

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0050.14
(77 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
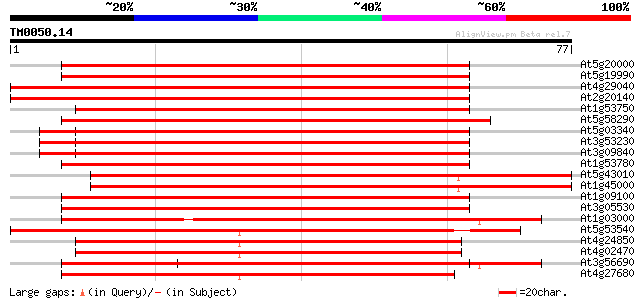
At5g20000 26S proteasome AAA-ATPase subunit RPT6a - like protein 106 2e-24
At5g19990 26S proteasome AAA-ATPase subunit RPT6a 106 2e-24
At4g29040 26S proteasome AAA-ATPase subunit RPT2a 75 8e-15
At2g20140 26S proteasome subunit 4 75 8e-15
At1g53750 26S proteasome ATPase subunit 74 1e-14
At5g58290 26S proteasome AAA-ATPase subunit RPT3 (gb|AAF22523.1) 73 2e-14
At5g03340 transitional endoplasmic reticulum ATPase 73 3e-14
At3g53230 CDC48 - like protein 73 3e-14
At3g09840 putative transitional endoplasmic reticulum ATPase 73 3e-14
At1g53780 hypothetical protein 69 3e-13
At5g43010 26S proteasome AAA-ATPase subunit RPT4a (gb|AAF22524.1) 69 4e-13
At1g45000 26S proteasome AAA-ATPase subunit RPT4a like protein 69 4e-13
At1g09100 putative 26S protease regulatory subunit 6A 69 5e-13
At3g05530 26S proteasome AAA-ATPase subunit RPT5a 68 7e-13
At1g03000 putative peroxisome assembly factor-2 59 3e-10
At5g53540 26S proteasome regulatory particle chain RPT6-like pro... 59 4e-10
At4g24850 unknown protein (At4g24850) 57 1e-09
At4g02470 57 2e-09
At3g56690 calmodulin-binding protein 56 3e-09
At4g27680 unknown protein 56 4e-09
>At5g20000 26S proteasome AAA-ATPase subunit RPT6a - like protein
Length = 419
Score = 106 bits (264), Expect = 2e-24
Identities = 51/56 (91%), Positives = 53/56 (94%)
Query: 8 ILQVIEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIRVSG 63
I +VIE+PIKHPELFES GIAQPKGVLLYGPPGT KTLLARAVAHHTDCTFIRVSG
Sbjct: 173 IKEVIELPIKHPELFESLGIAQPKGVLLYGPPGTGKTLLARAVAHHTDCTFIRVSG 228
>At5g19990 26S proteasome AAA-ATPase subunit RPT6a
Length = 419
Score = 106 bits (264), Expect = 2e-24
Identities = 51/56 (91%), Positives = 53/56 (94%)
Query: 8 ILQVIEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIRVSG 63
I +VIE+PIKHPELFES GIAQPKGVLLYGPPGT KTLLARAVAHHTDCTFIRVSG
Sbjct: 173 IKEVIELPIKHPELFESLGIAQPKGVLLYGPPGTGKTLLARAVAHHTDCTFIRVSG 228
>At4g29040 26S proteasome AAA-ATPase subunit RPT2a
Length = 443
Score = 74.7 bits (182), Expect = 8e-15
Identities = 35/63 (55%), Positives = 46/63 (72%)
Query: 1 LASCLSYILQVIEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIR 60
L + + I + +E+P+ HPEL+E GI PKGV+LYG PGT KTLLA+AVA+ T TF+R
Sbjct: 193 LEAQIQEIKEAVELPLTHPELYEDIGIKPPKGVILYGEPGTGKTLLAKAVANSTSATFLR 252
Query: 61 VSG 63
V G
Sbjct: 253 VVG 255
>At2g20140 26S proteasome subunit 4
Length = 443
Score = 74.7 bits (182), Expect = 8e-15
Identities = 35/63 (55%), Positives = 46/63 (72%)
Query: 1 LASCLSYILQVIEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIR 60
L + + I + +E+P+ HPEL+E GI PKGV+LYG PGT KTLLA+AVA+ T TF+R
Sbjct: 193 LEAQIQEIKEAVELPLTHPELYEDIGIKPPKGVILYGEPGTGKTLLAKAVANSTSATFLR 252
Query: 61 VSG 63
V G
Sbjct: 253 VVG 255
>At1g53750 26S proteasome ATPase subunit
Length = 426
Score = 74.3 bits (181), Expect = 1e-14
Identities = 36/54 (66%), Positives = 41/54 (75%)
Query: 10 QVIEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIRVSG 63
+V+E+P+ HPE F GI PKGVL YGPPGT KTLLARAVA+ TD FIRV G
Sbjct: 182 EVVELPMLHPEKFVKLGIDPPKGVLCYGPPGTGKTLLARAVANRTDACFIRVIG 235
>At5g58290 26S proteasome AAA-ATPase subunit RPT3 (gb|AAF22523.1)
Length = 408
Score = 73.2 bits (178), Expect = 2e-14
Identities = 33/59 (55%), Positives = 44/59 (73%)
Query: 8 ILQVIEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIRVSGKVY 66
I + +E+P+ H EL++ GI P+GVLLYGPPGT KT+LA+AVA+HT FIRV G +
Sbjct: 167 IREAVELPLTHHELYKQIGIDPPRGVLLYGPPGTGKTMLAKAVANHTTAAFIRVVGSEF 225
>At5g03340 transitional endoplasmic reticulum ATPase
Length = 810
Score = 72.8 bits (177), Expect = 3e-14
Identities = 30/59 (50%), Positives = 47/59 (78%)
Query: 5 LSYILQVIEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIRVSG 63
++ I +++E+P++HP+LF+S G+ PKG+LLYGPPG+ KTL+ARAVA+ T F ++G
Sbjct: 216 MAQIRELVELPLRHPQLFKSIGVKPPKGILLYGPPGSGKTLIARAVANETGAFFFCING 274
Score = 62.8 bits (151), Expect = 3e-11
Identities = 27/54 (50%), Positives = 37/54 (68%)
Query: 10 QVIEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIRVSG 63
+ ++ P++HPE FE G++ KGVL YGPPG KTLLA+A+A+ FI V G
Sbjct: 494 ETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLAKAIANECQANFISVKG 547
>At3g53230 CDC48 - like protein
Length = 815
Score = 72.8 bits (177), Expect = 3e-14
Identities = 30/59 (50%), Positives = 47/59 (78%)
Query: 5 LSYILQVIEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIRVSG 63
++ I +++E+P++HP+LF+S G+ PKG+LLYGPPG+ KTL+ARAVA+ T F ++G
Sbjct: 217 MAQIRELVELPLRHPQLFKSIGVKPPKGILLYGPPGSGKTLIARAVANETGAFFFCING 275
Score = 62.4 bits (150), Expect = 4e-11
Identities = 26/54 (48%), Positives = 37/54 (68%)
Query: 10 QVIEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIRVSG 63
+ ++ P++HPE FE G++ KGVL YGPPG KTLLA+A+A+ FI + G
Sbjct: 495 ETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLAKAIANECQANFISIKG 548
>At3g09840 putative transitional endoplasmic reticulum ATPase
Length = 809
Score = 72.8 bits (177), Expect = 3e-14
Identities = 30/59 (50%), Positives = 47/59 (78%)
Query: 5 LSYILQVIEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIRVSG 63
++ I +++E+P++HP+LF+S G+ PKG+LLYGPPG+ KTL+ARAVA+ T F ++G
Sbjct: 216 MAQIRELVELPLRHPQLFKSIGVKPPKGILLYGPPGSGKTLIARAVANETGAFFFCING 274
Score = 62.8 bits (151), Expect = 3e-11
Identities = 27/54 (50%), Positives = 37/54 (68%)
Query: 10 QVIEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIRVSG 63
+ ++ P++HPE FE G++ KGVL YGPPG KTLLA+A+A+ FI V G
Sbjct: 494 ETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLAKAIANECQANFISVKG 547
>At1g53780 hypothetical protein
Length = 464
Score = 69.3 bits (168), Expect = 3e-13
Identities = 34/56 (60%), Positives = 41/56 (72%)
Query: 8 ILQVIEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIRVSG 63
I +V+E+P+ HPE F GI PKGVL YGPPG+ KTL+ARAVA+ T FIRV G
Sbjct: 217 IREVVELPMLHPEKFVRLGIDPPKGVLCYGPPGSGKTLVARAVANRTGACFIRVVG 272
>At5g43010 26S proteasome AAA-ATPase subunit RPT4a (gb|AAF22524.1)
Length = 399
Score = 68.9 bits (167), Expect = 4e-13
Identities = 35/67 (52%), Positives = 44/67 (65%), Gaps = 1/67 (1%)
Query: 12 IEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIR-VSGKVYSLYS 70
IE+P+ +PELF GI PKGVLLYGPPGT KTLLARA+A + D F++ VS + Y
Sbjct: 155 IELPLMNPELFLRVGIKPPKGVLLYGPPGTGKTLLARAIASNIDANFLKVVSSAIIDKYI 214
Query: 71 ASSTHYL 77
S +
Sbjct: 215 GESARLI 221
>At1g45000 26S proteasome AAA-ATPase subunit RPT4a like protein
Length = 399
Score = 68.9 bits (167), Expect = 4e-13
Identities = 35/67 (52%), Positives = 44/67 (65%), Gaps = 1/67 (1%)
Query: 12 IEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIR-VSGKVYSLYS 70
IE+P+ +PELF GI PKGVLLYGPPGT KTLLARA+A + D F++ VS + Y
Sbjct: 155 IELPLMNPELFLRVGIKPPKGVLLYGPPGTGKTLLARAIASNIDANFLKVVSSAIIDKYI 214
Query: 71 ASSTHYL 77
S +
Sbjct: 215 GESARLI 221
>At1g09100 putative 26S protease regulatory subunit 6A
Length = 423
Score = 68.6 bits (166), Expect = 5e-13
Identities = 31/56 (55%), Positives = 42/56 (74%)
Query: 8 ILQVIEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIRVSG 63
+++ I +P+ H E FE GI PKGVLLYGPPGT KTL+ARA A T+ TF++++G
Sbjct: 182 LVEAIVLPMTHKEQFEKLGIRPPKGVLLYGPPGTGKTLMARACAAQTNATFLKLAG 237
>At3g05530 26S proteasome AAA-ATPase subunit RPT5a
Length = 424
Score = 68.2 bits (165), Expect = 7e-13
Identities = 30/56 (53%), Positives = 42/56 (74%)
Query: 8 ILQVIEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIRVSG 63
+++ I +P+ H E FE G+ PKGVLLYGPPGT KTL+ARA A T+ TF++++G
Sbjct: 183 LVEAIVLPMTHKERFEKLGVRPPKGVLLYGPPGTGKTLMARACAAQTNATFLKLAG 238
>At1g03000 putative peroxisome assembly factor-2
Length = 941
Score = 59.3 bits (142), Expect = 3e-10
Identities = 31/67 (46%), Positives = 44/67 (65%), Gaps = 2/67 (2%)
Query: 8 ILQVIEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIRVSG-KVY 66
IL +++P+ H +LF S G+ + GVLLYGPPGT KTLLA+AVA F+ V G ++
Sbjct: 670 ILDTVQLPLLHKDLFSS-GLRKRSGVLLYGPPGTGKTLLAKAVATECSLNFLSVKGPELI 728
Query: 67 SLYSASS 73
++Y S
Sbjct: 729 NMYIGES 735
>At5g53540 26S proteasome regulatory particle chain RPT6-like
protein
Length = 403
Score = 58.9 bits (141), Expect = 4e-10
Identities = 33/71 (46%), Positives = 46/71 (64%), Gaps = 3/71 (4%)
Query: 1 LASCLSYILQVIEVPIKHPELFESPGIAQP-KGVLLYGPPGTRKTLLARAVAHHTDCTFI 59
L S + +++ +P+K PELF + P KGVLLYGPPGT KT+LA+A+A ++ FI
Sbjct: 92 LESIKQALYELVILPLKRPELFAYGKLLGPQKGVLLYGPPGTGKTMLAKAIARESEAVFI 151
Query: 60 RVSGKVYSLYS 70
V KV +L S
Sbjct: 152 NV--KVSNLMS 160
>At4g24850 unknown protein (At4g24850)
Length = 442
Score = 57.4 bits (137), Expect = 1e-09
Identities = 26/54 (48%), Positives = 39/54 (72%), Gaps = 1/54 (1%)
Query: 10 QVIEVPIKHPELFESPGIAQP-KGVLLYGPPGTRKTLLARAVAHHTDCTFIRVS 62
+++ +P++ PELF + +P KG+LL+GPPGT KT+LA+AVA D FI +S
Sbjct: 154 ELVMLPLQRPELFCKGELTKPCKGILLFGPPGTGKTMLAKAVAKEADANFINIS 207
>At4g02470
Length = 371
Score = 56.6 bits (135), Expect = 2e-09
Identities = 25/54 (46%), Positives = 39/54 (71%), Gaps = 1/54 (1%)
Query: 10 QVIEVPIKHPELFESPGIAQP-KGVLLYGPPGTRKTLLARAVAHHTDCTFIRVS 62
+++ +P++ PELF+ + +P KG+LL+GPPGT KT+LA+AVA FI +S
Sbjct: 83 ELVMLPLQRPELFDKGQLTKPTKGILLFGPPGTGKTMLAKAVATEAGANFINIS 136
>At3g56690 calmodulin-binding protein
Length = 1022
Score = 56.2 bits (134), Expect = 3e-09
Identities = 24/56 (42%), Positives = 35/56 (61%)
Query: 8 ILQVIEVPIKHPELFESPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIRVSG 63
+++ +E P KH + F+ G P G+L++GPPG KTL+ARAVA F+ V G
Sbjct: 736 LMEAVEWPQKHQDAFKRIGTRPPSGILMFGPPGCSKTLMARAVASEAKLNFLAVKG 791
Score = 45.8 bits (107), Expect = 4e-06
Identities = 24/51 (47%), Positives = 31/51 (60%), Gaps = 1/51 (1%)
Query: 24 SPGIAQPKGVLLYGPPGTRKTLLARAVAHHTDCTFIRVSG-KVYSLYSASS 73
S G+ KGVL++GPPGT KT LAR A H+ F V+G ++ S Y S
Sbjct: 412 SLGLRPTKGVLIHGPPGTGKTSLARTFARHSGVNFFSVNGPEIISQYLGES 462
>At4g27680 unknown protein
Length = 398
Score = 55.8 bits (133), Expect = 4e-09
Identities = 27/55 (49%), Positives = 38/55 (69%), Gaps = 1/55 (1%)
Query: 8 ILQVIEVPIKHPELFESPGIAQP-KGVLLYGPPGTRKTLLARAVAHHTDCTFIRV 61
+ +++ +P+K PELF + P KGVLLYGPPGT KT+LA+A+A + FI V
Sbjct: 96 LYELVILPLKRPELFAYGKLLGPQKGVLLYGPPGTGKTMLAKAIAKESGAVFINV 150
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.138 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,769,374
Number of Sequences: 26719
Number of extensions: 58922
Number of successful extensions: 291
Number of sequences better than 10.0: 115
Number of HSP's better than 10.0 without gapping: 111
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 164
Number of HSP's gapped (non-prelim): 128
length of query: 77
length of database: 11,318,596
effective HSP length: 53
effective length of query: 24
effective length of database: 9,902,489
effective search space: 237659736
effective search space used: 237659736
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0050.14


