPIKRRSDEGSITEVVGSNDAE 216
++ + TP +RS E I S+ +
Sbjct: 372 DDSIDQATTPSSKRSGETCILPADNSSSTK 401
>At5g09850 unknown protein
Length = 353
Score = 33.9 bits (76), Expect = 0.072
Identities = 29/118 (24%), Positives = 58/118 (48%), Gaps = 17/118 (14%)
Query: 106 LMNKKGNEILEKMEKIGSMEHPPQEIVDLVHK----EFLFKVEGKNICGRWFN-----PA 156
L + + I+E EK+ + + +V+L+ + F+ + GR N P+
Sbjct: 119 LFDDEQKSIVEIKEKLEDPDLSEESLVELLQNLEDMDITFQALQETDIGRHVNRVRKHPS 178
Query: 157 FNVLKICKDQQIISKFKTKTITKSVKRNTP-----PPILEDSNTPIKRRSDEGSITEV 209
NV ++ K Q++ K+K +T+ + VK N P P ++ D ++P+++ GS +V
Sbjct: 179 NNVRRLAK--QLVKKWK-ETVDEWVKFNQPGDLEPPSLIADEDSPVQKALHNGSRQQV 233
>At5g08020 replication factor A - like protein
Length = 604
Score = 31.6 bits (70), Expect = 0.36
Identities = 23/119 (19%), Positives = 50/119 (41%), Gaps = 3/119 (2%)
Query: 28 QQRTIFIIYATIKKIESTNDWYYHDCE-CNS-AAYSDGAQYYCEKCNKHVTP-TIRYKLL 84
++ F A I I+ +Y C+ CN + + Y+CE C K ++RY +
Sbjct: 443 EKPVFFSTRAYISFIKPDQTMWYRACKTCNKKVTEAMDSGYWCESCQKKDQECSLRYIMA 502
Query: 85 LNVADDTASATFTVFYREGCYLMNKKGNEILEKMEKIGSMEHPPQEIVDLVHKEFLFKV 143
+ V+D T + F E ++ +++ + + G + ++ + LF++
Sbjct: 503 VKVSDSTGETWLSAFNDEAEKIIGCTADDLNDLKSEEGEVNEFQTKLKEATWSSHLFRI 561
>At2g07630 putative replication protein A1
Length = 458
Score = 31.2 bits (69), Expect = 0.47
Identities = 31/138 (22%), Positives = 50/138 (35%), Gaps = 40/138 (28%)
Query: 35 IYATIKKIESTNDWYYHDCECNSAAYSDGAQYYCEKCNKHVTPTI--------------- 79
+ TI I+ WYY C+KCNK VT +
Sbjct: 256 VLCTIYDIDRDWSWYY---------------IVCKKCNKKVTKVVMTSLKAQLWCETCRA 300
Query: 80 -------RYKLLLNVADDTASATFTVFYREGCYLMNKKGNEILE-KMEKIGSMEHPPQEI 131
RYKL + V D + +F ++ N +L+ ++I + P I
Sbjct: 301 PITNVLARYKLHVKVMDSSGEMKLMLFDAMSSEIVGCPANNLLDGSFDEIEDPDILPDSI 360
Query: 132 VDLVHK--EFLFKVEGKN 147
+L+ K ++L VE +N
Sbjct: 361 KNLIGKTYQYLVCVENEN 378
>At2g06510 replication protein A1 like
Length = 640
Score = 31.2 bits (69), Expect = 0.47
Identities = 20/97 (20%), Positives = 41/97 (41%), Gaps = 2/97 (2%)
Query: 52 DCECNSAAYSDGA-QYYCEKCNKHVTPT-IRYKLLLNVADDTASATFTVFYREGCYLMNK 109
D +CN G ++ C++CN+ RY L + + D T T F G +M
Sbjct: 509 DKQCNKKVTRSGTNRWLCDRCNQESDECDYRYLLQVQIQDHTGLTWITAFQETGEEIMGC 568
Query: 110 KGNEILEKMEKIGSMEHPPQEIVDLVHKEFLFKVEGK 146
++ ++ E + + D + +++ K++ K
Sbjct: 569 PAKKLYAMKYELEKEEEFAEIVRDRLFHQYMLKLKIK 605
>At4g21420 hypothetical protein
Length = 229
Score = 28.9 bits (63), Expect = 2.3
Identities = 24/107 (22%), Positives = 42/107 (38%), Gaps = 22/107 (20%)
Query: 45 TNDWYYHDCECNSAAYSDGAQYYCEKCNKHVTPTIRYKLLLNVADDTASATFTVFYREGC 104
T+ W D + S + CEK NKHV + V + + + YRE
Sbjct: 132 TDLWLEGDAKIIMDIISRRGRLRCEKTNKHV------NYIKVVMPELNNCVLSHVYRE-- 183
Query: 105 YLMNKKGNEILEKMEKIGSM--------EHPPQEIVDLVHKEFLFKV 143
GN + +K+ K+G PP+ ++ ++H + K+
Sbjct: 184 ------GNRVADKLAKLGHQFQDPKVWRVQPPEIVLPIMHDDAKGKI 224
>At5g04850 unknown protein
Length = 235
Score = 28.1 bits (61), Expect = 3.9
Identities = 16/45 (35%), Positives = 24/45 (52%), Gaps = 2/45 (4%)
Query: 175 KTITKSVKRNTPPPILEDSNTPIKRRSDEGSITEVVGSNDAEMFK 219
K I + PPP ++D++ I +R D S+ E V DAE+ K
Sbjct: 2 KRIFGAKNNKEPPPSIQDASDRINKRGD--SVEEKVKRLDAELCK 44
>At4g18860 hypothetical protein
Length = 97
Score = 28.1 bits (61), Expect = 3.9
Identities = 21/68 (30%), Positives = 33/68 (47%), Gaps = 5/68 (7%)
Query: 156 AFNVLKICKDQQIISKFKTKTITKSVKRNTPPPILE-DSNTPIK----RRSDEGSITEVV 210
A N+LK C D I+K +S N PP L DS++ + S+ G+ T V+
Sbjct: 21 AENLLKCCIDVMTITKHNLALQFRSFITNYPPKALVFDSSSAVSDLGFTESETGASTRVL 80
Query: 211 GSNDAEMF 218
G + + +F
Sbjct: 81 GKSASWIF 88
>At5g38500 putative protein
Length = 411
Score = 27.7 bits (60), Expect = 5.1
Identities = 27/143 (18%), Positives = 61/143 (41%), Gaps = 21/143 (14%)
Query: 80 RYKLLLNVADDTASATFTVFYREGC------YLMNKKGNEILEKMEKIGSMEHPPQEIVD 133
R +LL +D+ S TV + ++ KK + ++++ +K + + D
Sbjct: 28 RRRLLAEKREDSKSQKKTVSEEDDSEKRFLSHVPRKKRSSLVKRQQKPNGVSTSSSSLPD 87
Query: 134 LVHKEFLFKVEGKNICGRWFNPAFNVLKICKDQQIISKFKTKTITKSVKRNTPPPILEDS 193
L ++ E K NP+F +C ++Q + K K++ I + E++
Sbjct: 88 LNQIPIDYETETKQ------NPSFIERLVCDEEQRVKKGKSRIIWEEE---------EEA 132
Query: 194 NTPIKRRSDEGSITEVVGSNDAE 216
+ ++R E ++ + VG + +
Sbjct: 133 DEDSEKRLFEKNLMKFVGHSQQQ 155
>At3g21930 hypothetical protein
Length = 300
Score = 27.7 bits (60), Expect = 5.1
Identities = 9/36 (25%), Positives = 17/36 (47%)
Query: 31 TIFIIYATIKKIESTNDWYYHDCECNSAAYSDGAQY 66
T ++ + + TN + +H C N Y G++Y
Sbjct: 44 TQLLLVRNVSSLNLTNSYLHHKCVVNQGKYKPGSKY 79
>At3g21920 hypothetical protein
Length = 278
Score = 27.7 bits (60), Expect = 5.1
Identities = 9/36 (25%), Positives = 17/36 (47%)
Query: 31 TIFIIYATIKKIESTNDWYYHDCECNSAAYSDGAQY 66
T ++ + + TN + +H C N Y G++Y
Sbjct: 22 TQLLLVRNVSSLNLTNSYLHHKCVVNQGKYKPGSKY 57
>At1g63330 unknown protein
Length = 558
Score = 27.7 bits (60), Expect = 5.1
Identities = 22/111 (19%), Positives = 44/111 (38%), Gaps = 26/111 (23%)
Query: 1 MVKGKCLYREVQEQCTILKSLPTK-VQPQQRTIFIIYATIKKIESTNDWYYHDCECNSAA 59
++ C YR V + + K + TK ++P + Y+++ C C+
Sbjct: 190 IIDSLCKYRHVDDALNLFKEMETKGIRPN----VVTYSSLIS-----------CLCSYGR 234
Query: 60 YSDGAQYYCEKCNKHVTPTIRYKLLLNVADDTASATFTVFYREGCYLMNKK 110
+SD +Q + K + P + T +A F +EG ++ +K
Sbjct: 235 WSDASQLLSDMIEKKINPNL----------VTFNALIDAFVKEGKFVEAEK 275
>At1g62590 putative membrane-associated salt-inducible protein
(AL021637); similar to EST gb|AA728420
Length = 634
Score = 27.7 bits (60), Expect = 5.1
Identities = 22/111 (19%), Positives = 44/111 (38%), Gaps = 26/111 (23%)
Query: 1 MVKGKCLYREVQEQCTILKSLPTK-VQPQQRTIFIIYATIKKIESTNDWYYHDCECNSAA 59
++ C YR V + + K + TK ++P + Y+++ C C+
Sbjct: 266 IIDSLCKYRHVDDALNLFKEMETKGIRPN----VVTYSSLIS-----------CLCSYGR 310
Query: 60 YSDGAQYYCEKCNKHVTPTIRYKLLLNVADDTASATFTVFYREGCYLMNKK 110
+SD +Q + K + P + T +A F +EG ++ +K
Sbjct: 311 WSDASQLLSDMIEKKINPNL----------VTFNALIDAFVKEGKFVEAEK 351
>At1g30040 unknown protein
Length = 341
Score = 27.7 bits (60), Expect = 5.1
Identities = 28/110 (25%), Positives = 43/110 (38%), Gaps = 17/110 (15%)
Query: 57 SAAYSDGAQYYCEKCNKHVTPT--IRYKLLLNVAD-------DTASATFTVFYREGCYLM 107
SA + Q + E +++ + YK+L VA+ DT S + C +
Sbjct: 127 SAVFRQTPQIFRESVEEYMKEIKEVSYKVLEMVAEELGIEPRDTLSKMLRDEKSDSCLRL 186
Query: 108 NK--KGNEILEKMEKIGSMEHPPQEIVDLVHKEFLFKVEGKNIC---GRW 152
N E EKM K+G EH +I+ ++ G IC G W
Sbjct: 187 NHYPAAEEEAEKMVKVGFGEHTDPQIISVLRSN---NTAGLQICVKDGSW 233
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,333,210
Number of Sequences: 26719
Number of extensions: 221532
Number of successful extensions: 741
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 732
Number of HSP's gapped (non-prelim): 31
length of query: 226
length of database: 11,318,596
effective HSP length: 96
effective length of query: 130
effective length of database: 8,753,572
effective search space: 1137964360
effective search space used: 1137964360
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0051a.1


