

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0050.12
(143 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
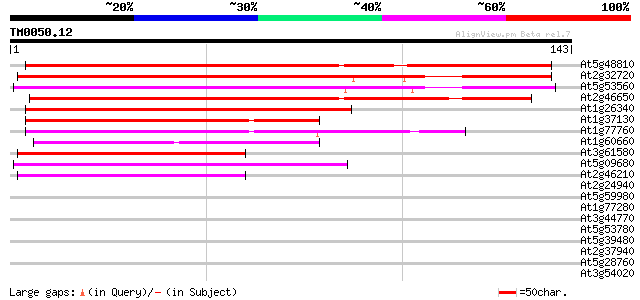
Score E
Sequences producing significant alignments: (bits) Value
At5g48810 cytochrome b5 (dbj|BAA74840.1) 115 8e-27
At2g32720 putative cytochrome b5 111 2e-25
At5g53560 cytochrome b5 (dbj|BAA74839.1) 98 1e-21
At2g46650 putative cytochrome b5 96 7e-21
At1g26340 cytochrome b5 like protein 96 9e-21
At1g37130 nitrate reductase (At1g37130) 75 9e-15
At1g77760 nitrate reductase 1 (NR1) 70 4e-13
At1g60660 Putative Cytochrome B5 59 9e-10
At3g61580 delta-8 sphingolipid desaturase (sld1) 53 5e-08
At5g09680 unknown protein 53 6e-08
At2g46210 putative fatty acid desaturase/cytochrome b5 fusion pr... 51 2e-07
At2g24940 putative steroid binding protein 32 0.15
At5g59980 putative protein 30 0.34
At1g77280 hypothetical protein 30 0.57
At3g44770 hypothetical protein 28 2.2
At5g53780 putative protein 27 4.9
At5g39480 hypothetical protein 26 6.4
At2g37940 unknown protein 26 6.4
At5g28760 putative protein 26 8.3
At3g54020 unknown protein 26 8.3
>At5g48810 cytochrome b5 (dbj|BAA74840.1)
Length = 140
Score = 115 bits (288), Expect = 8e-27
Identities = 60/134 (44%), Positives = 86/134 (63%), Gaps = 4/134 (2%)
Query: 5 RVFTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAVGHS 64
+VFTLS+V+QH S KDCW+VI+G+V DVTKFL +HPGGD+V+L GKD T +F+ VGHS
Sbjct: 6 KVFTLSEVSQHSSAKDCWIVIDGKVYDVTKFLDDHPGGDEVILTSTGKDATDDFEDVGHS 65
Query: 65 KEAQNLVLKYQVGILQGATVQEIDVVHKEPNSKQEMSAFVIKEDAESKSVALFEFFVPLV 124
A+ ++ +Y VG + ATV + P S + A ++ + + L +F VPL+
Sbjct: 66 STAKAMLDEYYVGDIDTATV-PVKAKFVPPTSTK---AVATQDKSSDFVIKLLQFLVPLL 121
Query: 125 VATLYFGYRCLTRT 138
+ L FG R T+T
Sbjct: 122 ILGLAFGIRYYTKT 135
>At2g32720 putative cytochrome b5
Length = 134
Score = 111 bits (277), Expect = 2e-25
Identities = 59/139 (42%), Positives = 86/139 (61%), Gaps = 12/139 (8%)
Query: 3 ERRVFTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAVG 62
E ++FTLS+V++H DCW+VING+V +VTKFL +HPGGDDVLL GKD T +F+ VG
Sbjct: 4 EAKIFTLSEVSEHNQAHDCWIVINGKVYNVTKFLEDHPGGDDVLLSSTGKDATDDFEDVG 63
Query: 63 HSKEAQNLVLKYQVGILQGATVQE--IDVVHKEPNSKQE-MSAFVIKEDAESKSVALFEF 119
HS+ A+ ++ +Y VG + T+ + K+P+ Q+ S F+IK L +F
Sbjct: 64 HSESAREMMEQYYVGEIDPTTIPKKVKYTPPKQPHYNQDKTSEFIIK---------LLQF 114
Query: 120 FVPLVVATLYFGYRCLTRT 138
VPL + L G R T++
Sbjct: 115 LVPLAILGLAVGIRIYTKS 133
>At5g53560 cytochrome b5 (dbj|BAA74839.1)
Length = 134
Score = 98.2 bits (243), Expect = 1e-21
Identities = 50/141 (35%), Positives = 85/141 (59%), Gaps = 12/141 (8%)
Query: 2 AERRVFTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAV 61
++R+V + +V++H KDCW++I+G+V DVT F+ +HPGGD+VLL GKD T +F+ V
Sbjct: 3 SDRKVLSFEEVSKHNKTKDCWLIISGKVYDVTPFMDDHPGGDEVLLSSTGKDATNDFEDV 62
Query: 62 GHSKEAQNLVLKYQVGILQGATV--QEIDVVHKEPNSKQEMS-AFVIKEDAESKSVALFE 118
GHS A++++ KY +G + ++V V ++P Q+ + F+IK + +
Sbjct: 63 GHSDTARDMMDKYFIGEIDSSSVPATRTYVAPQQPAYNQDKTPEFIIK---------ILQ 113
Query: 119 FFVPLVVATLYFGYRCLTRTD 139
F VP+++ L R T+ D
Sbjct: 114 FLVPILILGLALVVRHYTKKD 134
>At2g46650 putative cytochrome b5
Length = 132
Score = 95.9 bits (237), Expect = 7e-21
Identities = 48/128 (37%), Positives = 79/128 (61%), Gaps = 4/128 (3%)
Query: 6 VFTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAVGHSK 65
+ + VA+HK DCW++I+G+V D++ F+ EHPGGD+VLL V GKD + +F+ V HSK
Sbjct: 4 LISFHDVAKHKCKNDCWILIHGKVYDISTFMDEHPGGDNVLLAVTGKDASIDFEDVNHSK 63
Query: 66 EAQNLVLKYQVGILQGATVQEIDVVHKEPNSKQEMSAFVIKEDAESKSVALFEFFVPLVV 125
+A+ L+ KY +G + +TV + + P K+ +A KE++ K L + +PL++
Sbjct: 64 DAKELMKKYCIGDVDQSTV-PVTQQYIPPWEKESTAAETTKEESGKK---LLIYLIPLLI 119
Query: 126 ATLYFGYR 133
+ F R
Sbjct: 120 LGVAFALR 127
>At1g26340 cytochrome b5 like protein
Length = 135
Score = 95.5 bits (236), Expect = 9e-21
Identities = 41/83 (49%), Positives = 61/83 (73%)
Query: 5 RVFTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAVGHS 64
+++++ + A H DCWVVI+G+V DV+ ++ EHPGGDDVLL VAGKD T +F+ GHS
Sbjct: 6 KLYSMEEAATHNKQDDCWVVIDGKVYDVSSYMDEHPGGDDVLLAVAGKDATDDFEDAGHS 65
Query: 65 KEAQNLVLKYQVGILQGATVQEI 87
K+A+ L+ KY +G L +++ EI
Sbjct: 66 KDARELMEKYFIGELDESSLPEI 88
>At1g37130 nitrate reductase (At1g37130)
Length = 917
Score = 75.5 bits (184), Expect = 9e-15
Identities = 30/75 (40%), Positives = 55/75 (73%), Gaps = 1/75 (1%)
Query: 5 RVFTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAVGHS 64
+++++S+V +H S CW++++G + D T+FL++HPGG D +L AG D T+EF+A+ HS
Sbjct: 543 KMYSMSEVKKHNSADSCWIIVHGHIYDCTRFLMDHPGGSDSILINAGTDCTEEFEAI-HS 601
Query: 65 KEAQNLVLKYQVGIL 79
+A+ ++ Y++G L
Sbjct: 602 DKAKKMLEDYRIGEL 616
>At1g77760 nitrate reductase 1 (NR1)
Length = 917
Score = 70.1 bits (170), Expect = 4e-13
Identities = 39/114 (34%), Positives = 67/114 (58%), Gaps = 5/114 (4%)
Query: 5 RVFTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAVGHS 64
+++++S+V +H + W++++G + D T+FL +HPGG D +L AG D T+EF+A+ HS
Sbjct: 546 KMYSISEVRKHNTADSAWIIVHGHIYDCTRFLKDHPGGTDSILINAGTDCTEEFEAI-HS 604
Query: 65 KEAQNLVLKYQVG--ILQGATVQEIDVVHKEPNSKQEMSAFVIKEDAESKSVAL 116
+A+ L+ Y++G I G VH N ++ IKE K++AL
Sbjct: 605 DKAKKLLEDYRIGELITTGYDSSPNVSVHGASNFGPLLAP--IKELTPQKNIAL 656
>At1g60660 Putative Cytochrome B5
Length = 121
Score = 58.9 bits (141), Expect = 9e-10
Identities = 26/73 (35%), Positives = 43/73 (58%), Gaps = 1/73 (1%)
Query: 7 FTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAVGHSKE 66
++ S+VA H DCW++I +V D+T ++ EHPGG D +L+ AG D T F H+
Sbjct: 49 YSKSEVAVHNKRNDCWIIIKDKVYDITSYVEEHPGG-DAILDHAGDDSTDGFFGPQHATR 107
Query: 67 AQNLVLKYQVGIL 79
+++ + +G L
Sbjct: 108 VFDMIEDFYIGEL 120
>At3g61580 delta-8 sphingolipid desaturase (sld1)
Length = 449
Score = 53.1 bits (126), Expect = 5e-08
Identities = 22/58 (37%), Positives = 36/58 (61%)
Query: 3 ERRVFTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDA 60
E++ T + +H + D W+ I G+V +V+ ++ HPGGD V+L + G+DVT F A
Sbjct: 6 EKKYITNEDLKKHNKSGDLWIAIQGKVYNVSDWIKTHPGGDTVILNLVGQDVTDAFIA 63
>At5g09680 unknown protein
Length = 211
Score = 52.8 bits (125), Expect = 6e-08
Identities = 24/85 (28%), Positives = 47/85 (55%)
Query: 2 AERRVFTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAV 61
+ +R+ + +V +H++ W V+ GRV +++ ++ HPGG D+L++ G+D T F+
Sbjct: 124 SNKRLIPMDEVKKHRTGDSMWTVLKGRVYNISPYMNFHPGGVDMLMKAVGRDGTLLFNKY 183
Query: 62 GHSKEAQNLVLKYQVGILQGATVQE 86
L+ K VG+L V++
Sbjct: 184 HAWVNVDILLEKCLVGVLDDTKVKK 208
>At2g46210 putative fatty acid desaturase/cytochrome b5 fusion
protein
Length = 449
Score = 50.8 bits (120), Expect = 2e-07
Identities = 22/58 (37%), Positives = 35/58 (59%)
Query: 3 ERRVFTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDA 60
++R T + +H D W+ I G+V DV+ ++ HPGG+ +L +AG+DVT F A
Sbjct: 6 KKRYVTSEDLKKHNKPGDLWISIQGKVYDVSDWVKSHPGGEAAILNLAGQDVTDAFIA 63
>At2g24940 putative steroid binding protein
Length = 100
Score = 31.6 bits (70), Expect = 0.15
Identities = 25/88 (28%), Positives = 43/88 (48%), Gaps = 11/88 (12%)
Query: 7 FTLSQVAQHKS---NKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAVGH 63
FT Q++Q+ +K +V I GRV DVT + G D + AGKD ++ +
Sbjct: 3 FTAEQLSQYNGTDESKPIYVAIKGRVFDVTTGKSFYGSGGDYSM-FAGKDASRALGKMSK 61
Query: 64 SKEAQNLVLKYQVGILQGATVQEIDVVH 91
++E + L+G T +EI+ ++
Sbjct: 62 NEEDVS-------PSLEGLTEKEINTLN 82
>At5g59980 putative protein
Length = 680
Score = 30.4 bits (67), Expect = 0.34
Identities = 27/113 (23%), Positives = 49/113 (42%), Gaps = 15/113 (13%)
Query: 5 RVFTLSQVAQHKSNK--DCWVVING------RVLDVTKFLIEHPGGDDVLLEVAGKDVTK 56
+ F + V HKS+K D V++G RV D+ G + V A K +
Sbjct: 308 KAFDATNVVAHKSSKAIDFTSVLDGLPKHGFRVKDIV-------GTESVTQPSAAKVIDT 360
Query: 57 EFDAVGHSKEAQNLVLKYQVGILQGATVQEIDVVHKEPNSKQEMSAFVIKEDA 109
+ + E + + + T+ +ID++ E ++K E + V+KE+A
Sbjct: 361 QVHSSNQVSELRMATASSDDNLREIETISQIDMLMSEDDNKVEPTTNVLKEEA 413
>At1g77280 hypothetical protein
Length = 781
Score = 29.6 bits (65), Expect = 0.57
Identities = 25/81 (30%), Positives = 39/81 (47%), Gaps = 13/81 (16%)
Query: 2 AERRVFTLSQVAQHKSNKDCWVVI--NGRVL-----DVTKFLIEHPGGDDV----LLEVA 50
A R ++++ K +KDCWV+ NG++L + + + G +DV LL V
Sbjct: 158 AIRSSASVAKYIAKKLSKDCWVIAVNNGKILFQKEGSPSSTINQSQGKEDVRRITLLNVL 217
Query: 51 GKDVT--KEFDAVGHSKEAQN 69
+ VT K V HS+E N
Sbjct: 218 QRSVTLNKTTKVVSHSEEDSN 238
>At3g44770 hypothetical protein
Length = 505
Score = 27.7 bits (60), Expect = 2.2
Identities = 18/63 (28%), Positives = 27/63 (42%), Gaps = 4/63 (6%)
Query: 8 TLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAVGHSKEA 67
T A H K CW+V N T+F ++ +L + K K D GH ++
Sbjct: 65 TRENCALHTKTKSCWIVENYLTKPATRFCVDWVNMS--MLYIYPKPKKKSTD--GHLRKM 120
Query: 68 QNL 70
+NL
Sbjct: 121 KNL 123
>At5g53780 putative protein
Length = 376
Score = 26.6 bits (57), Expect = 4.9
Identities = 12/34 (35%), Positives = 22/34 (64%), Gaps = 2/34 (5%)
Query: 59 DAVGHSKEAQNLVLKYQVGILQGATVQEIDVVHK 92
D VG S EA+N+++ +++ L + I+V+HK
Sbjct: 55 DKVGESPEAENIIMSFKLFDLSKEEI--IEVIHK 86
>At5g39480 hypothetical protein
Length = 568
Score = 26.2 bits (56), Expect = 6.4
Identities = 18/63 (28%), Positives = 30/63 (47%), Gaps = 1/63 (1%)
Query: 49 VAGKDVTKEFDAVGHSKEAQNLVLKYQVGILQGATVQEIDVVHK-EPNSKQEMSAFVIKE 107
+ G D + F G KE L + +GI + V ++DVV + E +S E+ +E
Sbjct: 144 ICGFDGSNGFFLHGRDKEGSCLYPGFVMGIEKSCNVLQLDVVPRQEKSSCNEIERGASRE 203
Query: 108 DAE 110
+ E
Sbjct: 204 EGE 206
>At2g37940 unknown protein
Length = 305
Score = 26.2 bits (56), Expect = 6.4
Identities = 16/36 (44%), Positives = 18/36 (49%)
Query: 22 WVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKE 57
W +N V + K L E P VLL V KD TKE
Sbjct: 230 WYTVNLVVFCLDKKLPELPDRTAVLLPVISKDRTKE 265
>At5g28760 putative protein
Length = 520
Score = 25.8 bits (55), Expect = 8.3
Identities = 9/27 (33%), Positives = 16/27 (58%)
Query: 1 MAERRVFTLSQVAQHKSNKDCWVVING 27
M + L + AQ + +++CW V+NG
Sbjct: 117 MTQGMCMLLLRTAQTEMDRECWFVVNG 143
>At3g54020 unknown protein
Length = 305
Score = 25.8 bits (55), Expect = 8.3
Identities = 19/57 (33%), Positives = 25/57 (43%), Gaps = 7/57 (12%)
Query: 1 MAERRVFTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKE 57
+A R+ +T+ V W +N V + K L E P LL V KD TKE
Sbjct: 217 IASRKHYTVDVVV-------AWYTVNLVVFFLDKKLPELPDRTTALLPVISKDRTKE 266
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,957,079
Number of Sequences: 26719
Number of extensions: 111240
Number of successful extensions: 295
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 277
Number of HSP's gapped (non-prelim): 22
length of query: 143
length of database: 11,318,596
effective HSP length: 89
effective length of query: 54
effective length of database: 8,940,605
effective search space: 482792670
effective search space used: 482792670
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0050.12


