

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0050.4
(236 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
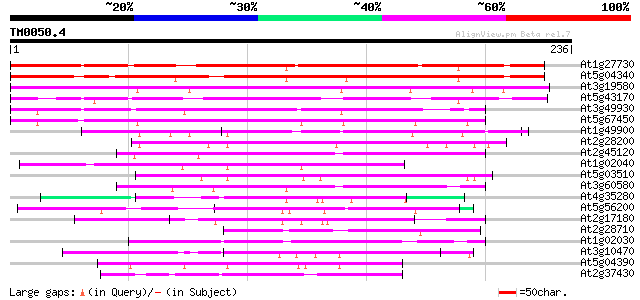
Score E
Sequences producing significant alignments: (bits) Value
At1g27730 salt-tolerance zinc finger protein like 226 8e-60
At5g04340 putative c2h2 zinc finger transcription factor 197 4e-51
At3g19580 zinc finger protein, putative 188 2e-48
At5g43170 Cys2/His2-type zinc finger protein 3 (dbj|BAA85109.1) 177 4e-45
At3g49930 zinc-finger-like protein 162 1e-40
At5g67450 Cys2/His2-type zinc finger protein 1 (dbj|BAA85108.1) 160 7e-40
At1g49900 Cys2/His2-type zinc finger protein, putative 100 5e-22
At2g28200 putative zinc-finger protein 100 9e-22
At2g45120 putative C2H2-type zinc finger protein 93 1e-19
At1g02040 hypothetical protein 91 5e-19
At5g03510 unknown protein 90 9e-19
At3g60580 zinc finger protein - like 88 3e-18
At4g35280 putative zinc-finger protein 87 1e-17
At5g56200 unknown protein 82 2e-16
At2g17180 putative C2H2-type zinc finger protein 81 4e-16
At2g28710 putative C2H2-type zinc finger protein 81 6e-16
At1g02030 unknown protein 79 2e-15
At3g10470 unknown protein 79 2e-15
At5g04390 zinc finger transcription factor-like protein 75 4e-14
At2g37430 putative C2H2-type zinc finger protein 74 9e-14
>At1g27730 salt-tolerance zinc finger protein like
Length = 227
Score = 226 bits (576), Expect = 8e-60
Identities = 127/233 (54%), Positives = 156/233 (66%), Gaps = 23/233 (9%)
Query: 1 MAMEALKSPTTAAPTFTPFQEPNHSYMVDAPWAKRKRSKRSRTDSHHNHASCTEEEYLAL 60
MA+EAL SP A+P F++ + + V+ W K KRSKRSR+D HH + TEEEYLA
Sbjct: 1 MALEALTSPRLASPIPPLFEDSSVFHGVEH-WTKGKRSKRSRSDFHHQNL--TEEEYLAF 57
Query: 61 CLIMLARGSTAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHRK----- 115
CL++LAR + P P EKLSYKCSVC+KTF SYQALGGHKASHRK
Sbjct: 58 CLMLLARDNR--------QPPPPPAVEKLSYKCSVCDKTFSSYQALGGHKASHRKNLSQT 109
Query: 116 LAGAAAEDHSTSSAVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCHYEGGGGASST 175
L+G +DHSTSSA TTS+ + G GK H C+IC KSFP+GQALGGHKRCHYEG +T
Sbjct: 110 LSGGG-DDHSTSSATTTSAVTTGSGKSHVCTICNKSFPSGQALGGHKRCHYEGNNNI-NT 167
Query: 176 ATATASEGVGST---HSHQRNFDLNLPAFPDFSASKFFVEEEVSSPLPSKKPR 225
++ + SEG GST S R FDLN+P P+FS ++EV SP+P+KKPR
Sbjct: 168 SSVSNSEGAGSTSHVSSSHRGFDLNIPPIPEFSMVN--GDDEVMSPMPAKKPR 218
>At5g04340 putative c2h2 zinc finger transcription factor
Length = 238
Score = 197 bits (501), Expect = 4e-51
Identities = 121/245 (49%), Positives = 149/245 (60%), Gaps = 34/245 (13%)
Query: 1 MAMEALKSPTTAAPTFTPFQEPNHSYMVDAPWAKRKRSKRSRTDSHHNHASCTEEEYLAL 60
MA+E L SP ++P T FQ+ + +K KRSKRSR S + S TE+EY+AL
Sbjct: 1 MALETLTSPRLSSPMPTLFQDSALGFH----GSKGKRSKRSR--SEFDRQSLTEDEYIAL 54
Query: 61 CLIMLARG----------STAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHK 110
CL++LAR S++ +P L P P+ YKCSVC+K F SYQALGGHK
Sbjct: 55 CLMLLARDGDRNRDLDLPSSSSSPPLLPPLPTPI------YKCSVCDKAFSSYQALGGHK 108
Query: 111 ASHRK----LAGAAAEDHSTSSAVTTSSASNGGG---KVHQCSICQKSFPTGQALGGHKR 163
ASHRK A ++ STSSA+TTS S GGG K H CSIC KSF TGQALGGHKR
Sbjct: 109 ASHRKSFSLTQSAGGDELSTSSAITTSGISGGGGGSVKSHVCSICHKSFATGQALGGHKR 168
Query: 164 CHYEGGGGASSTATATASEGVGST---HSHQRNFDLNLPAFPDFSASKFFVEEEVSSPLP 220
CHYEG G +++ + SE VGST S R FDLN+P P+FS +EEV SP+P
Sbjct: 169 CHYEGKNGGGVSSSVSNSEDVGSTSHVSSGHRGFDLNIPPIPEFSMVN--GDEEVMSPMP 226
Query: 221 SKKPR 225
+KK R
Sbjct: 227 AKKLR 231
>At3g19580 zinc finger protein, putative
Length = 273
Score = 188 bits (478), Expect = 2e-48
Identities = 114/257 (44%), Positives = 145/257 (56%), Gaps = 30/257 (11%)
Query: 1 MAMEALKSPTTAAPTFTPFQEPNHSYMVDAPWAKRKRSKRSRTDSHHNHASC-------- 52
MA+EA+ +PT++ ++ + + PW KRKRSKR R+ S + +S
Sbjct: 1 MALEAMNTPTSSFTRIETKEDLMNDAVFIEPWLKRKRSKRQRSHSPSSSSSSPPRSRPKS 60
Query: 53 -----TEEEYLALCLIMLARGSTAVTP--KLTLSRPAPVTAEKLSYKCSVCEKTFPSYQA 105
TEEEYLALCL+MLA+ + T + + S P ++ L YKC+VCEK FPSYQA
Sbjct: 61 QNQDLTEEEYLALCLLMLAKDQPSQTRFHQQSQSLTPPPESKNLPYKCNVCEKAFPSYQA 120
Query: 106 LGGHKASHRKLAGAAAEDHSTSSAVTTSSASNG-------GGKVHQCSICQKSFPTGQAL 158
LGGHKASHR + S T S G GK+H+CSIC K FPTGQAL
Sbjct: 121 LGGHKASHRIKPPTVISTTADDSTAPTISIVAGEKHPIAASGKIHECSICHKVFPTGQAL 180
Query: 159 GGHKRCHYE---GGGGASSTATATASEGVGSTHSHQRN----FDLNLPAFPDFSA-SKFF 210
GGHKRCHYE GGGG + + + S V ST S +R+ DLNLPA P+ S
Sbjct: 181 GGHKRCHYEGNLGGGGGGGSKSISHSGSVSSTVSEERSHRGFIDLNLPALPELSLHHNPI 240
Query: 211 VEEEVSSPLPSKKPRLL 227
V+EE+ SPL KKP LL
Sbjct: 241 VDEEILSPLTGKKPLLL 257
>At5g43170 Cys2/His2-type zinc finger protein 3 (dbj|BAA85109.1)
Length = 193
Score = 177 bits (449), Expect = 4e-45
Identities = 113/231 (48%), Positives = 131/231 (55%), Gaps = 44/231 (19%)
Query: 1 MAMEALKSPTTAAPTFTPFQEPNHSYMVDAPWAK-RKRSKRSRTDSHHNHASCTEEEYLA 59
MA+EAL SP ++P V+ W K +KRSKRSR+D HHNH TEEEYLA
Sbjct: 1 MALEALNSPRLV-------EDPLRFNGVEQ-WTKCKKRSKRSRSDLHHNHR-LTEEEYLA 51
Query: 60 LCLIMLARGSTAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHRKLAGA 119
CL++LAR + + AEK SYKC VC KTF SYQALGGHKASHR L G
Sbjct: 52 FCLMLLARDGGDLD--------SVTVAEKPSYKCGVCYKTFSSYQALGGHKASHRSLYGG 103
Query: 120 AAEDHSTSSAVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCHYEGGGGASSTATAT 179
D ST S K H CS+C KSF TGQALGGHKRCHY+GG +
Sbjct: 104 GENDKSTPSTAV---------KSHVCSVCGKSFATGQALGGHKRCHYDGG--------VS 146
Query: 180 ASEGVGST----HSHQRNFDLNLPAFPDFSASKFFVEEEVSSPLPSKKPRL 226
SEGVGST S R FDLN+ FS ++EV SP+ +KKPRL
Sbjct: 147 NSEGVGSTSHVSSSSHRGFDLNIIPVQGFSP-----DDEVMSPMATKKPRL 192
>At3g49930 zinc-finger-like protein
Length = 215
Score = 162 bits (411), Expect = 1e-40
Identities = 99/210 (47%), Positives = 124/210 (58%), Gaps = 27/210 (12%)
Query: 1 MAMEALKSPT-----TAAPTFTPFQEPNHSYMVDAPWAKRKRSKRSRTDSHHNHASCTEE 55
MA++ L SPT TA P F + +++ W KRKR+KR R D + S EE
Sbjct: 1 MALDTLNSPTSTTTTTAPPPFLRCLDETEPENLES-WTKRKRTKRHRIDQPNPPPS--EE 57
Query: 56 EYLALCLIMLARGSTAV-TPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHR 114
EYLALCL+MLARGS+ +P +P++ + YKCSVC K+FPSYQALGGHK SHR
Sbjct: 58 EYLALCLLMLARGSSDHHSPPSDHHSLSPLSDHQKDYKCSVCGKSFPSYQALGGHKTSHR 117
Query: 115 KLAGAAAEDHSTSSAVTTSSASNG----GGKVHQCSICQKSFPTGQALGGHKRCHYEGGG 170
K ++S + + SNG GK H CSIC KSFP+GQALGGHKRCHY+GG
Sbjct: 118 KPVSVDV-NNSNGTVTNNGNISNGLVGQSGKTHNCSICFKSFPSGQALGGHKRCHYDGGN 176
Query: 171 GASSTATATASEGVGSTHSHQRNFDLNLPA 200
G S+ +SH+ FDLNLPA
Sbjct: 177 GNSN-----------GDNSHK--FDLNLPA 193
>At5g67450 Cys2/His2-type zinc finger protein 1 (dbj|BAA85108.1)
Length = 245
Score = 160 bits (404), Expect = 7e-40
Identities = 101/239 (42%), Positives = 124/239 (51%), Gaps = 41/239 (17%)
Query: 1 MAMEALKSPT---TAAPTFTPFQEPNHSYMVDAPWAKRKRSKRSRTDSHHNHASC----- 52
MA+E L SPT TA P +E + WAKRKR+KR R D H +
Sbjct: 1 MALETLNSPTATTTARPLLRYREEMEPENLEQ--WAKRKRTKRQRFDHGHQNQETNKNLP 58
Query: 53 TEEEYLALCLIMLARGSTAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKAS 112
+EEEYLALCL+MLARGS +P L ++ YKC+VC K+F SYQALGGHK S
Sbjct: 59 SEEEYLALCLLMLARGSAVQSPPLPPLPSRASPSDHRDYKCTVCGKSFSSYQALGGHKTS 118
Query: 113 HRKLAGAAA---------EDHSTSSAVTTSSASNGG------GKVHQCSICQKSFPTGQA 157
HRK + HS S +V + N G GK+H CSIC KSF +GQA
Sbjct: 119 HRKPTNTSITSGNQELSNNSHSNSGSVVINVTVNTGNGVSQSGKIHTCSICFKSFASGQA 178
Query: 158 LGGHKRCHYEGG------GGASSTATATASEGVGSTHSHQ----------RNFDLNLPA 200
LGGHKRCHY+GG G +S++ A V + + R FDLNLPA
Sbjct: 179 LGGHKRCHYDGGNNGNGNGSSSNSVELVAGSDVSDVDNERWSEESAIGGHRGFDLNLPA 237
>At1g49900 Cys2/His2-type zinc finger protein, putative
Length = 917
Score = 100 bits (250), Expect = 5e-22
Identities = 76/227 (33%), Positives = 96/227 (41%), Gaps = 42/227 (18%)
Query: 31 PWAKRKRSKRSRTDSHHNHASCT---EEEYLALCLIMLA------RGSTAVTP-----KL 76
PW R KR R + +S + E + AL L+ + + T P K
Sbjct: 111 PWLDSFRLKRQRLGDDEDPSSSSSPSEVDIAALTLLQFSCDRRHPQTQTLTRPQPQTHKT 170
Query: 77 TLSRPAPVTAEKLS--------YKCSVCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSS 128
L RP P + +KCS+CEK F SYQALGGHKASH A + +
Sbjct: 171 QLQRPPPQLQSQTQTAPPKSDLFKCSICEKVFTSYQALGGHKASHSIKAAQLENAGADAG 230
Query: 129 AVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCHYEG--GGGASSTATATASEGVGS 186
T S + GK+H+C IC FPTGQALGGHKR HYEG GG A S
Sbjct: 231 EKTRSKMLSPSGKIHKCDICHVLFPTGQALGGHKRRHYEGLLGGHKRGNDEAVLKLSPNS 290
Query: 187 THS------------------HQRNFDLNLPAFPDFSASKFFVEEEV 215
S H+RN D +P+ +S S V + V
Sbjct: 291 NKSVVTKVLDAEQSLRASDNIHKRNQDEVVPSRDKWSLSNVSVVKNV 337
Score = 88.6 bits (218), Expect = 3e-18
Identities = 53/131 (40%), Positives = 71/131 (53%), Gaps = 7/131 (5%)
Query: 90 SYKCSVCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTTSSASNGGGKVHQCSICQ 149
SY+C+VC + PSYQALGGHKASHR +++T + + G K+H+CSIC
Sbjct: 749 SYQCNVCGRELPSYQALGGHKASHRTKPPV---ENATGEKMRPKKLAPSG-KIHKCSICH 804
Query: 150 KSFPTGQALGGHKRCHYEG--GGGASSTATATASEGVGSTHSHQRNFDLNLPAFPDFSAS 207
+ F TGQ+LGGHKR HYEG G S S+G + S + ++P P S
Sbjct: 805 REFSTGQSLGGHKRLHYEGVLRGHKRSQEKEAVSQGDKLSPSGNGSVVTHVPD-PKQSRK 863
Query: 208 KFFVEEEVSSP 218
V +V SP
Sbjct: 864 GLIVINKVPSP 874
Score = 37.0 bits (84), Expect = 0.009
Identities = 22/68 (32%), Positives = 32/68 (46%), Gaps = 4/68 (5%)
Query: 80 RPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHRK--LAG--AAAEDHSTSSAVTTSSA 135
RP + +KCS+C + F + Q+LGGHK H + L G + E + S S +
Sbjct: 787 RPKKLAPSGKIHKCSICHREFSTGQSLGGHKRLHYEGVLRGHKRSQEKEAVSQGDKLSPS 846
Query: 136 SNGGGKVH 143
NG H
Sbjct: 847 GNGSVVTH 854
>At2g28200 putative zinc-finger protein
Length = 284
Score = 100 bits (248), Expect = 9e-22
Identities = 77/201 (38%), Positives = 95/201 (46%), Gaps = 43/201 (21%)
Query: 52 CT-EEEYLALCLIMLARGSTAVTPKLTLSRPA--PVTAEKLS---YKCSVCEKTFPSYQA 105
CT EEE +A+CLIMLARG+ +P L SR +++E S Y+C C +TF S+QA
Sbjct: 68 CTQEEEDMAICLIMLARGTVLPSPDLKNSRKIHQKISSENSSFYVYECKTCNRTFSSFQA 127
Query: 106 LGGHKASHRKLAGAAAEDHSTSSAVTTSSAS-----------------------NGGGKV 142
LGGH+ASH+K + E SSAS N KV
Sbjct: 128 LGGHRASHKKPRTSTEEKTRLPLTQPKSSASEEGQNSHFKVSGSALASQASNIINKANKV 187
Query: 143 HQCSICQKSFPTGQALGGHKRCHYEGGGGASS-TATATASE---------GVGSTHSHQR 192
H+CSIC F +GQALGGH R H S ATA S +G + QR
Sbjct: 188 HECSICGSEFTSGQALGGHMRRHRTAVTTISPVAATAEVSRNSTEEEIEINIGRSMEQQR 247
Query: 193 NF---DLNLPA-FPDFSASKF 209
+ DLNLPA D SKF
Sbjct: 248 KYLPLDLNLPAPEDDLRESKF 268
>At2g45120 putative C2H2-type zinc finger protein
Length = 314
Score = 93.2 bits (230), Expect = 1e-19
Identities = 60/166 (36%), Positives = 75/166 (45%), Gaps = 14/166 (8%)
Query: 46 HHNHAS-CTEEEYLALCLIMLARGSTAVTPKLTL----------SRPAPVTAEKLSYKCS 94
HH+ AS T EE LA CLIML+R K S + + +KC
Sbjct: 138 HHSSASDTTTEEDLAFCLIMLSRDKWKQQKKKKQRVEEDETDHDSEDYKSSKSRGRFKCE 197
Query: 95 VCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTTSSASNGGGKVHQCSICQKSFPT 154
C K F SYQALGGH+ASH+K + + KVH+C IC + F +
Sbjct: 198 TCGKVFKSYQALGGHRASHKKNKACMTKTEQVETEYVLGVKEK---KVHECPICFRVFTS 254
Query: 155 GQALGGHKRCHYEGGGGASSTATATASEGVGSTHSHQRNFDLNLPA 200
GQALGGHKR H G + + + QR DLNLPA
Sbjct: 255 GQALGGHKRSHGSNIGAGRGLSVSQIVQIEEEVSVKQRMIDLNLPA 300
Score = 33.9 bits (76), Expect = 0.077
Identities = 17/42 (40%), Positives = 20/42 (47%)
Query: 143 HQCSICQKSFPTGQALGGHKRCHYEGGGGASSTATATASEGV 184
++C C KSF G+ALGGH R H T A E V
Sbjct: 4 YKCRFCFKSFINGRALGGHMRSHMLTLSAERCVITGEAEEEV 45
Score = 32.0 bits (71), Expect = 0.29
Identities = 13/23 (56%), Positives = 16/23 (69%)
Query: 91 YKCSVCEKTFPSYQALGGHKASH 113
YKC C K+F + +ALGGH SH
Sbjct: 4 YKCRFCFKSFINGRALGGHMRSH 26
>At1g02040 hypothetical protein
Length = 324
Score = 90.9 bits (224), Expect = 5e-19
Identities = 57/191 (29%), Positives = 87/191 (44%), Gaps = 32/191 (16%)
Query: 5 ALKSPTTAAPTFTPFQEPNHSYMVDAPWAKRKRSKRSRTDSHHNHASCTEEEYLALCLIM 64
A+ SP F+ Q + PW KR + T HH + S E+E LA CL++
Sbjct: 61 AMNSPCIDFVMFSRGQHDEDNMSRRPPW---KRERSMSTQHHHLNLSPNEDEELANCLVL 117
Query: 65 LARGSTA----VTPKLTLSRPAPVTAEKLS--YKCSVCEKTFPSYQALGGHKASHRKLAG 118
L+ A + + V +K + ++C C+K F S+QALGGH+ASH+K+ G
Sbjct: 118 LSNSGDAHGGDQHKQHGHGKGKTVKKQKTAQVFQCKACKKVFTSHQALGGHRASHKKVKG 177
Query: 119 AAA-----------------------EDHSTSSAVTTSSASNGGGKVHQCSICQKSFPTG 155
A ++ +T+ + H+C+IC + F +G
Sbjct: 178 CFASQDKEEEEEEEYKEDDDDNDEDEDEEEDEEDKSTAHIARKRSNAHECTICHRVFSSG 237
Query: 156 QALGGHKRCHY 166
QALGGHKRCH+
Sbjct: 238 QALGGHKRCHW 248
>At5g03510 unknown protein
Length = 292
Score = 90.1 bits (222), Expect = 9e-19
Identities = 68/192 (35%), Positives = 94/192 (48%), Gaps = 42/192 (21%)
Query: 54 EEEYLALCLIMLARGSTAVTPKLTLS--------RPAPVTAEKLS----YKCSVCEKTFP 101
E+E +A CLI+L++G A + LS PV + L Y+C C+K+F
Sbjct: 69 EDEDMANCLILLSQGHQAKSSDDHLSMQRMGFFSNKKPVASLGLGLDGVYQCKTCDKSFH 128
Query: 102 SYQALGGHKASHRKLAGAAAE----DHSTSSA------------------VTTSSASNGG 139
S+QALGGH+ASH+K A+ + T+SA VT+S S
Sbjct: 129 SFQALGGHRASHKKPKLGASVFKCVEKKTASASTVETVEAGAVGSFLSLQVTSSDGSKKP 188
Query: 140 GKVHQCSICQKSFPTGQALGGHKRCHYEGGGGASSTATATASEGVGSTHSHQ-----RNF 194
K H+CSIC+ F +GQALGGH R H A++T+ + S H H+ +NF
Sbjct: 189 EKTHECSICKAEFSSGQALGGHMRRHRGLTINANATSAIKTAISSSSHHHHEESIRPKNF 248
Query: 195 ---DLNLPAFPD 203
DLNLPA D
Sbjct: 249 LQLDLNLPAPED 260
>At3g60580 zinc finger protein - like
Length = 288
Score = 88.2 bits (217), Expect = 3e-18
Identities = 59/170 (34%), Positives = 80/170 (46%), Gaps = 23/170 (13%)
Query: 46 HHNHASCTEEEYLALCLIMLAR-------GSTAVTPKLTLSRPAPV------TAEKLSYK 92
H + + T EE LA CL+ML+R + V ++ + K YK
Sbjct: 115 HSSASDTTTEEDLAFCLMMLSRDKWKKNKSNKEVVEEIETEEESEGYNKINRATTKGRYK 174
Query: 93 CSVCEKTFPSYQALGGHKASHRK--LAGAAAEDHSTSSAVTTSSASNGGGKVHQCSICQK 150
C C K F SYQALGGH+ASH+K ++ E S + + ++H+C IC +
Sbjct: 175 CETCGKVFKSYQALGGHRASHKKNRVSNNKTEQRSETEYDNVVVVAK---RIHECPICLR 231
Query: 151 SFPTGQALGGHKRCHYEGGGGASSTATATASEGVGSTHSHQRNFDLNLPA 200
F +GQALGGHKR H G + +E V QR DLNLPA
Sbjct: 232 VFASGQALGGHKRSHGVGNLSVNQQRRVHRNESV-----KQRMIDLNLPA 276
Score = 38.1 bits (87), Expect = 0.004
Identities = 21/52 (40%), Positives = 27/52 (51%)
Query: 90 SYKCSVCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTTSSASNGGGK 141
SYKC VC K+F + +ALGGH SH + + S S T S S+ K
Sbjct: 3 SYKCRVCFKSFVNGKALGGHMRSHMSNSHEEEQRPSQLSYETESDVSSSDPK 54
Score = 35.0 bits (79), Expect = 0.035
Identities = 13/23 (56%), Positives = 17/23 (73%)
Query: 143 HQCSICQKSFPTGQALGGHKRCH 165
++C +C KSF G+ALGGH R H
Sbjct: 4 YKCRVCFKSFVNGKALGGHMRSH 26
>At4g35280 putative zinc-finger protein
Length = 284
Score = 86.7 bits (213), Expect = 1e-17
Identities = 48/122 (39%), Positives = 72/122 (58%), Gaps = 21/122 (17%)
Query: 54 EEEYLALCLIMLARGSTAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASH 113
E+ +A CL+ML+ G+ P+ + E+ ++C C+K F S+QALGGH+ASH
Sbjct: 138 EDHEVASCLLMLSNGT-----------PSSSSIER--FECGGCKKVFGSHQALGGHRASH 184
Query: 114 RKLAGAAAEDHSTSS--AVTTSSASNGGGKV------HQCSICQKSFPTGQALGGHKRCH 165
+ + G A + T V+TSS + GK+ H+C+IC + F +GQALGGH RCH
Sbjct: 185 KNVKGCFAITNVTDDPMTVSTSSGHDHQGKILTFSGHHKCNICFRVFSSGQALGGHMRCH 244
Query: 166 YE 167
+E
Sbjct: 245 WE 246
Score = 45.8 bits (107), Expect = 2e-05
Identities = 46/213 (21%), Positives = 83/213 (38%), Gaps = 53/213 (24%)
Query: 14 PTFTPFQEPNHSYMVDAPWAKRKRSKRSRTDSHHNHASCTEEEYLALCLIMLARGSTAVT 73
P+ T PN + + + + + KR++T + + +S +A
Sbjct: 18 PSNTSNPNPNLQFALSSSYDHSPKKKRTKTVASSSSSS----------------PKSASK 61
Query: 74 PKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHRK------------------ 115
PK T +P P A K++ C+ C + F S++AL GH H +
Sbjct: 62 PKYT-KKPDP-NAPKITRPCTECGRKFWSWKALFGHMRCHPERQWRGINPPPNYRVPTAA 119
Query: 116 ------------LAGAAAEDHSTSSAV---TTSSASNGGGKVHQCSICQKSFPTGQALGG 160
++ + EDH +S + + + S+ + +C C+K F + QALGG
Sbjct: 120 SSKQLNQILPNWVSFMSEEDHEVASCLLMLSNGTPSSSSIERFECGGCKKVFGSHQALGG 179
Query: 161 HKRCH--YEGGGGASSTATATASEGVGSTHSHQ 191
H+ H +G ++ + S H HQ
Sbjct: 180 HRASHKNVKGCFAITNVTDDPMTVSTSSGHDHQ 212
>At5g56200 unknown protein
Length = 493
Score = 82.0 bits (201), Expect = 2e-16
Identities = 67/209 (32%), Positives = 92/209 (43%), Gaps = 44/209 (21%)
Query: 4 EALKSPTTAAPTFTPFQEPNHSYMVDAPWAKRKR----SKRSRTDSHHNHASCTEEEYLA 59
EA + A + + E + MV K+KR K S T +HH+H +
Sbjct: 271 EAAANLNVAETSASRSVEEKYLEMVKKRKKKQKRLSEMEKESSTSTHHDH------QLDQ 324
Query: 60 LCLIMLARGSTAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHRKL--- 116
+ ++ G A + + C C K+F SYQALGGH+ASH K+
Sbjct: 325 VVVVAAEEGGGA--------------GAREKHVCVTCNKSFSSYQALGGHRASHNKVKIL 370
Query: 117 ------AGAAAEDHSTSSAVT---------TSSASNGGGKVHQCSICQKSFPTGQALGGH 161
A A A T + +T TS +S+ G H C+IC KSF TGQALGGH
Sbjct: 371 ENHQARANAEASLLGTEAIITGLASAQGTNTSLSSSHNGD-HVCNICHKSFSTGQALGGH 429
Query: 162 KRCHYEGG-GGASSTATATASEGVGSTHS 189
KRCH+ + T TA+ V +T S
Sbjct: 430 KRCHWTAPVSTVAPTTVPTAAPTVPATAS 458
Score = 41.6 bits (96), Expect = 4e-04
Identities = 37/128 (28%), Positives = 50/128 (38%), Gaps = 24/128 (18%)
Query: 87 EKLSYKCSVCEKTFPSYQALGGHKASH----RKLAGAAAEDHSTSSAVTTSSASNGGGKV 142
E+ + C C K F S +ALGGHK H RK + S V S G
Sbjct: 75 EEKKHICCECGKRFVSGKALGGHKRIHVLETRKF--SMMRPKMVSGMV---GRSERGDLE 129
Query: 143 HQCSICQKSFPTGQALGGHKRCHYEGG---------------GGASSTATATASEGVGST 187
C +C K F + +AL GH R H + G G +SS+ E + S
Sbjct: 130 VACCVCYKKFTSMKALYGHMRFHPDRGWKGVLPPPPLLPHPLGNSSSSTLDHDDEFISSD 189
Query: 188 HSHQRNFD 195
+ +FD
Sbjct: 190 YDDDDDFD 197
Score = 36.6 bits (83), Expect = 0.012
Identities = 16/28 (57%), Positives = 18/28 (64%)
Query: 138 GGGKVHQCSICQKSFPTGQALGGHKRCH 165
G K H C C K F +G+ALGGHKR H
Sbjct: 74 GEEKKHICCECGKRFVSGKALGGHKRIH 101
Score = 34.3 bits (77), Expect = 0.059
Identities = 16/50 (32%), Positives = 25/50 (50%)
Query: 93 CSVCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTTSSASNGGGKV 142
C++C K+F + QALGGHK H + + +A T A+ +V
Sbjct: 413 CNICHKSFSTGQALGGHKRCHWTAPVSTVAPTTVPTAAPTVPATASSSQV 462
Score = 27.7 bits (60), Expect = 5.5
Identities = 17/48 (35%), Positives = 21/48 (43%), Gaps = 7/48 (14%)
Query: 73 TPKLTLSRPAPVTAE-------KLSYKCSVCEKTFPSYQALGGHKASH 113
T K ++ RP V+ L C VC K F S +AL GH H
Sbjct: 105 TRKFSMMRPKMVSGMVGRSERGDLEVACCVCYKKFTSMKALYGHMRFH 152
>At2g17180 putative C2H2-type zinc finger protein
Length = 270
Score = 81.3 bits (199), Expect = 4e-16
Identities = 52/184 (28%), Positives = 79/184 (42%), Gaps = 37/184 (20%)
Query: 28 VDAPWAKRKRSKRSRTDSHHNHASCTEEEYLALCLIMLARGSTAVTPKLTLSRPAPVTAE 87
++ P ++R + S + EE +A CL+M+A G P +
Sbjct: 95 INPPSNFKRRINSNAASSSSSWDPSEEEHNIASCLLMMANGDV----------PTRSSEV 144
Query: 88 KLSYKCSVCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTT-----------SSAS 136
+ ++C C+K F S+QALGGH+A+H+ + G A + T S
Sbjct: 145 EERFECDGCKKVFGSHQALGGHRATHKDVKGCFANKNITEDPPPPPPQEIVDQDKGKSVK 204
Query: 137 NGGGKVHQCSICQKSFPTGQALGGHKRCHYEGGGGASSTATATASEGVGSTHSHQRNFDL 196
G H+C+IC + F +GQALGGH RCH+E + R DL
Sbjct: 205 LVSGMNHRCNICSRVFSSGQALGGHMRCHWE----------------KDQEENQVRGIDL 248
Query: 197 NLPA 200
N+PA
Sbjct: 249 NVPA 252
Score = 42.0 bits (97), Expect = 3e-04
Identities = 38/139 (27%), Positives = 60/139 (42%), Gaps = 41/139 (29%)
Query: 68 GSTAVTPKLTLSRPAPVT-----AEKLSYKCSVCEKTFPSYQALGGHKASH--------- 113
GS++ +P RP PVT A +++ C+ C K F S +AL GH H
Sbjct: 42 GSSSSSP-----RPKPVTQPDPDASQIARPCTECGKQFGSLKALFGHMRCHPERQWRGIN 96
Query: 114 ------RKLAGAAA----------EDHSTSSAV------TTSSASNGGGKVHQCSICQKS 151
R++ AA E+H+ +S + + S+ + +C C+K
Sbjct: 97 PPSNFKRRINSNAASSSSSWDPSEEEHNIASCLLMMANGDVPTRSSEVEERFECDGCKKV 156
Query: 152 FPTGQALGGHKRCHYEGGG 170
F + QALGGH+ H + G
Sbjct: 157 FGSHQALGGHRATHKDVKG 175
Score = 30.0 bits (66), Expect = 1.1
Identities = 19/69 (27%), Positives = 25/69 (35%)
Query: 99 TFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTTSSASNGGGKVHQCSICQKSFPTGQAL 158
T PSY K + + S T + C+ C K F + +AL
Sbjct: 21 TLPSYNQNPRRKRTKLTNNEVGSSSSSPRPKPVTQPDPDASQIARPCTECGKQFGSLKAL 80
Query: 159 GGHKRCHYE 167
GH RCH E
Sbjct: 81 FGHMRCHPE 89
>At2g28710 putative C2H2-type zinc finger protein
Length = 156
Score = 80.9 bits (198), Expect = 6e-16
Identities = 47/114 (41%), Positives = 55/114 (48%), Gaps = 14/114 (12%)
Query: 91 YKCSVCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTTSSASNGGGKVHQCSICQK 150
+ C C K FPS+QALGGH+ASHR+ AA E H+ S H+C IC
Sbjct: 34 FACKTCNKEFPSFQALGGHRASHRR--SAALEGHAPPSPKRVKPVK------HECPICGA 85
Query: 151 SFPTGQALGGHKRCHYEGGGG------ASSTATATASEGVGSTHSHQRNFDLNL 198
F GQALGGH R H G GG A +TA T + G DLNL
Sbjct: 86 EFAVGQALGGHMRKHRGGSGGGGGRSLAPATAPVTMKKSGGGNGKRVLCLDLNL 139
Score = 38.1 bits (87), Expect = 0.004
Identities = 17/60 (28%), Positives = 27/60 (44%)
Query: 81 PAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTTSSASNGGG 140
P+P + + ++C +C F QALGGH HR +G + + +GGG
Sbjct: 68 PSPKRVKPVKHECPICGAEFAVGQALGGHMRKHRGGSGGGGGRSLAPATAPVTMKKSGGG 127
Score = 34.7 bits (78), Expect = 0.045
Identities = 13/25 (52%), Positives = 16/25 (64%)
Query: 141 KVHQCSICQKSFPTGQALGGHKRCH 165
+V C C K FP+ QALGGH+ H
Sbjct: 32 RVFACKTCNKEFPSFQALGGHRASH 56
>At1g02030 unknown protein
Length = 267
Score = 79.3 bits (194), Expect = 2e-15
Identities = 55/150 (36%), Positives = 71/150 (46%), Gaps = 15/150 (10%)
Query: 51 SCTEEEYLALCLIMLARGSTAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHK 110
+ T EE +AL L++L+R + + K ++C CEK F SYQALGGH+
Sbjct: 121 AATTEEDVALSLMLLSRDKWEKEEEESDEERWKKKRNKW-FECETCEKVFKSYQALGGHR 179
Query: 111 ASHRKLAGAAAEDHSTSSAVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCHYEGGG 170
ASH+K AE S H+C IC K F +GQALGGHKR H
Sbjct: 180 ASHKK---KIAETDQLGSDELKKKKKKSTSSHHECPICAKVFTSGQALGGHKRSH----- 231
Query: 171 GASSTATATASEGVGSTHSHQRNFDLNLPA 200
AS+ T G+ + DLNLPA
Sbjct: 232 -ASANNEFTRRSGIIIS-----LIDLNLPA 255
Score = 38.1 bits (87), Expect = 0.004
Identities = 14/23 (60%), Positives = 17/23 (73%)
Query: 143 HQCSICQKSFPTGQALGGHKRCH 165
H+C +C KSF G+ALGGH R H
Sbjct: 5 HKCKLCWKSFANGRALGGHMRSH 27
Score = 34.7 bits (78), Expect = 0.045
Identities = 15/40 (37%), Positives = 25/40 (62%)
Query: 91 YKCSVCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSSAV 130
+KC +C K+F + +ALGGH SH + ++ S SS++
Sbjct: 5 HKCKLCWKSFANGRALGGHMRSHMLIHPLPSQPESYSSSM 44
>At3g10470 unknown protein
Length = 398
Score = 79.0 bits (193), Expect = 2e-15
Identities = 45/118 (38%), Positives = 61/118 (51%), Gaps = 27/118 (22%)
Query: 91 YKCSVCEKTFPSYQALGGHKASHRKLAGA----AAEDHSTS-----------SAVTTSSA 135
Y+C C++TFPS+QALGGH+ASH+K A + DH S + +TT+
Sbjct: 182 YQCKTCDRTFPSFQALGGHRASHKKPKAAMGLHSNHDHKKSNYDDAVSLHLNNVLTTTPN 241
Query: 136 SN------------GGGKVHQCSICQKSFPTGQALGGHKRCHYEGGGGASSTATATAS 181
+N KVH+C IC F +GQALGGH R H A++ +TAT S
Sbjct: 242 NNSNHRSLVVYGKGSNNKVHECGICGAEFTSGQALGGHMRRHRGAVVAAAAASTATVS 299
Score = 57.0 bits (136), Expect = 9e-09
Identities = 44/175 (25%), Positives = 80/175 (45%), Gaps = 24/175 (13%)
Query: 23 NHSYMVDAPWAKRKRSKRSRTDSHHNHASCTEEEYLALCLIMLARGSTAVTPKLTLSRPA 82
N++ +++ + S S ++ + E++ +A CLI+LA+G + P L +P
Sbjct: 73 NNNTLINGVTSSSSASSSSNNNATLKATADEEDQDMANCLILLAQGHSL--PHL---QPQ 127
Query: 83 PVTAEKLSYKCSVCEKTFPSYQALGGHKAS-HRKLAGAAAEDHSTSSAVTTSSASNGGGK 141
P ++ + SYQ G + + +R + E S++ T G
Sbjct: 128 PHPQQQT-------RQLMMSYQDSGNNNNNAYRSSSRRFLETSSSNGTTTNGGGGRAGYY 180
Query: 142 VHQCSICQKSFPTGQALGGHKRCHYEGGGGASSTATATASEGVGSTHSHQR-NFD 195
V+QC C ++FP+ QALGGH+ H + A+ G+ S H H++ N+D
Sbjct: 181 VYQCKTCDRTFPSFQALGGHRASHKK----------PKAAMGLHSNHDHKKSNYD 225
Score = 37.0 bits (84), Expect = 0.009
Identities = 31/112 (27%), Positives = 47/112 (41%), Gaps = 9/112 (8%)
Query: 45 SHHNHASCTEEEYLALCLIMLARGSTAVTPKLTLSRPAPVTAEKLS----YKCSVCEKTF 100
S+H+H ++ ++L L TP + + V K S ++C +C F
Sbjct: 215 SNHDHKKSNYDDAVSLHL----NNVLTTTPNNNSNHRSLVVYGKGSNNKVHECGICGAEF 270
Query: 101 PSYQALGGHKASHR-KLAGAAAEDHSTSSAVTTSSASNGGGKVHQCSICQKS 151
S QALGGH HR + AAA +T S + +N + S Q S
Sbjct: 271 TSGQALGGHMRRHRGAVVAAAAASTATVSVAAIPATANTALSLSPMSFDQMS 322
>At5g04390 zinc finger transcription factor-like protein
Length = 362
Score = 74.7 bits (182), Expect = 4e-14
Identities = 54/176 (30%), Positives = 81/176 (45%), Gaps = 48/176 (27%)
Query: 38 SKRSRTDSHHNH----ASCTEEEYLALCLIMLARGSTAV-------------TPKLTLSR 80
S + S +NH A E++ +A CLI+LA+G + T + T R
Sbjct: 76 SSSASWSSQNNHTLKAAEDEEDQDIANCLILLAQGHSLPHNNHHLPNSNNNNTYRFTSRR 135
Query: 81 PAPVTAEKLS-------YKCSVCEKTFPSYQALGGHKASHRKLAGAA------------A 121
++ Y+C C++TFPS+QALGGH+ASH+K A+ A
Sbjct: 136 FLETSSSNSGGKAGYYVYQCKTCDRTFPSFQALGGHRASHKKPKAASFYSNLDLKKNTYA 195
Query: 122 ED-----HSTSSAVTTSSASN-------GGGKVHQCSICQKSFPTGQALGGHKRCH 165
D H+T++ +++ + KVH+C IC F +GQALGGH R H
Sbjct: 196 NDAVSLVHTTTTVFKNNNSRSLVVYGKASKNKVHECGICGAEFTSGQALGGHMRRH 251
Score = 32.7 bits (73), Expect = 0.17
Identities = 17/46 (36%), Positives = 24/46 (51%), Gaps = 3/46 (6%)
Query: 91 YKCSVCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTTSSAS 136
++C +C F S QALGGH HR GA + VT ++A+
Sbjct: 229 HECGICGAEFTSGQALGGHMRRHR---GAVVVPAVIAPTVTVATAA 271
>At2g37430 putative C2H2-type zinc finger protein
Length = 178
Score = 73.6 bits (179), Expect = 9e-14
Identities = 46/127 (36%), Positives = 67/127 (52%), Gaps = 12/127 (9%)
Query: 39 KRSRTDSHHNHASCTEEEYLALCLIMLARGSTAVTPKLTLSRPAPVTAEKLSYKCSVCEK 98
KR R+D + + +A CL++LA+ T++ ++ L++ ++C C K
Sbjct: 2 KRERSDFEESLKNID----IAKCLMILAQ--TSMVKQIGLNQHTESHTSN-QFECKTCNK 54
Query: 99 TFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTTSSASNGGGKVHQCSICQKSFPTGQAL 158
F S+QALGGH+ASH+K + V S G H+CSIC +SF TGQAL
Sbjct: 55 RFSSFQALGGHRASHKKPKLTVEQKD-----VKHLSNDYKGNHFHKCSICSQSFGTGQAL 109
Query: 159 GGHKRCH 165
GGH R H
Sbjct: 110 GGHMRRH 116
Score = 35.0 bits (79), Expect = 0.035
Identities = 13/24 (54%), Positives = 18/24 (74%)
Query: 91 YKCSVCEKTFPSYQALGGHKASHR 114
+KCS+C ++F + QALGGH HR
Sbjct: 94 HKCSICSQSFGTGQALGGHMRRHR 117
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.126 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,634,193
Number of Sequences: 26719
Number of extensions: 241394
Number of successful extensions: 980
Number of sequences better than 10.0: 91
Number of HSP's better than 10.0 without gapping: 74
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 709
Number of HSP's gapped (non-prelim): 189
length of query: 236
length of database: 11,318,596
effective HSP length: 96
effective length of query: 140
effective length of database: 8,753,572
effective search space: 1225500080
effective search space used: 1225500080
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0050.4


