

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0048.9
(1366 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
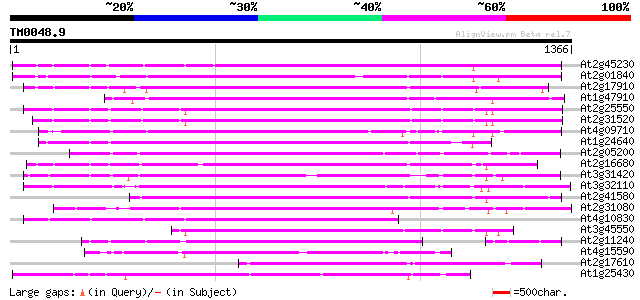
Score E
Sequences producing significant alignments: (bits) Value
At2g45230 putative non-LTR retroelement reverse transcriptase 784 0.0
At2g01840 putative non-LTR retroelement reverse transcriptase 703 0.0
At2g17910 putative non-LTR retroelement reverse transcriptase 701 0.0
At1g47910 reverse transcriptase, putative 679 0.0
At2g25550 putative non-LTR retroelement reverse transcriptase 669 0.0
At2g31520 putative non-LTR retroelement reverse transcriptase 665 0.0
At4g09710 RNA-directed DNA polymerase -like protein 657 0.0
At1g24640 hypothetical protein 613 e-175
At2g05200 putative non-LTR retroelement reverse transcriptase 604 e-173
At2g16680 putative non-LTR retroelement reverse transcriptase 577 e-164
At3g31420 hypothetical protein 577 e-164
At3g32110 non-LTR reverse transcriptase, putative 560 e-159
At2g41580 putative non-LTR retroelement reverse transcriptase 557 e-158
At2g31080 putative non-LTR retroelement reverse transcriptase 551 e-157
At4g10830 putative protein 537 e-152
At3g45550 putative protein 518 e-146
At2g11240 pseudogene 499 e-141
At4g15590 reverse transcriptase like protein 459 e-129
At2g17610 putative non-LTR retroelement reverse transcriptase 447 e-125
At1g25430 hypothetical protein 425 e-119
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 784 bits (2024), Expect = 0.0
Identities = 463/1368 (33%), Positives = 713/1368 (51%), Gaps = 48/1368 (3%)
Query: 6 MRLLSWNCRGLGNLEAVRALRGLIHSQAPDIVFLMETKLFSFEMQR---HRGMGGLSNIF 62
MR+LSWNC+G+GN VR LR + P+++FL ETK ++ H G L +
Sbjct: 1 MRILSWNCQGVGNTPTVRHLREIRGLYFPEVIFLCETKKRRNYLENVVGHLGFFDLHTVE 60
Query: 63 PVQCGRNRAGGLCLLWRSEVEVTIINASLHHILFTVVHSDSVTVPMQVYALYGFPELHLK 122
P+ ++GGL L+W+ V++ ++ + I ++ D + +YG P +
Sbjct: 61 PI----GKSGGLALMWKDSVQIKVLQSDKRLIDALLIWQDK---EFYLTCIYGEPVQAER 113
Query: 123 DRTWEMIRSVRPMAPIPWLCIGDFNDILSPSDKLGGAPPDLARLQVATQACADCGLQRVD 182
WE + + PW+ GDFN+++ PS+K+GG + Q CGL V+
Sbjct: 114 GELWERLTRLGLSRSGPWMLTGDFNELVDPSEKIGGPARKESSCLEFRQMLNSCGLWEVN 173
Query: 183 FSGPSFTWTNNRVGPGRVDERLDYALINQAWVNLWPVSSVSHLIRHQSDHNPVVLHCGSR 242
SG F+W NR V RLD + NQAW+ L+P + ++L + SDH+P++ +
Sbjct: 174 HSGYQFSWYGNR-NDELVQCRLDRTVANQAWMELFPQAKATYLQKICSDHSPLI----NN 228
Query: 243 RTETQRHRTRMFRFEEVWLESGEECAEIISEGWSN----TNLSILSRINLVGRSLDSWGR 298
+ F++++ W++ E +++ WS TN ++ +I R + W R
Sbjct: 229 LVGDNWRKWAGFKYDKRWVQR-EGFKDLLCNFWSQQSTKTNALMMEKIASCRREISKWKR 287
Query: 299 EKYGELPKRIKEARAFLQRLQEEVQTEQVVRATREAEKNLDVLLKQEEIVWSQRSRANWL 358
RI+E + L +++ ++ R +K L EE W ++SR W+
Sbjct: 288 VSKPSSAVRIQELQFKLDAATKQIPFDR--RELARLKKELSQEYNNEEQFWQEKSRIMWM 345
Query: 359 KHGDRNTKFFHSKATQRRKRNLIEEIQDDNGRKFVRDADIARVLTSYFQEIFSTCNPTG- 417
++GDRNTK+FH+ RR +N I+++ D+ GR++ D D+ RV +YF+++F++ G
Sbjct: 346 RNGDRNTKYFHAATKNRRAQNRIQKLIDEEGREWTSDEDLGRVAEAYFKKLFAS-EDVGY 404
Query: 418 -IEEVATLVDGRVTDAHRRILSTPFTREDVEEALFQMHPTKAPGLDGFPALFYQKFWPIV 476
+EE+ L V+D L P T+E+V+ A F ++P K PG DG YQ+FW +
Sbjct: 405 TVEELENLTP-LVSDQMNNNLLAPITKEEVQRATFSINPHKCPGPDGMNGFLYQQFWETM 463
Query: 477 GDDISEFCLQVLQ-GDISPGMVNQTLIVLIPKVKKAVFANQYRPISLCNVIFKIISKTIA 535
GD I+E + G I GM N+T I LIPK+ KA +RPISLCNVI+K+I K +A
Sbjct: 464 GDQITEMVQAFFRSGSIEEGM-NKTNICLIPKILKAEKMTDFRPISLCNVIYKVIGKLMA 522
Query: 536 NRMKLILHDLISEAQSAFVPGRLITDNALIAFECFHYMKKRISGRSGMMALKLDMSKAYD 595
NR+K IL LISE Q+AFV GRLI+DN LIA E H + +A+K D+SKAYD
Sbjct: 523 NRLKKILPSLISETQAAFVKGRLISDNILIAHELLHALSSNNKCSEEFIAIKTDISKAYD 582
Query: 596 RVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVNFAVMLNGNPQPTFAPHRGLRQGDPLSPY 655
RVEWPFL + +GF HW+ L+M CV +V + V++NG P P RGLRQGDPLSPY
Sbjct: 583 RVEWPFLEKAMRGLGFADHWIRLIMECVKSVRYQVLINGTPHGEIIPSRGLRQGDPLSPY 642
Query: 656 LFILCGEAFSALIQKSISTSALHGIKISRSAPVISHLLFADDSVLFARATVQEAECIKNI 715
LF++C E ++Q + + + G+K++R AP ISHLLFADDS+ + + + I I
Sbjct: 643 LFVICTEMLVKMLQSAEQKNQITGLKVARGAPPISHLLFADDSMFYCKVNDEALGQIIRI 702
Query: 716 LATYERASGQKINLEKSMLSVSRNVPENCFVDLQLLLGVKAVDSYDKYLGLPTIIGKSKT 775
+ Y ASGQ++N KS + +++ E ++ LG++ YLGLP SK
Sbjct: 703 IEEYSLASGQRVNYLKSSIYFGKHISEERRCLVKRKLGIEREGGEGVYLGLPESFQGSKV 762
Query: 776 QIFRFVKERVWKKLKGWKEQTLSRAGREVLIKSVAQAIPSYVMSCFVLPDGLCNEIESMI 835
++K+R+ KK+ GW+ LS G+E+L+K+VA A+P+Y MSCF +P +C +IES++
Sbjct: 763 ATLSYLKDRLGKKVLGWQSNFLSPGGKEILLKAVAMALPTYTMSCFKIPKTICQQIESVM 822
Query: 836 SRFYWGGDASKRGIHWVKWKTMCQPKHVGGLGFRDFKSFNLALVAKNWWRIYNYPDSLLG 895
+ F+W RG+HW W + +PK VGGLGF++ ++FN+AL+ K WR+ DSL+
Sbjct: 823 AEFWWKNKKEGRGLHWKAWCHLSRPKAVGGLGFKEIEAFNIALLGKQLWRMITEKDSLMA 882
Query: 896 KVFKSVYFSSGRMECAKKGYRPSYAWSRILKTSAMIQKGSCWRIGEGSRVRIWEDNWL-- 953
KVFKS YFS A G RPS+AW I + +I++G IG G + +W D W+
Sbjct: 883 KVFKSRYFSKSDPLNAPLGSRPSFAWKSIYEAQVLIKQGIRAVIGNGETINVWTDPWIGA 942
Query: 954 --ANGPPVNFRQDVVDELGLTK---VADLMLSGNRGWNVPLIEWTFCPATASRIMSVPLP 1008
A R +V + V DL+L R WN L+ F T I+++
Sbjct: 943 KPAKAAQAVKRSHLVSQYAANSIHVVKDLLLPDGRDWNWNLVSLLFPDNTQENILALRPG 1002
Query: 1009 RQPESDQLFWLGTADGLYSVKTGYEFLQEVVAQ-DSPSSSLSRGLDQTLWKRFWKAPSMP 1067
+ D+ W + G YSVK+GY + E++ Q ++P L LD ++++ WK P
Sbjct: 1003 GKETRDRFTWEYSRSGHYSVKSGYWVMTEIINQRNNPQEVLQPSLD-PIFQQIWKLDVPP 1061
Query: 1068 RVREVSWRVCTGALPVRTKLRQRGVDVDSSCPLCGLEDETVDHLFLGCNITRGCWFASPM 1127
++ WR L V + L R + + SC C ETV+HL C R W SP+
Sbjct: 1062 KIHHFLWRCVNNCLSVASNLAYRHLAREKSCVRCPSHGETVNHLLFKCPFARLTWAISPL 1121
Query: 1128 -----GVRVDPSLKMVDFLTMVIRDM--DLEVVACVQRFLFAIWEARNKHIFEEKLFCIA 1180
G + + + + V + + + A + L+ +W+ RN +F+ + F
Sbjct: 1122 PAPPGGEWAESLFRNMHHVLSVHKSQPEESDHHALIPWILWRLWKNRNDLVFKGREFTAP 1181
Query: 1181 GVLDRAASLV---SQSTLPFTLAASQEKDNLKRWRRPAENVVKVNVDAAVGQD-RFAGFG 1236
V+ +A + + P S +D +W+ P+ VK N D A +D G G
Sbjct: 1182 QVILKATEDMDAWNNRKEPQPQVTSSTRDRCVKWQPPSHGWVKCNTDGAWSKDLGNCGVG 1241
Query: 1237 LVARDHAGEVLAAAAKYPIMVLSPTVAEALSLRWAMDLAIQLGFRRVQFETDCLLLQQAW 1296
V R+H G +L + S E +LRWA+ + +RRV FE+D L
Sbjct: 1242 WVLRNHTGRLLWLGLRALPSQQSVLETEVEALRWAVLSLSRFNYRRVIFESDSQYLVSLI 1301
Query: 1297 KKVSGNSYLFSIIKDCHKLVVFFDHVDLTFVRREGNSCADYLARNASS 1344
+ L I+D L+ F+ V F RREGN+ AD AR + S
Sbjct: 1302 QNEMDIPSLAPRIQDIRNLLRHFEEVKFQFTRREGNNVADRTARESLS 1349
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 703 bits (1815), Expect = 0.0
Identities = 428/1368 (31%), Positives = 686/1368 (49%), Gaps = 69/1368 (5%)
Query: 6 MRLLSWNCRGLGNLEAVRALRGLIHSQAPDIVFLMETKL---FSFEMQRHRGMGGLSNIF 62
MR+ WNC+GLG VR L + D++FL+ETK ++ ++ G + I
Sbjct: 363 MRVGFWNCQGLGQPLTVRRLEEVQRVYFLDMLFLIETKQQDNYTRDLGVKMGFEDMCIIS 422
Query: 63 PVQCGRNRAGGLCLLWRSEVEVTIINASLHHILFTVVHSDSVTVPMQVYALYGFPELHLK 122
P R +GGL + W+ + + +I+ H + ++ + + +YG P +
Sbjct: 423 P----RGLSGGLVVYWKKHLSIQVIS---HDVRLVDLYVEYKNFNFYLSCIYGHPIPSER 475
Query: 123 DRTWEMIRSVRPMAPIPWLCIGDFNDILSPSDKLGGAPPDLARLQVATQACADCGLQRVD 182
WE ++ V PW+ GDFN+IL+ ++K GG + LQ T C ++ +
Sbjct: 476 HHLWEKLQRVSAHRSGPWMMCGDFNEILNLNEKKGGRRRSIGSLQNFTNMINCCNMKDLK 535
Query: 183 FSGPSFTWTNNRVGPGRVDERLDYALINQAWVNLWPVSSVSHLIRHQSDHNPVVLHCGSR 242
G ++W R ++ LD IN W +P L SDH PV++
Sbjct: 536 SKGNPYSWVGKRQNE-TIESCLDRVFINSDWQASFPAFETEFLPIAGSDHAPVIIDIAEE 594
Query: 243 RTETQ---RHRTRMFRFEEVWLESGEECAEIISEGW----SNTNLSILSRINLVGRSLDS 295
+ R+ R F+FE+ + + GW S+++ +++ + L
Sbjct: 595 VCTKRGQFRYDRRHFQFEDF--------VDSVQRGWNRGRSDSHGGYYEKLHCCRQELAK 646
Query: 296 WGREKYGELPKRIKEARAFLQRLQEE-VQTEQVVRATREAEKNLDVLLKQEEIVWSQRSR 354
W R ++I+ + + + + Q + R+ +L+ + EE+ W +SR
Sbjct: 647 WKRRTKTNTAEKIETLKYRVDAAERDHTLPHQTILRLRQ---DLNQAYRDEELYWHLKSR 703
Query: 355 ANWLKHGDRNTKFFHSKATQRRKRNLIEEIQDDNGRKFVRDADIARVLTSYFQEIFSTCN 414
W+ GDRNT FF++ R+ RN I+ I D G + RD I +V +YF ++F+T
Sbjct: 704 NRWMLLGDRNTMFFYASTKLRKSRNRIKAITDAQGIENFRDDTIGKVAENYFADLFTTTQ 763
Query: 415 PTGIEEVATLVDGRVTDAHRRILSTPFTREDVEEALFQMHPTKAPGLDGFPALFYQKFWP 474
+ EE+ + + +VT+ L T ++V +A+F + +APG DGF A FY W
Sbjct: 764 TSDWEEIISGIAPKVTEQMNHELLQSVTDQEVRDAVFAIGADRAPGFDGFTAAFYHHLWD 823
Query: 475 IVGDDISEFCLQVLQGDISPGMVNQTLIVLIPKVKKAVFANQYRPISLCNVIFKIISKTI 534
++G+D+ + D+ +NQT I LIPK+ + YRPISLC +KIISK +
Sbjct: 824 LIGNDVCLMVRHFFESDVMDNQINQTQICLIPKIIDPKHMSDYRPISLCTASYKIISKIL 883
Query: 535 ANRMKLILHDLISEAQSAFVPGRLITDNALIAFECFHYMKKRISGRSGMMALKLDMSKAY 594
R+K L D+IS++Q+AFVPG+ I+DN L+A E H +K R +SG +A+K D+SKAY
Sbjct: 884 IKRLKQCLGDVISDSQAAFVPGQNISDNVLVAHELLHSLKSRRECQSGYVAVKTDISKAY 943
Query: 595 DRVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVNFAVMLNGNPQPTFAPHRGLRQGDPLSP 654
DRVEW FL V+ +GF WV +M CVT+V++ V++NG+P P RG+RQGDPLSP
Sbjct: 944 DRVEWNFLEKVMIQLGFAPRWVKWIMTCVTSVSYEVLINGSPYGKIFPSRGIRQGDPLSP 1003
Query: 655 YLFILCGEAFSALIQKSISTSALHGIKISRSAPVISHLLFADDSVLFARATVQEAECIKN 714
YLF+ C E S +++K+ +HG+KI++ ISHLLFADDS+ F RA+ Q E +
Sbjct: 1004 YLFLFCAEVLSNMLRKAEVNKQIHGMKITKDCLAISHLLFADDSLFFCRASNQNIEQLAL 1063
Query: 715 ILATYERASGQKINLEKSMLSVSRNVPENCFVDLQLLLGVKAVDSYDKYLGLPTIIGKSK 774
I YE ASGQKIN KS + + +P L LLG+ V KYLGLP +G+ K
Sbjct: 1064 IFKKYEEASGQKINYAKSSIIFGQKIPTMRRQRLHRLLGIDNVRGGGKYLGLPEQLGRRK 1123
Query: 775 TQIFRFVKERVWKKLKGWKEQTLSRAGREVLIKSVAQAIPSYVMSCFVLPDGLCNEIESM 834
++F ++ +V ++ +GW LS AG+E++IK++A A+P Y M+CF+LP +CNEI S+
Sbjct: 1124 VELFEYIVTKVKERTEGWAYNYLSPAGKEIVIKAIAMALPVYSMNCFLLPTLICNEINSL 1183
Query: 835 ISRFYWGGDASKRGIHWVKWKTMCQPKHVGGLGFRDFKSFNLALVAKNWWRIYNYPDSLL 894
I+ F+WG ++ G LGF+D FN AL+AK WRI P SLL
Sbjct: 1184 ITAFWWG------------------KENEGDLGFKDLHQFNRALLAKQAWRILTNPQSLL 1225
Query: 895 GKVFKSVYFSSGRMECAKKGYRPSYAWSRILKTSAMIQKGSCWRIGEGSRVRIWEDNWLA 954
+++K +Y+ + A KG SY W+ I + ++Q+G R+G+G +IWED WL
Sbjct: 1226 ARLYKGLYYPNTTYLRANKGGHASYGWNSIQEGKLLLQQGLRVRLGDGQTTKIWEDPWLP 1285
Query: 955 NGPPVNFRQDVVDELGLTKVADLMLSGNRGWNVPLIEWTFCPATASRIMSVPLPRQPESD 1014
PP R ++DE KVADL R W+ + E P S+ L D
Sbjct: 1286 TLPPRPARGPILDE--DMKVADLWRENKREWDPVIFEGVLNPEDQQLAKSLYLSNYAARD 1343
Query: 1015 QLFWLGTADGLYSVKTGYEFLQEVVAQDSPSSSLSRGLDQTLWKRFWKAPSMPRVREVSW 1074
W T + Y+V++GY V + + G D L + W+ P+++ W
Sbjct: 1344 SYKWAYTRNTQYTVRSGYWVATHVNLTEEEIINPLEG-DVPLKQEIWRLKITPKIKHFIW 1402
Query: 1075 RVCTGALPVRTKLRQRGVDVDSSCPLCGLEDETVDHLFLGCNITRGCWFASPMG-----V 1129
R +GAL T+LR R + D +C C DET++H+ C+ + W ++
Sbjct: 1403 RCLSGALSTTTQLRNRNIPADPTCQRCCNADETINHIIFTCSYAQVVWRSANFSGSNRLC 1462
Query: 1130 RVDPSLKMVDFLTMVIRDMDLEVVACVQRF--LFAIWEARNKHIFEEK---LFCIAGVLD 1184
D + + + ++ +L ++ + F ++ +W++RN+++F++ + +A +
Sbjct: 1463 FTDNLEENIRLILQGKKNQNLPILNGLMPFWIMWRLWKSRNEYLFQQLDRFPWKVAQKAE 1522
Query: 1185 RAA-----SLVSQSTLPFTLAASQEK--DNLKRWRRPAENVVKVNVDAAVGQDR-FAGFG 1236
+ A ++V+ + + A S ++ K+W P E +K N D+ Q R + G
Sbjct: 1523 QEATEWVETMVNDTAISHNTAQSNDRPLSRSKQWSSPPEGFLKCNFDSGYVQGRDYTSTG 1582
Query: 1237 LVARDHAGEVLAAAAKYPIMVLSPTVAEALSLRWAMDLAIQLGFRRVQFETDCLLLQQAW 1296
+ RD G VL + S AEAL A+ + G+ V FE D L L
Sbjct: 1583 WILRDCNGRVLHSGCAKLQQSYSALQAEALGFLHALQMVWIRGYCYVWFEGDNLELTNLI 1642
Query: 1297 KKVSGNSYLFSIIKDCHKLVVFFDHVDLTFVRREGNSCADYLARNASS 1344
K + L +++ D + + +V RE N AD L + A+S
Sbjct: 1643 NKTEDHHLLETLLYDIRFWMTKLPFSSIGYVNRERNLAADKLTKYANS 1690
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 701 bits (1810), Expect = 0.0
Identities = 437/1311 (33%), Positives = 669/1311 (50%), Gaps = 60/1311 (4%)
Query: 34 PDIVFLMETKL---FSFEMQRHRGMGGLSNIFPVQCGRNRAGGLCLLWRSEVEVTIINAS 90
PDI+FLMETK F +++ G + + P R+GGL + W+S +E+ + A
Sbjct: 7 PDILFLMETKNSQDFVYKVFCWLGYDFIHTVEP----EGRSGGLAIFWKSHLEIEFLYAD 62
Query: 91 LHHILFTVVHSDSVTVPMQVYALYGFPELHLKDRTWEMIRSVRPMAPIPWLCIGDFNDIL 150
+ + V + V + +YG P H++ + WE + S+ W IGDFNDI
Sbjct: 63 KNLMDLQVSSRNKVWF---ISCVYGLPVTHMRPKLWEHLNSIGLKRAEAWCLIGDFNDIR 119
Query: 151 SPSDKLGGAPPDLARLQVATQACADCGLQRVDFSGPSFTWTNNRVGPGRVDERLDYALIN 210
S +KLGG + Q +C + + +G SFTW NR V +LD N
Sbjct: 120 SNDEKLGGPRRSPSSFQCFEHMLLNCSMHELGSTGNSFTWGGNR-NDQWVQCKLDRCFGN 178
Query: 211 QAWVNLWPVSSVSHLIRHQSDHNPVVLHCGSRRTETQRHRTRMFRFEEVWLESGEECAEI 270
AW +++P + L + SDH PV++ + T FR+++ L+ C E+
Sbjct: 179 PAWFSIFPNAHQWFLEKFGSDHRPVLV----KFTNDNELFRGQFRYDKR-LDDDPYCIEV 233
Query: 271 ISEGWSN-----TNLSILSRINLVGRSLDSWGREKYGELPKRIKEARAFLQRLQEEVQTE 325
I W++ T+ S S I R++ W RIK RL++++ E
Sbjct: 234 IHRSWNSAMSQGTHSSFFSLIEC-RRAISVWKHSSDTNAQSRIK-------RLRKDLDAE 285
Query: 326 QVVRAT-----REAEKNLDVLLKQEEIVWSQRSRANWLKHGDRNTKFFHSKATQRRKRNL 380
+ ++ + L + EE+ W Q+SR WL GD+NT FFH+ R +N
Sbjct: 286 KSIQIPCWPRIEYIKDQLSLAYGDEELFWRQKSRQKWLAGGDKNTGFFHATVHSERLKNE 345
Query: 381 IEEIQDDNGRKFVRDADIARVLTSYFQEIFSTCNPTGIEEVATLVDGRVTDAHRRILSTP 440
+ + D+N ++F R++D ++ +S+F+ +F++ + +VT L
Sbjct: 346 LSFLLDENDQEFTRNSDKGKIASSFFENLFTSTYILTHNNHLEGLQAKVTSEMNHNLIQE 405
Query: 441 FTREDVEEALFQMHPTKAPGLDGFPALFYQKFWPIVGDDISEFCLQVLQGDISPGMVNQT 500
T +V A+F ++ APG DGF ALF+Q+ W +V I + + P N T
Sbjct: 406 VTELEVYNAVFSINKESAPGPDGFTALFFQQHWDLVKHQILTEIFGFFETGVLPQDWNHT 465
Query: 501 LIVLIPKVKKAVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVPGRLIT 560
I LIPK+ + RPISLC+V++KIISK + R+K L ++S QSAFVP RLI+
Sbjct: 466 HICLIPKITSPQRMSDLRPISLCSVLYKIISKILTQRLKKHLPAIVSTTQSAFVPQRLIS 525
Query: 561 DNALIAFECFHYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVM 620
DN L+A E H ++ MA K DMSKAYDRVEWPFL +++T +GF W++ +M
Sbjct: 526 DNILVAHEMIHSLRTNDRISKEHMAFKTDMSKAYDRVEWPFLETMMTALGFNNKWISWIM 585
Query: 621 NCVTTVNFAVMLNGNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHGI 680
NCVT+V+++V++NG P P RG+RQGDPLSP LF+LC EA ++ K+ + GI
Sbjct: 586 NCVTSVSYSVLINGQPYGHIIPTRGIRQGDPLSPALFVLCTEALIHILNKAEQAGKITGI 645
Query: 681 KISRSAPVISHLLFADDSVLFARATVQEAECIKNILATYERASGQKINLEKSMLSVSRNV 740
+ ++HLLFADD++L +AT QE E + L+ Y + SGQ INL KS ++ +NV
Sbjct: 646 QFQDKKVSVNHLLFADDTLLMCKATKQECEELMQCLSQYGQLSGQMINLNKSAITFGKNV 705
Query: 741 PENCFVDLQLLLGVKAVDSYDKYLGLPTIIGKSKTQIFRFVKERVWKKLKGWKEQTLSRA 800
++ G+ KYLGLP + SK +F F+KE++ +L GW +TLS+
Sbjct: 706 DIQIKDWIKSRSGISLEGGTGKYLGLPECLSGSKRDLFGFIKEKLQSRLTGWYAKTLSQG 765
Query: 801 GREVLIKSVAQAIPSYVMSCFVLPDGLCNEIESMISRFYWGGDASKRGIHWVKWKTMCQP 860
G+EVL+KS+A A+P YVMSCF LP LC ++ +++ F+W KR IHW+ W+ + P
Sbjct: 766 GKEVLLKSIALALPVYVMSCFKLPKNLCQKLTTVMMDFWWNSMQQKRKIHWLSWQRLTLP 825
Query: 861 KHVGGLGFRDFKSFNLALVAKNWWRIYNYPDSLLGKVFKSVYFSSGRMECAKKGYRPSYA 920
K GG GF+D + FN AL+AK WR+ SL +VF+S YFS+ A +G RPSYA
Sbjct: 826 KDQGGFGFKDLQCFNQALLAKQAWRVLQEKGSLFSRVFQSRYFSNSDFLSATRGSRPSYA 885
Query: 921 WSRILKTSAMIQKGSCWRIGEGSRVRIWEDNWLANGP---PVNFRQDVVDELGLTKVADL 977
W IL ++ +G IG G + +W D WL +G P+N R+ + +L KV+ L
Sbjct: 886 WRSILFGRELLMQGLRTVIGNGQKTFVWTDKWLHDGSNRRPLNRRRFINVDL---KVSQL 942
Query: 978 MLSGNRGWNVPLIEWTFCPATASRIMSVPLPRQPESDQLFWLGTADGLYSVKTGYEFLQE 1037
+ +R WN+ ++ F P I+ P + D WL + +GLYSVKTGYEFL +
Sbjct: 943 IDPTSRNWNLNMLRDLF-PWKDVEIILKQRPLFFKEDSFCWLHSHNGLYSVKTGYEFLSK 1001
Query: 1038 VVAQDSPSSSLSRGLDQTLWKRFWKAPSMPRVREVSWRVCTGALPVRTKLRQRGVDVDSS 1097
V + + +L+ + W + P++R W+ GA+PV +LR RG+ D
Sbjct: 1002 QVHHRLYQEAKVKPSVNSLFDKIWNLHTAPKIRIFLWKALHGAIPVEDRLRTRGIRSDDG 1061
Query: 1098 CPLCGLEDETVDHLFLGCNITRGCW---FASPMGVRVDPSL-----KMVDFLTMVIRDMD 1149
C +C E+ET++H+ C + R W S G S+ +++D D+
Sbjct: 1062 CLMCDTENETINHILFECPLARQVWAITHLSSAGSEFSNSVYTNMSRLIDLTQQ--NDLP 1119
Query: 1150 LEVVACVQRFLFAIWEARNKHIFEEKLFCIAGVLDRAASLVSQSTLPFTLAASQEKD-NL 1208
+ L+ +W+ RN +FE K ++D+A + T + EK +
Sbjct: 1120 HHLRFVSPWILWFLWKNRNALLFEGKGSITTTLVDKAYEAYHEWFSAQTHMQNDEKHLKI 1179
Query: 1209 KRWRRPAENVVKVNVDAAVG-QDRFAGFGLVARDHAGEVLAAAAKYPIMVLSPTVAEALS 1267
+W P +K N+ A Q F+G V RD G+VL + + V SP A+ S
Sbjct: 1180 TKWCPPLPGELKCNIGFAWSKQHHFSGASWVVRDSQGKVLLHSRRSFNEVHSPYSAKIRS 1239
Query: 1268 LRWAMDLAIQLGFRRVQFETDCLLLQQA------WKKVSGN-SYLFSIIKD 1311
WA++ F RV F + + QA W + G+ S L S KD
Sbjct: 1240 WEWALESMTHHHFDRVIFASSTHEIIQALHKPHEWPLLLGDISELLSFTKD 1290
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 679 bits (1752), Expect = 0.0
Identities = 406/1151 (35%), Positives = 604/1151 (52%), Gaps = 55/1151 (4%)
Query: 230 SDHNPVVLHCGSRRTETQRHRTRMFRFEEVWLESGEECAEIISEGWSNTNL----SILSR 285
SDH+PV+ + +++ FRF++ W+ + E IS+GW+ + + +
Sbjct: 4 SDHSPVIATIADKIPRGKQN----FRFDKRWIGK-DGLLEAISQGWNLDSGFREGQFVEK 58
Query: 286 INLVGRSLDSW-------GREKYGELPKRIKEARAFLQRLQEEVQTEQVVRATREAEKNL 338
+ R++ W GR+ +L + A+ R +EE+ TE +R L
Sbjct: 59 LTNCRRAISKWRKSLIPFGRQTIEDLKAELDVAQRDDDRSREEI-TELTLR--------L 109
Query: 339 DVLLKQEEIVWSQRSRANWLKHGDRNTKFFHSKATQRRKRNLIEEIQDDNGRKFVRDADI 398
+ EE W Q+SR+ W+K GD N+KFFH+ QRR RN I + D+NG + D DI
Sbjct: 110 KEAYRDEEQYWYQKSRSLWMKLGDNNSKFFHALTKQRRARNRITGLHDENGIWSIEDDDI 169
Query: 399 ARVLTSYFQEIFSTCNPTGIEEVATLVDGRVTDAHRRILSTPFTREDVEEALFQMHPTKA 458
+ SYFQ +F+T NP +E V +TD +L+ T +V ALF +HP KA
Sbjct: 170 QNIAVSYFQNLFTTANPQVFDEALGEVQVLITDRINDLLTADATECEVRAALFMIHPEKA 229
Query: 459 PGLDGFPALFYQKFWPIVGDDISEFCLQVLQGDISPGMVNQTLIVLIPKVKKAVFANQYR 518
PG DG ALF+QK W I+ D+ LQ + +N T I LIPK ++ + R
Sbjct: 230 PGPDGMTALFFQKSWAIIKSDLLSLVNSFLQEGVFDKRLNTTNICLIPKTERPTRMTELR 289
Query: 519 PISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVPGRLITDNALIAFECFHYMKKRIS 578
PISLCNV +K+ISK + R+K +L +LISE QSAFV GRLI+DN LIA E FH ++ S
Sbjct: 290 PISLCNVGYKVISKILCQRLKTVLPNLISETQSAFVDGRLISDNILIAQEMFHGLRTNSS 349
Query: 579 GRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVNFAVMLNGNPQP 638
+ MA+K DMSKAYD+VEW F+ ++L MGF W++ +M C+TTV + V++NG P+
Sbjct: 350 CKDKFMAIKTDMSKAYDQVEWNFIEALLRKMGFCEKWISWIMWCITTVQYKVLINGQPKG 409
Query: 639 TFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHGIKISRSAPVISHLLFADDS 698
P RGLRQGDPLSPYLFILC E A I+K+ + + GIK++ +P +SHLLFADDS
Sbjct: 410 LIIPERGLRQGDPLSPYLFILCTEVLIANIRKAERQNLITGIKVATPSPAVSHLLFADDS 469
Query: 699 VLFARATVQEAECIKNILATYERASGQKINLEKSMLSVSRNVPENCFVDLQLLLGVKAVD 758
+ F +A ++ I IL YE SGQ+IN KS + V ++ D++L+LG+ +
Sbjct: 470 LFFCKANKEQCGIILEILKQYESVSGQQINFSKSSIQFGHKVEDSIKADIKLILGIHNLG 529
Query: 759 SYDKYLGLPTIIGKSKTQIFRFVKERVWKKLKGWKEQTLSRAGREVLIKSVAQAIPSYVM 818
YLGLP +G SKT++F FV++R+ ++ GW + LS+ G+EV+IKSVA +P YVM
Sbjct: 530 GMGSYLGLPESLGGSKTKVFSFVRDRLQSRINGWSAKFLSKGGKEVMIKSVAATLPRYVM 589
Query: 819 SCFVLPDGLCNEIESMISRFYWGGDASKRGIHWVKWKTMCQPKHVGGLGFRDFKSFNLAL 878
SCF LP + +++ S +++F+W + RG+HW+ W +C K GGLGFR+ FN AL
Sbjct: 590 SCFRLPKAITSKLTSAVAKFWWSSNGDSRGMHWMAWDKLCSSKSDGGLGFRNVDDFNSAL 649
Query: 879 VAKNWWRIYNYPDSLLGKVFKSVYFSSGRMECAKKGYRPSYAWSRILKTSAMIQKGSCWR 938
+AK WR+ PDSL KVFK YF + K Y PSY W ++ +++ KG R
Sbjct: 650 LAKQLWRLITAPDSLFAKVFKGRYFRKSNPLDSIKSYSPSYGWRSMISARSLVYKGLIKR 709
Query: 939 IGEGSRVRIWEDNWL-ANGP-PVNFRQDVVDELGLTKVADLMLSGNRGWNVPLIEWTFCP 996
+G G+ + +W D W+ A P P + +VD KV L+ S + WN+ L++ F P
Sbjct: 710 VGSGASISVWNDPWIPAQFPRPAKYGGSIVDPS--LKVKSLIDSRSNFWNIDLLKELFDP 767
Query: 997 ATASRIMSVPLPRQPESDQLFWLGTADGLYSVKTGYEFLQEVVAQDSPSSSLSRGLDQTL 1056
I ++P+ D L W T G Y+VK+GY D + G D T
Sbjct: 768 EDVPLISALPIGNPNMEDTLGWHFTKAGNYTVKSGYH----TARLDLNEGTTLIGPDLTT 823
Query: 1057 WKRF-WKAPSMPRVREVSWRVCTGALPVRTKLRQRGVDVDSSCPLCGLEDETVDHLFLGC 1115
K + WK P++R W++ +G +PV LR+RG+ D C CG +E+++H C
Sbjct: 824 LKAYIWKVQCPPKLRHFLWQILSGCVPVSENLRKRGILCDKGCVSCGASEESINHTLFQC 883
Query: 1116 NITRGCWFAS--PMGVRVDPSLKMVDFLTMVIRDMDLEVVACVQRFL-FAIWEARNKHIF 1172
+ R W S P + PS + L + + V + ++ + IW+ARN+ +F
Sbjct: 884 HPARQIWALSQIPTAPGIFPSNSIFTNLDHLFWRIPSGVDSAPYPWIIWYIWKARNEKVF 943
Query: 1173 EE-----KLFCIAGVLDRAASLVSQSTLPFTLAASQEKDNLKRWRRPAENVV----KVNV 1223
E + V + + +Q L S D+ R R +++ + +
Sbjct: 944 ENVDKDPMEILLLAVKEAQSWQEAQVELHSERHGSLSIDSRIRVRDVSQDTTFSGFRCFI 1003
Query: 1224 DAA-VGQDRFAGFGLVARDHAGEVLAAAAKYPIMVLSPTVAEALSLRWAMDLAIQLGFRR 1282
D + D+F+G G GE A LSP E +L WAM I +
Sbjct: 1004 DGSWKASDQFSGTGWFCLSSLGESPTMGAANVRRSLSPLHTEMEALLWAMKCMIGADNQN 1063
Query: 1283 VQFETDC----LLLQQAWKKVSGNSYLFSIIKDCHKLVVFFDHVDLTFVRREGNSCADYL 1338
V F TDC ++ + + + YL + D + F + L+ + R N AD L
Sbjct: 1064 VAFFTDCSDLVKMVSSPTEWPAFSVYLEELQSDREE----FTNFSLSLISRSANVKADKL 1119
Query: 1339 ARNASSYPDSV 1349
AR + P V
Sbjct: 1120 ARKIRTVPHHV 1130
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 669 bits (1725), Expect = 0.0
Identities = 424/1337 (31%), Positives = 668/1337 (49%), Gaps = 46/1337 (3%)
Query: 35 DIVFLMETKLFSFEMQRHRGMGGLSNIFPVQCGRNRAGGLCLLWRSEVEVTIINASLHHI 94
DI+FL+ET ++ + G N+ Q R+GGL L+W++ V +++I+ I
Sbjct: 411 DILFLVETLNQCDKVCKLAYDLGFPNVI-TQPPNGRSGGLALMWKNNVSLSLISQDERLI 469
Query: 95 LFTVVHSDSVTVPMQVYALYGFPELHLKDRTWEMIRSVRPMAPIPWLCIGDFNDILSPSD 154
V ++ + +YG P + + W+ + + WL +GDFN+ILS ++
Sbjct: 470 DSHVTFNNK---SFYLSCVYGHPTQSERHQLWQTLEHISDNRNAEWLLVGDFNEILSNAE 526
Query: 155 KLGGAPPDLARLQVATQACADCGLQRVDFSGPSFTWTNNRVGPGRVDERLDYALINQAWV 214
K+GG + + + C ++ + G F+W R V LD IN AW
Sbjct: 527 KIGGPMREEWTFRNFRNMVSHCDIEDMRSKGDRFSWVGER-HTHTVKCCLDRVFINSAWT 585
Query: 215 NLWPVSSVSHLIRHQSDHNPVVLHCGSRRTETQRHRTRMFRFEEVWLESGEECAEIISEG 274
+P + + L SDH PV++H E+ R+++FRF+ ++ I+
Sbjct: 586 ATFPYAEIEFLDFTGSDHKPVLVHFN----ESFPRRSKLFRFDNRLIDI-PTFKRIVQTS 640
Query: 275 WSNTNLS----ILSRINLVGRSLDSWGREKYGELPKRIKEARAFLQRLQEEVQTEQVVRA 330
W S I RI+ +++ +RIK+ ++ L R E T +V R
Sbjct: 641 WRTNRNSRSTPITERISSCRQAMARLKHASNLNSEQRIKKLQSSLNRAMES--TRRVDRQ 698
Query: 331 T-REAEKNLDVLLKQEEIVWSQRSRANWLKHGDRNTKFFHSKATQRRKRNLIEEIQDDNG 389
+ +++L EEI W Q+SR W+K GD+NT +FH+ R +N + I DD G
Sbjct: 699 LIPQLQESLAKAFSDEEIYWKQKSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQG 758
Query: 390 RKFVRDADIARVLTSYFQEIFSTCNPTGIEEVATLVD-----GRVTDAHRRILSTPFTRE 444
R F D +I +F IFST GI+ + +D VT+ L+ F+
Sbjct: 759 RMFTGDKEIGNHAQDFFTNIFST---NGIK--VSPIDFADFKSTVTNTVNLDLTKEFSDT 813
Query: 445 DVEEALFQMHPTKAPGLDGFPALFYQKFWPIVGDDISEFCLQVLQGDISPGMVNQTLIVL 504
++ +A+ Q+ KAPG DG A FY+ W IVG D+ + + +N T I +
Sbjct: 814 EIYDAICQIGDDKAPGPDGLTARFYKNCWDIVGYDVILEVKKFFETSFMKPSINHTNICM 873
Query: 505 IPKVKKAVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVPGRLITDNAL 564
IPK+ + YRPI+LCNV++K+ISK + NR+K L+ ++S++Q+AF+PGR+I DN +
Sbjct: 874 IPKITNPTTLSDYRPIALCNVLYKVISKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVM 933
Query: 565 IAFECFHYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVMNCVT 624
IA E H +K R MA+K D+SKAYDRVEW FL + + GF W+ +M V
Sbjct: 934 IAHEVMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCNKWIGWIMAAVK 993
Query: 625 TVNFAVMLNGNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHGIKISR 684
+V+++V++NG+P P RG+RQGDPLSPYLFILCG+ S LI S+ L G++I
Sbjct: 994 SVHYSVLINGSPHGYITPTRGIRQGDPLSPYLFILCGDILSHLINGRASSGDLRGVRIGN 1053
Query: 685 SAPVISHLLFADDSVLFARATVQEAECIKNILATYERASGQKINLEKSMLSVSRNVPENC 744
AP I+HL FADDS+ F +A V+ + +K++ YE SGQKIN++KSM++ V +
Sbjct: 1054 GAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSMITFGSRVYGST 1113
Query: 745 FVDLQLLLGVKAVDSYDKYLGLPTIIGKSKTQIFRFVKERVWKKLKGWKEQTLSRAGREV 804
L+ +L + KYLGLP G+ K ++F ++ +RV K+ W + LS AG+E+
Sbjct: 1114 QSRLKQILEIPNQGGGGKYLGLPEQFGRKKKEMFEYIIDRVKKRTSTWSARFLSPAGKEI 1173
Query: 805 LIKSVAQAIPSYVMSCFVLPDGLCNEIESMISRFYWGGDASKRGIHWVKWKTMCQPKHVG 864
++KSVA A+P Y MSCF LP G+ +EIES++ F+W +++RGI WV WK + K G
Sbjct: 1174 MLKSVALAMPVYAMSCFKLPKGIVSEIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEG 1233
Query: 865 GLGFRDFKSFNLALVAKNWWRIYNYPDSLLGKVFKSVYFSSGRMECAKKGYRPSYAWSRI 924
GLGFRD FN AL+AK WR+ YP+SL +V K+ YF + AK + SY W+ +
Sbjct: 1234 GLGFRDLAKFNDALLAKQAWRLIQYPNSLFARVMKARYFKDVSILDAKVRKQQSYGWASL 1293
Query: 925 LKTSAMIQKGSCWRIGEGSRVRIWEDNWLANGPPVNFR-QDVVDELGLTKVADLMLSGNR 983
L A+++KG+ IG+G +RI DN + + PP ++ E+ + + +
Sbjct: 1294 LDGIALLKKGTRHLIGDGQNIRIGLDNIVDSHPPRPLNTEETYKEMTINNLFE-RKGSYY 1352
Query: 984 GWNVPLIEWTFCPATASRIMSVPLPRQPESDQLFWLGTADGLYSVKTGYEFLQEVVAQDS 1043
W+ I + I + L + + D++ W G Y+V++GY L + +
Sbjct: 1353 FWDDSKISQFVDQSDHGFIHRIYLAKSKKPDKIIWNYNTTGEYTVRSGYWLLTHDPSTNI 1412
Query: 1044 PSSSLSRGLDQTLWKRFWKAPSMPRVREVSWRVCTGALPVRTKLRQRGVDVDSSCPLCGL 1103
P+ + G L R W P MP+++ WR + AL +L RG+ +D SCP C
Sbjct: 1413 PAINPPHG-SIDLKTRIWNLPIMPKLKHFLWRALSQALATTERLTTRGMRIDPSCPRCHR 1471
Query: 1104 EDETVDHLFLGCNITRGCWFASPMGVRVDPSLKMVDFLTMVIRDMDLEVVACVQRF---- 1159
E+E+++H C W S + + L DF + ++ + F
Sbjct: 1472 ENESINHALFTCPFATMAWRLSDSSL-IRNQLMSNDFEENISNILNFVQDTTMSDFHKLL 1530
Query: 1160 ----LFAIWEARNKHIFEE------KLFCIAGVLDRAASLVSQSTLPFTLAASQEKDNLK 1209
++ IW+ARN +F + K A +QS Q +N
Sbjct: 1531 PVWLIWRIWKARNNVVFNKFRESPSKTVLSAKAETHDWLNATQSHKKTPSPTRQIAENKI 1590
Query: 1210 RWRRPAENVVKVNVDAAVGQDRF-AGFGLVARDHAGEVLAAAAKYPIMVLSPTVAEALSL 1268
WR P VK N DA + A G + R+H G ++ + +P AE +L
Sbjct: 1591 EWRNPPATYVKCNFDAGFDVQKLEATGGWIIRNHYGTPISWGSMKLAHTSNPLEAETKAL 1650
Query: 1269 RWAMDLAIQLGFRRVQFETDCLLLQQAWKKVSGNSYLFSIIKDCHKLVVFFDHVDLTFVR 1328
A+ G+ +V E DC L +S +S L + ++D F + F+R
Sbjct: 1651 LAALQQTWIRGYTQVFMEGDCQTLINLINGISFHSSLANHLEDISFWANKFASIQFGFIR 1710
Query: 1329 REGNSCADYLARNASSY 1345
++GN A LA+ +Y
Sbjct: 1711 KKGNKLAHVLAKYGCTY 1727
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 665 bits (1717), Expect = 0.0
Identities = 418/1315 (31%), Positives = 656/1315 (49%), Gaps = 46/1315 (3%)
Query: 57 GLSNIFPVQCGRNRAGGLCLLWRSEVEVTIINASLHHILFTVVHSDSVTVPMQVYALYGF 116
G N+ Q R+GGL L+W++ V +++I+ I V ++ + +YG
Sbjct: 207 GFPNVI-TQPPNGRSGGLALMWKNNVSLSLISQDERLIDSHVTFNNK---SFYLSCVYGH 262
Query: 117 PELHLKDRTWEMIRSVRPMAPIPWLCIGDFNDILSPSDKLGGAPPDLARLQVATQACADC 176
P + + W+ + + WL +GDFN+ILS ++K+GG + + + C
Sbjct: 263 PTQSERHQLWQTLEHISDNRNAEWLLVGDFNEILSNAEKIGGPMREEWTFRNFRNMVSHC 322
Query: 177 GLQRVDFSGPSFTWTNNRVGPGRVDERLDYALINQAWVNLWPVSSVSHLIRHQSDHNPVV 236
++ + G F+W R V LD IN AW +P + L SDH PV+
Sbjct: 323 DIEDMRSKGDRFSWVGER-HTHTVKCCLDRVFINSAWTATFPYAETEFLDFTGSDHKPVL 381
Query: 237 LHCGSRRTETQRHRTRMFRFEEVWLESGEECAEIISEGWSNTNLS----ILSRINLVGRS 292
+H E+ R+++FRF+ ++ I+ W S I RI+ ++
Sbjct: 382 VHFN----ESFPRRSKLFRFDNRLIDI-PTFKRIVQTSWRTNRNSRSTPITERISSCRQA 436
Query: 293 LDSWGREKYGELPKRIKEARAFLQRLQEEVQTEQVVRAT-REAEKNLDVLLKQEEIVWSQ 351
+ +RIK+ ++ L R E T +V R + +++L EEI W Q
Sbjct: 437 MARLKHASNLNSEQRIKKLQSSLNRAMES--TRRVDRQLIPQLQESLAKAFSDEEIYWKQ 494
Query: 352 RSRANWLKHGDRNTKFFHSKATQRRKRNLIEEIQDDNGRKFVRDADIARVLTSYFQEIFS 411
+SR W+K GD+NT +FH+ R +N + I DD GR F D +I +F IFS
Sbjct: 495 KSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQGRMFTGDKEIGNHAQDFFTNIFS 554
Query: 412 TCNPTGIEEVATLVD-----GRVTDAHRRILSTPFTREDVEEALFQMHPTKAPGLDGFPA 466
T GI+ + +D VT+ L+ F+ ++ +A+ Q+ KAPG DG A
Sbjct: 555 T---NGIK--VSPIDFADFKSTVTNTVNLDLTKEFSDTEIYDAICQIGDDKAPGPDGLTA 609
Query: 467 LFYQKFWPIVGDDISEFCLQVLQGDISPGMVNQTLIVLIPKVKKAVFANQYRPISLCNVI 526
FY+ W IVG D+ + + +N T I +IPK+ + YRPI+LCNV+
Sbjct: 610 RFYKNCWDIVGYDVILEVKKFFETSFMKPSINHTNICMIPKITNPTTLSDYRPIALCNVL 669
Query: 527 FKIISKTIANRMKLILHDLISEAQSAFVPGRLITDNALIAFECFHYMKKRISGRSGMMAL 586
+K+ISK + NR+K L+ ++S++Q+AF+PGR+I DN +IA E H +K R MA+
Sbjct: 670 YKVISKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVMIAHEVMHSLKVRKRVSKTYMAV 729
Query: 587 KLDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVNFAVMLNGNPQPTFAPHRGL 646
K D+SKAYDRVEW FL + + GF W+ +M V +V+++V++NG+P P RG+
Sbjct: 730 KTDVSKAYDRVEWDFLETTMRLFGFCNKWIGWIMAAVKSVHYSVLINGSPHGYITPTRGI 789
Query: 647 RQGDPLSPYLFILCGEAFSALIQKSISTSALHGIKISRSAPVISHLLFADDSVLFARATV 706
RQGDPLSPYLFILCG+ S LI S+ L G++I AP I+HL FADDS+ F +A V
Sbjct: 790 RQGDPLSPYLFILCGDILSHLINGRASSGDLRGVRIGNGAPAITHLQFADDSLFFCQANV 849
Query: 707 QEAECIKNILATYERASGQKINLEKSMLSVSRNVPENCFVDLQLLLGVKAVDSYDKYLGL 766
+ + +K++ YE SGQKIN++KSM++ V + L+ +L + KYLGL
Sbjct: 850 RNCQALKDVFDVYEYYSGQKINVQKSMITFGSRVYGSTQSKLKQILEIPNQGGGGKYLGL 909
Query: 767 PTIIGKSKTQIFRFVKERVWKKLKGWKEQTLSRAGREVLIKSVAQAIPSYVMSCFVLPDG 826
P G+ K ++F ++ +RV K+ W + LS AG+E+++KSVA A+P Y MSCF LP G
Sbjct: 910 PEQFGRKKKEMFEYIIDRVKKRTSTWSARFLSPAGKEIMLKSVALAMPVYAMSCFKLPKG 969
Query: 827 LCNEIESMISRFYWGGDASKRGIHWVKWKTMCQPKHVGGLGFRDFKSFNLALVAKNWWRI 886
+ +EIES++ F+W +++RGI WV WK + K GGLGFRD FN AL+AK WR+
Sbjct: 970 IVSEIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWRL 1029
Query: 887 YNYPDSLLGKVFKSVYFSSGRMECAKKGYRPSYAWSRILKTSAMIQKGSCWRIGEGSRVR 946
YP+SL +V K+ YF + AK + SY W+ +L A+++KG+ IG+G +R
Sbjct: 1030 IQYPNSLFARVMKARYFKDVSILDAKVRKQQSYGWASLLDGIALLKKGTRHLIGDGQNIR 1089
Query: 947 IWEDNWLANGPPVNFR-QDVVDELGLTKVADLMLSGNRGWNVPLIEWTFCPATASRIMSV 1005
I DN + + PP ++ E+ + + + W+ I + I +
Sbjct: 1090 IGLDNIVDSHPPRPLNTEETYKEMTINNLFE-RKGSYYFWDDSKISQFVDQSDHGFIHRI 1148
Query: 1006 PLPRQPESDQLFWLGTADGLYSVKTGYEFLQEVVAQDSPSSSLSRGLDQTLWKRFWKAPS 1065
L + + D++ W G Y+V++GY L + + P+ + G L R W P
Sbjct: 1149 YLAKSKKPDKIIWNYNTTGEYTVRSGYWLLTHDPSTNIPAINPPHG-SIDLKTRIWNLPI 1207
Query: 1066 MPRVREVSWRVCTGALPVRTKLRQRGVDVDSSCPLCGLEDETVDHLFLGCNITRGCWFAS 1125
MP+++ WR + AL +L RG+ +D CP C E+E+++H C W+ S
Sbjct: 1208 MPKLKHFLWRALSQALATTERLTTRGMRIDPICPRCHRENESINHALFTCPFATMAWWLS 1267
Query: 1126 PMGVRVDPSLKMVDFLTMVIRDMDLEVVACVQRF--------LFAIWEARNKHIFEE--- 1174
+ + L DF + ++ + F ++ IW+ARN +F +
Sbjct: 1268 DSSL-IRNQLMSNDFEENISNILNFVQDTTMSDFHKLLPVWLIWRIWKARNNVVFNKFRE 1326
Query: 1175 ---KLFCIAGVLDRAASLVSQSTLPFTLAASQEKDNLKRWRRPAENVVKVNVDAAVGQDR 1231
K A +QS Q +N WR P VK N DA +
Sbjct: 1327 SPSKTVLSAKAETHDWLNATQSHKKTPSPTRQIAENKIEWRNPPATYVKCNFDAGFDVQK 1386
Query: 1232 F-AGFGLVARDHAGEVLAAAAKYPIMVLSPTVAEALSLRWAMDLAIQLGFRRVQFETDCL 1290
A G + R+H G ++ + +P AE +L A+ G+ +V E DC
Sbjct: 1387 LEATGGWIIRNHYGTPISWGSMKLAHTSNPLEAETKALLAALQQTWIRGYTQVFMEGDCQ 1446
Query: 1291 LLQQAWKKVSGNSYLFSIIKDCHKLVVFFDHVDLTFVRREGNSCADYLARNASSY 1345
L +S +S L + ++D F + F+RR+GN A LA+ +Y
Sbjct: 1447 TLINLINGISFHSSLANHLEDISFWANKFASIQFGFIRRKGNKLAHVLAKYGCTY 1501
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 657 bits (1694), Expect = 0.0
Identities = 422/1298 (32%), Positives = 643/1298 (49%), Gaps = 88/1298 (6%)
Query: 71 AGGLCLLWRSEVEVTIINASLHHILFTVVHSDSVTVPMQVYALYGFPELHLKDRTWEMIR 130
+GGL L W+ VEV I+ A+ + I D+ +V W+ I
Sbjct: 31 SGGLALYWKENVEVEILEAAPNFI-------DNRSV------------------FWDKIS 65
Query: 131 SVRPMAPIPWLCIGDFNDILSPSDKLGGAPPDLARLQVATQACADCGLQRVDFSGPSFTW 190
S+ WL GDFNDIL S+K GG + GL ++ +G S +W
Sbjct: 66 SLGAQRSSAWLLTGDFNDILDNSEKQGGPLRWEGFFLAFRSFVSQNGLWDINHTGNSLSW 125
Query: 191 TNNRVGPGRVDERLDYALINQAWVNLWPVSSVSHLIRHQSDHNPVVLHCGSRRTETQRHR 250
R + RLD AL N +W L+P+S +L SDH P+V + G+ + R
Sbjct: 126 RGTRYSHF-IKSRLDRALGNCSWSELFPMSKCEYLRFEGSDHRPLVTYFGAPPLK----R 180
Query: 251 TRMFRFEEVWLESGEECAEIISEGWSNTNL-SILSRINLVGRSLDSWGREKYGELPKRIK 309
++ FRF+ L EE ++ E W S+L +I+ +S+ W +E+ K IK
Sbjct: 181 SKPFRFDRR-LREKEEIRALVKEVWELARQDSVLYKISRCRQSIIKWTKEQNSNSAKAIK 239
Query: 310 EARAFLQR-LQEEVQTEQVVRATREAEKNLDVLLKQEEIVWSQRSRANWLKHGDRNTKFF 368
+A+ L+ L ++ ++ + + L+ +QEE+ W Q SR WL GDRN +F
Sbjct: 240 KAQQALESALSADIPDPSLIGSITQ---ELEAAYRQEELFWKQWSRVQWLNSGDRNKGYF 296
Query: 369 HSKATQRRKRNLIEEIQDDNGRKFVRDADIARVLTSYFQEIFSTCNPTGIEEVATLVDGR 428
H+ RR N + I+D +G++F + IA ++SYFQ IF+T N + ++ V +
Sbjct: 297 HATTRTRRMLNNLSVIEDGSGQEFHEEEQIASTISSYFQNIFTTSNNSDLQVVQEALSPI 356
Query: 429 VTDAHRRILSTPFTREDVEEALFQMHPTKAPGLDGFPALFYQKFWPIVGDDISEFCLQVL 488
++ L + +++EALF + KAPG DGF A F+ +W I+ D+S
Sbjct: 357 ISSHCNEELIKISSLLEIKEALFSISADKAPGPDGFSASFFHAYWDIIEADVSRDIRSFF 416
Query: 489 QGDISPGMVNQTLIVLIPKVKKAVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISE 548
+N+T + LIPK+ + YRPI+LCNV +KI++K + R++ L +LIS
Sbjct: 417 VDSCLSPRLNETHVTLIPKISAPRKVSDYRPIALCNVQYKIVAKILTRRLQPWLSELISL 476
Query: 549 AQSAFVPGRLITDNALIAFECFHYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTH 608
QSAFVPGR I DN LI E H+++ + + MA+K DMSKAYDR++W FL+ VL
Sbjct: 477 HQSAFVPGRAIADNVLITHEILHFLRVSGAKKYCSMAIKTDMSKAYDRIKWNFLQEVLMR 536
Query: 609 MGFPVHWVNLVMNCVTTVNFAVMLNGNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALI 668
+GF W+ VM CV TV+++ ++NG+PQ + P RGLRQGDPLSPYLFILC E S L
Sbjct: 537 LGFHDKWIRWVMQCVCTVSYSFLINGSPQGSVVPSRGLRQGDPLSPYLFILCTEVLSGLC 596
Query: 669 QKSISTSALHGIKISRSAPVISHLLFADDSVLFARATVQEAECIKNILATYERASGQKIN 728
+K+ + GI+++R +P ++HLLFADD++ F + + NIL YE ASGQ IN
Sbjct: 597 RKAQEKGVMVGIRVARGSPQVNHLLFADDTMFFCKTNPTCCGALSNILKKYELASGQSIN 656
Query: 729 LEKSMLSVSRNVPENCFVDLQLLLGVKAVDSYDKYLGLPTIIGKSKTQIFRFVKERVWKK 788
L KS ++ S P++ ++L L + KYLGLP G+ K IF + +R+ ++
Sbjct: 657 LAKSAITFSSKTPQDIKRRVKLSLRIDNEGGIGKYLGLPEHFGRRKRDIFSSIVDRIRQR 716
Query: 789 LKGWKEQTLSRAGREVLIKSVAQAIPSYVMSCFVLPDGLCNEIESMISRFYWGGDASKRG 848
W + LS AG+++L+K+V ++PSY M CF LP LC +I+S+++RF+W KR
Sbjct: 717 SHSWSIRFLSSAGKQILLKAVLSSMPSYAMMCFKLPASLCKQIQSVLTRFWWDSKPDKRK 776
Query: 849 IHWVKWKTMCQPKHVGGLGFRDFKSFNLALVAKNWWRIYNYPDSLLGKVFKSVYF-SSGR 907
+ WV W + P + GGLGFR+ + AK WRI P SLL +V Y +S
Sbjct: 777 MAWVSWDKLTLPINEGGLGFREIE-------AKLSWRILKEPHSLLSRVLLGKYCNTSSF 829
Query: 908 MECAKKGYRPSYAWSRILKTSAMIQKGSCWRIGEGSRVRIWEDNWLAN-------GPPVN 960
M+C+ S+ W IL +++KG W IG+G + +W + WL+ GPP
Sbjct: 830 MDCSASPSFASHGWRGILAGRDLLRKGLGWSIGQGDSINVWTEAWLSPSSPQTPIGPPTE 889
Query: 961 FRQDVVDELGLTKVADLMLSGNRGWNVPLIEWTFCPATASRIMSVPLPRQPESDQLFWLG 1020
+D+ V DL+ + WNV I P +I + + P D L WL
Sbjct: 890 TNKDL-------SVHDLICHDVKSWNVEAIR-KHLPQYEDQIRKITINALPLQDSLVWLP 941
Query: 1021 TADGLYSVKTGYEFLQEVVAQDSPSSSLSRGLDQTLWKRFWKAPSMPRVREVSWRVCTGA 1080
G Y+ KTGY + P+S LD K WK + P+V+ W+ GA
Sbjct: 942 VKSGEYTTKTGYALAK---LNSFPASQ----LDFNWQKNIWKIHTSPKVKHFLWKAMKGA 994
Query: 1081 LPVRTKLRQRGVDVDSSCPLCGLEDETVDHLFLGCNITRGCWFASPM------GVRVDPS 1134
LPV L +R ++ + +C CG + E+ HL L C + W +P+ +
Sbjct: 995 LPVGEALSRRNIEAEVTCKRCG-QTESSLHLMLLCPYAKKVWELAPVLFNPSEATHSSVA 1053
Query: 1135 LKMVDFLTMV-IRDMDLEVVACVQRFLFAIWEARNKHIFE-----EKLFCIAGVLDRAAS 1188
L +VD MV + L L+ +W+ARN+ IF+ E+ + +LD A
Sbjct: 1054 LLLVDAKRMVALPPTGLGSAPLYPWLLWHLWKARNRLIFDNHSCSEEGLVLKAILDARAW 1113
Query: 1189 LVSQSTLPFTLAASQEKDNLKRWRRPAEN--VVKVNVDAAVGQDRFAGFGLVARDHAGEV 1246
+ +Q + + + P N V VDAA + G G +D
Sbjct: 1114 MEAQLLI-------HHPSPISDYPSPTPNLKVTSCFVDAAWTTSGYCGMGWFLQDPYKVK 1166
Query: 1247 LAAAAKYPIMVLSPTVAEALSLRWAMDLAIQLGFRRVQFETDCLLLQQAWKKVSGNSYLF 1306
+ V S +AE L++ A+ A+ G R++ +DC L L
Sbjct: 1167 IKENQSSSSFVGSALMAETLAVHLALVDALSTGVRQLNVFSDCKELISLLNSGKSIVELR 1226
Query: 1307 SIIKDCHKLVVFFDHVDLTFVRREGNSCADYLARNASS 1344
++ D +L V F H+ F+ R N AD LA++A S
Sbjct: 1227 GLLHDIRELSVSFTHLCFFFIPRLSNVVADSLAKSALS 1264
>At1g24640 hypothetical protein
Length = 1270
Score = 613 bits (1582), Expect = e-175
Identities = 374/1116 (33%), Positives = 575/1116 (51%), Gaps = 49/1116 (4%)
Query: 70 RAGGLCLLWRSEVEVTIINASLHHILFTVVHSDSVTVPMQVYALYGFPELHLKDRTWEMI 129
+ GGL LLW+S V+V + +++ V +V V +YG P+ + + WE I
Sbjct: 27 KCGGLALLWKSSVQVDLKFVD-KNLMDAQVQFGAVN--FCVSCVYGDPDRSKRSQAWERI 83
Query: 130 RSVRPMAPIPWLCIGDFNDILSPSDKLGGAPPDLARLQVATQACADCGLQRVDFSGPSFT 189
+ W GDFNDIL +K GG + + C L + G FT
Sbjct: 84 SRIGVGRRDKWCMFGDFNDILHNGEKNGGPRRSDLDCKAFNEMIKGCDLVEMPAHGNGFT 143
Query: 190 WTNNRVGPGRVDERLDYALINQAWVNLWPVSSVSHLIRHQSDHNPVVLHCGSRRTETQRH 249
W R G + RLD A N+ W +PVS+ + L SDH PV++ S +Q
Sbjct: 144 WAGRR-GDHWIQCRLDRAFGNKEWFCFFPVSNQTFLDFRGSDHRPVLIKLMS----SQDS 198
Query: 250 RTRMFRFEEVWLESGEECAEIISEGWSN----TNLSILSRINLVGRSLDSWGREKYGELP 305
FRF++ +L E+ E I WS TN+S+ R+ +SL SW ++
Sbjct: 199 YRGQFRFDKRFLFK-EDVKEAIIRTWSRGKHGTNISVADRLRACRKSLSSWKKQNNLNSL 257
Query: 306 KRIKEARAFLQRLQEEVQTEQVVRATREAEKNLDVLLKQEEIVWSQRSRANWLKHGDRNT 365
+I + A L++ Q V + + +K+L ++EE W Q+SR WL+ G+RN+
Sbjct: 258 DKINQLEAALEKEQSLVWP--IFQRVSVLKKDLAKAYREEEAYWKQKSRQKWLRSGNRNS 315
Query: 366 KFFHSKATQRRKRNLIEEIQDDNGRKFVRDADIARVLTSYFQEIFSTCNPTGIEEVATLV 425
K+FH+ Q R+R IE+++D NG +A V +YF +F + NP+G + + +
Sbjct: 316 KYFHAAVKQNRQRKRIEKLKDVNGNMQTSEAAKGEVAAAYFGNLFKSSNPSGFTDWFSGL 375
Query: 426 DGRVTDAHRRILSTPFTREDVEEALFQMHPTKAPGLDGFPALFYQKFWPIVGDDISEFCL 485
RV++ L + ++++EA+F + P APG DG ALF+Q +W VG+ ++
Sbjct: 376 VPRVSEVMNESLVGEVSAQEIKEAVFSIKPASAPGPDGMSALFFQHYWSTVGNQVTSEVK 435
Query: 486 QVLQGDISPGMVNQTLIVLIPKVKKAVFANQYRPISLCNVIFKIISKTIANRMKLILHDL 545
+ I P N T + LIPK + RPISLC+V++KIISK +A R++ L ++
Sbjct: 436 KFFADGIMPAEWNYTHLCLIPKTQHPTEMVDLRPISLCSVLYKIISKIMAKRLQPWLPEI 495
Query: 546 ISEAQSAFVPGRLITDNALIAFECFHYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSV 605
+S+ QSAFV RLITDN L+A E H +K S MA+K DMSKAYDRVEW +LRS+
Sbjct: 496 VSDTQSAFVSERLITDNILVAHELVHSLKVHPRISSEFMAVKSDMSKAYDRVEWSYLRSL 555
Query: 606 LTHMGFPVHWVNLVMNCVTTVNFAVMLNGNPQPTFAPHRGLRQGDPLSPYLFILCGEAFS 665
L +GF + WVN +M CV++V ++V++N P RGLRQGDPLSP+LF+LC E +
Sbjct: 556 LLSLGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRGLRQGDPLSPFLFVLCTEGLT 615
Query: 666 ALIQKSISTSALHGIKISRSAPVISHLLFADDSVLFARATVQEAECIKNILATYERASGQ 725
L+ K+ AL GI+ S + P++ HLLFADDS+ +A+ +++ ++ IL Y A+GQ
Sbjct: 616 HLLNKAQWEGALEGIQFSENGPMVHHLLFADDSLFLCKASREQSLVLQKILKVYGNATGQ 675
Query: 726 KINLEKSMLSVSRNVPENCFVDLQLLLGVKAVDSYDKYLGLPTIIGKSKTQIFRFVKERV 785
INL KS ++ V E ++ LG+ YLGLP SK + ++K+R+
Sbjct: 676 TINLNKSSITFGEKVDEQLKGTIRTCLGIFTEGGAGTYLGLPECFSGSKVDMLHYLKDRL 735
Query: 786 WKKLKGWKEQTLSRAGREVLIKSVAQAIPSYVMSCFVLPDGLCNEIESMISRFYWGGDAS 845
+KL W + LS+ G+EVL+KSVA A+P + MSCF LP C +ES ++ F+W
Sbjct: 736 KEKLDVWFTRCLSQGGKEVLLKSVALAMPVFAMSCFKLPITTCENLESAMASFWWDSCDH 795
Query: 846 KRGIHWVKWKTMCQPKHVGGLGFRDFKSFNLALVAKNWWRIYNYPDSLLGKVFKSVYFSS 905
R IHW W+ +C PK GGLGFRD +SFN AL+AK WR+ ++PD LL ++ KS YF +
Sbjct: 796 SRKIHWQSWERLCLPKDSGGLGFRDIQSFNQALLAKQAWRLLHFPDCLLSRLLKSRYFDA 855
Query: 906 GRMECAKKGYRPSYAWSRILKTSAMIQKGSCWRIGEGSRVRIWEDNWL-ANGPPVNFRQD 964
A RPS+ W IL ++ KG R+G+G+ + +W D W+ NG +R++
Sbjct: 856 TDFLDAALSQRPSFGWRSILFGRELLSKGLQKRVGDGASLFVWIDPWIDDNGFRAPWRKN 915
Query: 965 VVDELGLTKVADLMLSGNRGWNVPLIEWTFCPATASRIMSVPLPRQPESDQLFWLGTADG 1024
++ ++ L KV L+ W+ ++ F P RI ++ P ++D W G
Sbjct: 916 LIYDVTL-KVKALLNPRTGFWDEEVLHDLFLPEDILRIKAIK-PVISQADFFVWKLNKSG 973
Query: 1025 LYSVKTGYEFLQEVVAQDSPSSSLSRGLDQTLWKRFWKAPSMPRVREVSWRVCTGALPVR 1084
+SVK+ Y + +Q+ S + L + W + P+++ W+V
Sbjct: 974 DFSVKSAYWLAYQTKSQNLRSEVSMQPSTLGLKTQVWNLQTDPKIKIFLWKV-------- 1025
Query: 1085 TKLRQRGVDVDSSCPLCGLEDETVDHLFLGCNITRGCWFAS-----PMGVRVDPSLKMVD 1139
CG E+ +H C ++R W S P G ++
Sbjct: 1026 ----------------CGELGESTNHTLFLCPLSRQIWALSDYPFPPDGFSNGSIYSNIN 1069
Query: 1140 FL--TMVIRDMDLEVVACVQRFLFAIWEARNKHIFE 1173
L ++ + + L+ IW+ RN IFE
Sbjct: 1070 HLLENKDNKEWPINLRKIFPWILWRIWKNRNSFIFE 1105
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 604 bits (1558), Expect = e-173
Identities = 391/1204 (32%), Positives = 609/1204 (50%), Gaps = 40/1204 (3%)
Query: 147 NDILSPSDKLGGAPPDLARLQVATQACADCGLQRVDFSGPSFTWTNNRVGPGRVDERLDY 206
N+IL S+K GG P D + GL + +SG F+W R V +RLD
Sbjct: 36 NEILDNSEKRGGPPRDQGSFIDFRSFISKNGLWDLKYSGNPFSWRGMRYD-WFVRQRLDR 94
Query: 207 ALINQAWVNLWPVSSVSHLIRHQSDHNPVVLHCGSRRTETQRHRTRMFRFEEVWLESGEE 266
A+ N +W+ +P +L SDH P+V+ R + R FRF+ L +
Sbjct: 95 AMSNNSWLESFPSGRSEYLRFEGSDHRPLVVFVDEARVK----RRGQFRFDNR-LRDNDV 149
Query: 267 CAEIISEGWSNT-NLSILSRINLVGRSLDSWGREKYGELPKRIKEARAFLQRLQEEVQTE 325
+I E W+N + S+L+++N R + +W R + + I++ Q+ EE T
Sbjct: 150 VNALIQETWTNAGDASVLTKMNQCRREIINWTRLQNLNSAELIEKT----QKALEEALTA 205
Query: 326 QVVRATR--EAEKNLDVLLKQEEIVWSQRSRANWLKHGDRNTKFFHSKATQRRKRNLIEE 383
T L+ K EE W QRSR WL GDRNT +FH+ RR +N +
Sbjct: 206 DPPNPTTIGALTATLEHAYKLEEQFWKQRSRVLWLHSGDRNTGYFHAVTRNRRTQNRLTV 265
Query: 384 IQDDNGRKFVRDADIARVLTSYFQEIFSTCNPTGIEEVATLVDGRVTDAHRRILSTPFTR 443
++D NG + I+++++ YFQ+IF++ + V ++ V+ L+
Sbjct: 266 MEDINGVAQHEEHQISQIISGYFQQIFTSESDGDFSVVDEAIEPMVSQGDNDFLTRIPND 325
Query: 444 EDVEEALFQMHPTKAPGLDGFPALFYQKFWPIVGDDISEFCLQVLQGDISPGMVNQTLIV 503
E+V++A+F ++ +KAPG DGF A FY +W I+ D+ P +N+T I
Sbjct: 326 EEVKDAVFSINASKAPGPDGFTAGFYHSYWHIISTDVGREIRLFFTSKNFPRRMNETHIR 385
Query: 504 LIPKVKKAVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVPGRLITDNA 563
LIPK YRPI+LCN+ +KI++K + RM+LIL LISE QSAFVPGR+I+DN
Sbjct: 386 LIPKDLGPRKVADYRPIALCNIFYKIVAKIMTKRMQLILPKLISENQSAFVPGRVISDNV 445
Query: 564 LIAFECFHYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVMNCV 623
LI E H+++ + + MA+K DMSKAYDRVEW FL+ VL GF W++ V+ CV
Sbjct: 446 LITHEVLHFLRTSSAKKHCSMAVKTDMSKAYDRVEWDFLKKVLQRFGFHSIWIDWVLECV 505
Query: 624 TTVNFAVMLNGNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHGIKIS 683
T+V+++ ++NG PQ P RGLRQGDPLSP LFILC E S L ++ L G+++S
Sbjct: 506 TSVSYSFLINGTPQGKVVPTRGLRQGDPLSPCLFILCTEVLSGLCTRAQRLRQLPGVRVS 565
Query: 684 RSAPVISHLLFADDSVLFARATVQEAECIKNILATYERASGQKINLEKSMLSVSRNVPEN 743
+ P ++HLLFADD++ F+++ + + IL+ Y +ASGQ IN KS ++ S P +
Sbjct: 566 INGPRVNHLLFADDTMFFSKSDPESCNKLSEILSRYGKASGQSINFHKSSVTFSSKTPRS 625
Query: 744 CFVDLQLLLGVKAVDSYDKYLGLPTIIGKSKTQIFRFVKERVWKKLKGWKEQTLSRAGRE 803
++ +L ++ KYLGLP G+ K IF + +++ +K W + LS+AG++
Sbjct: 626 VKGQVKRILKIRKEGGTGKYLGLPEHFGRRKRDIFGAIIDKIRQKSHSWASRFLSQAGKQ 685
Query: 804 VLIKSVAQAIPSYVMSCFVLPDGLCNEIESMISRFYWGGDASKRGIHWVKWKTMCQPKHV 863
V++K+V ++P Y MSCF LP LC +I+S+++RF+W R WV W + PK+
Sbjct: 686 VMLKAVLASMPLYSMSCFKLPSALCRKIQSLLTRFWWDTKPDVRKTSWVAWSKLTNPKNA 745
Query: 864 GGLGFRDFKSFNLALVAKNWWRIYNYPDSLLGKVFKSVY-FSSGRMECAKKGYRPSYAWS 922
GGLGFRD + N +L+AK WR+ N P+SLL ++ Y SS MEC K +PS+ W
Sbjct: 746 GGLGFRDIERCNDSLLAKLGWRLLNSPESLLSRILLGKYCHSSSFMEC-KLPSQPSHGWR 804
Query: 923 RILKTSAMIQKGSCWRIGEGSRVRIWEDNWLANGPPVNFRQDVVDELGLTKVADLMLSGN 982
I+ ++++G W I G +V IW D WL+ P+ + E +V+ L+
Sbjct: 805 SIIAGREILKEGLGWLITNGEKVSIWNDPWLSISKPLVPIGPALREHQDLRVSALINQNT 864
Query: 983 RGWNVPLIEWTFCPATASRIMSVPLPRQPESDQLFWLGTADGLYSVKTGYEFLQEVVAQD 1042
W+ I P + I +P P D+L WL G Y+ ++GY + V +
Sbjct: 865 LQWDWNKIA-VILPNYENLIKQLPAPSSRGVDKLAWLPVKSGQYTSRSGYG-IASVASIP 922
Query: 1043 SPSSSLSRGLDQTLWK-RFWKAPSMPRVREVSWRVCTGALPVRTKLRQRGVDVDSSCPLC 1101
P + + W+ WK ++P+++ + W+ ALPV +L +R + ++C C
Sbjct: 923 IPQTQFN-------WQSNLWKLQTLPKIKHLMWKAAMEALPVGIQLVRRHISPSAACHRC 975
Query: 1102 GLEDETVDHLFLGCNITRGCWFASPM-GVRVDPSLKMVDFLTMVIRDMDLEVVACVQRFL 1160
G + T HLF C W +P+ V P M+D L+++ + + L L
Sbjct: 976 GAPESTT-HLFFHCEFAAQVWELAPLQETTVPPGSSMLDALSLLKKAIILPPTGVTSAAL 1034
Query: 1161 FAIWEARNKHIFEEKLFCIAGVLDRAASLVSQSTLPFTLAASQEKDNLKRWRRPAENVVK 1220
F W I+ + +LD A +Q LP T L R +
Sbjct: 1035 FP-WIC---GIYGKLGTMTRAILDALAWQSAQRCLPKTRNVVHPISQLPVLR----SGYF 1086
Query: 1221 VNVDAA-VGQDRFAGFGLVARDHAGEVLAAAAKYPI---MVLSPTVAEALSLRWAMDLAI 1276
VDAA + Q AG G V + A + A Y + S AEA +++ A+ A+
Sbjct: 1087 CFVDAAWIAQSSLAGSGWVFQS-ATALEKETATYSAGCRRLPSALSAEAWAIKSALLHAL 1145
Query: 1277 QLGFRRVQFETDCLLLQQAWKKVSGNSYLFSIIKDCHKLVVFFDHVDLTFVRREGNSCAD 1336
QLG + +D + A + ++ ++ + L V F + F+ R N+ AD
Sbjct: 1146 QLGRTDLMVLSDSKSVVDALTSNISINEIYGLLMEIRALRVSFHSLCFNFISRSANAIAD 1205
Query: 1337 YLAR 1340
A+
Sbjct: 1206 ATAK 1209
>At2g16680 putative non-LTR retroelement reverse transcriptase
Length = 1319
Score = 577 bits (1488), Expect = e-164
Identities = 379/1269 (29%), Positives = 610/1269 (47%), Gaps = 62/1269 (4%)
Query: 40 METKLFSFEMQRHRGMGGLSNIFPVQCGRNRAGGLCLLWRSEVEVTIINASLHHILFTVV 99
M + F ++ R G I PV ++GGL + W++ +E+ + + + V
Sbjct: 1 MNSDQFLLKVFRWLGYDYFHTIEPV----GKSGGLAIFWKNHLEIDFLFEDKNLLDLKVS 56
Query: 100 HSDSVTVPMQVYALYGFPELHLKDRTWEMIRSVRPMAPIPWLCIGDFNDILSPSDKLGGA 159
V +YG P LHL+ + + S+ W IGDFNDILS KLGG
Sbjct: 57 QGKKSWF---VSCVYGNPVLHLRYLLLDKLSSIGVQRNSAWCMIGDFNDILSNDGKLGGP 113
Query: 160 PPDLARLQVATQACADCGLQRVDFSGPSFTWTNNRVGPGRVDERLDYALINQAWVNLWPV 219
++ Q +C + ++ SG SFTW R + +LD N W ++
Sbjct: 114 SRLISSFQPFKNMLLNCDMHQMGSSGNSFTWGGTR-NDQWIQCKLDRCFGNSEWFTMFSN 172
Query: 220 SSVSHLIRHQSDHNPVVLHCGSRRTETQRHRTRMFRFEEVWLESGEECAEIISEGWSNTN 279
S L + S H PV+++ Q F +++ + E +CA W
Sbjct: 173 SHQWFLEKLGSHHRPVLVNF----VNDQEVFRGQFCYDKRFAED-PQCAASTLSSWIGNG 227
Query: 280 LSILS----RINLVGRSLDSWGREKYGELPKRIKEARAFLQRLQEEVQTEQVVRATREA- 334
+S +S R+ +++ W + RI R+ L +E +++Q +R +
Sbjct: 228 ISDVSSSMLRMVKCRKAISGWKKNSDFNAQNRILRLRSEL----DEEKSKQYPCWSRISV 283
Query: 335 -EKNLDVLLKQEEIVWSQRSRANWLKHGDRNTKFFHSKATQRRKRNLIEEIQDDNGRKFV 393
+ L V ++EE W +S+ WL GDRN+KFF + R +N + + D+NG +
Sbjct: 284 IQTQLGVAFREEESFWRLKSKDKWLFGGDRNSKFFQAMVKANRTKNSLRFLVDENGNEHT 343
Query: 394 RDADIARVLTSYFQEIFSTCNPTGIEEVATLVDGRVTDAHRRILSTPFTREDVEEALFQM 453
+ + + + YF+ +F + P + RV++ + L+ T ++ A+F +
Sbjct: 344 LNREKGNIASVYFENLFMSSYPANSQSALDGFKTRVSEEMNQELTQAVTELEIHSAVFSI 403
Query: 454 HPTKAPGLDGFPALFYQKFWPIVGDDISEFCLQVLQGDISPGMVNQTLIVLIPKVKKAVF 513
+ AP +K G D E L + + P N T + LIPK
Sbjct: 404 NVESAP----------EKLECCQGSDYIEI-LGFFETGVLPQEWNHTHLYLIPKFTNPQR 452
Query: 514 ANQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVPGRLITDNALIAFECFHYM 573
+ RPISLC+V++KIISK ++ ++K L ++S +QSAF RLI+DN LIA E H +
Sbjct: 453 MSDIRPISLCSVLYKIISKILSFKLKKHLPSIVSPSQSAFFAERLISDNILIAHEIVHSL 512
Query: 574 KKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVNFAVMLN 633
+ M K DMSKAYDRVEW FL+ +L +GF W++ +M CVT+V ++V++N
Sbjct: 513 RTNDKISKEFMVFKTDMSKAYDRVEWSFLQEILVALGFNDKWISWIMGCVTSVTYSVLIN 572
Query: 634 GNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHGIKISRSAPVISHLL 693
G P RG+RQGDP+SP+LF+LC EA ++Q++ ++ + GI+ + S P ++HLL
Sbjct: 573 GQHFGHITPERGIRQGDPISPFLFVLCTEALIHILQQAENSKKVSGIQFNGSGPSVNHLL 632
Query: 694 FADDSVLFARATVQEAECIKNILATYERASGQKINLEKSMLSVSRNVPENCFVDLQLLLG 753
F DD+ L RAT + E + L+ Y SGQ IN+EKS ++ V E+ ++ G
Sbjct: 633 FVDDTQLVCRATKSDCEQMMLCLSQYGHISGQLINVEKSSITFGVKVDEDTKRWIKNRSG 692
Query: 754 VKAVDSYDKYLGLPTIIGKSKTQIFRFVKERVWKKLKGWKEQTLSRAGREVLIKSVAQAI 813
+ KYLGLP + SK +F ++KE++ L GW ++TLS+ G+E+L+KS+A A+
Sbjct: 693 IHLEGGTGKYLGLPENLSGSKQDLFGYIKEKLQSHLSGWYDKTLSQGGKEILLKSIALAL 752
Query: 814 PSYVMSCFVLPDGLCNEIESMISRFYWGGDASKRGIHWVKWKTMCQPKHVGGLGFRDFKS 873
P Y+M+CF LP GLC ++ S++ F+W IHW+ K + PK +GG GF+D +
Sbjct: 753 PVYIMTCFRLPKGLCTKLTSVMMDFWWNSMEFSNKIHWIGGKKLTLPKSLGGFGFKDLQC 812
Query: 874 FNLALVAKNWWRIYNYPDSLLGKVFKSVYFSSGRMECAKKGYRPSYAWSRILKTSAMIQK 933
FN AL+AK WR+++ S++ ++FKS YF + A++G RPSY W IL ++
Sbjct: 813 FNQALLAKQAWRLFSDSKSIVSQIFKSRYFMNTDFLNARQGTRPSYTWRSILYGRELLNG 872
Query: 934 GSCWRIGEGSRVRIWEDNWLANG---PPVNFRQDVVDELGLTKVADLMLSGNRGWNVPLI 990
G IG G + +W D WL +G P+N + + KV+ L+ R WN+ +
Sbjct: 873 GLKRLIGNGEQTNVWIDKWLFDGHSRRPMNLHSLMNIHM---KVSHLIDPLTRNWNLKKL 929
Query: 991 EWTFCPATASRIMSVPLPRQPESDQLFWLGTADGLYSVKTGYEFLQEVVAQDSPSSSLSR 1050
F IM P D W GT +GLY+VK+GYE ++ +
Sbjct: 930 TELFHEKDVQLIMH-QRPLISSEDSYCWAGTNNGLYTVKSGYERSSRETFKNLFKEADVY 988
Query: 1051 GLDQTLWKRFWKAPSMPRVREVSWRVCTGALPVRTKLRQRGVDVDSSCPLCGLEDETVDH 1110
L+ + W ++P+++ W+ GAL V +LR RG+ C C E ET++H
Sbjct: 989 PSVNPLFDKVWSLETVPKIKVFMWKALKGALAVEDRLRSRGIRTADGCLFCKEEIETINH 1048
Query: 1111 LFLGCNITRGCWFASPMGVRVDPSLKMVDFLTMVIRDMDLEVVACVQRF----------- 1159
L C R W S + F T + +++ V+ Q F
Sbjct: 1049 LLFQCPFARQVWALSLI------QAPATGFGTSIFSNIN-HVIQNSQNFGIPRHMRTVSP 1101
Query: 1160 --LFAIWEARNKHIFEEKLFCIAGVLDRAASLVSQSTLPFTLAASQEKDNLKRWRRPAEN 1217
L+ IW+ RNK +F+ + ++ +A + ++ + +W P
Sbjct: 1102 WLLWEIWKNRNKTLFQGTGLTSSEIVAKAYEECNLWINAQEKSSGGVSPSEHKWNPPPAG 1161
Query: 1218 VVKVNVDAAVG-QDRFAGFGLVARDHAGEVLAAAAKYPIMVLSPTVAEALSLRWAMDLAI 1276
+K N+ A Q + AG V RD G+VL + + V S A+ S WA++
Sbjct: 1162 ELKCNIGVAWSRQKQLAGVSWVLRDSMGQVLLHSRRSYSQVYSLFDAKIKSWDWALESMD 1221
Query: 1277 QLGFRRVQF 1285
F +V F
Sbjct: 1222 HFHFDKVTF 1230
>At3g31420 hypothetical protein
Length = 1491
Score = 577 bits (1487), Expect = e-164
Identities = 400/1346 (29%), Positives = 618/1346 (45%), Gaps = 134/1346 (9%)
Query: 35 DIVFLMETKLFSFEMQRHRGMG---GLSNIFPVQCGRNRAGGLCLLWRSEVEVTIINASL 91
DI+ L ETK + R R +G G N V +GGL + W S V++ + ++
Sbjct: 210 DIICLSETKQ---QDDRIRDVGAELGFCNHVTVPPD-GLSGGLVVFWNSSVDIFLCFSNS 265
Query: 92 HHILFTVVHSDSVTVPMQVYALYGFPELHLKDRTWEMIRSVRPMAP-IPWLCIGDFNDIL 150
+ + +H S + +YG P + WE + + W +GDFN+IL
Sbjct: 266 NLV---DLHVKSNEGSFYLSFVYGHPNPSHRHHLWERLERLNTTRQGTAWFIMGDFNEIL 322
Query: 151 SPSDKLGGAPPDLARLQVATQACADCGLQRVDFSGPSFTWTNNRVGPGRVDERLDYALIN 210
S +K GG Q C + G F+W R G V LD + N
Sbjct: 323 SNREKRGGRLRPERTFQDFRNMVRGCNFTDLKSVGDRFSWAGKR-GDHHVTCSLDRTMAN 381
Query: 211 QAWVNLWPVSSVSHLIRHQSDHNPVVLHCGSRRTETQRHRTRMFRFEEVWLESGEECAEI 270
W L+P S + +SDH P+V + +++ E R F ++ L E ++
Sbjct: 382 NEWHTLFPESETVFMEYGESDHRPLVTNISAQKEE----RRGFFSYDSR-LTHKEGFKQV 436
Query: 271 ISEGWSNTNLSILSRINL------VGRSLDSWGREKYGELPKRIKEARAFLQRLQEEVQT 324
+ W N S +L +++ W + +RIK R +L + T
Sbjct: 437 VLNQWHRRNGSFEGDSSLNRKLVECRQAISRWKKNNRVNAEERIKIIR---HKLDRAIAT 493
Query: 325 EQVV-RATREAEKNLDVLLKQEEIVWSQRSRANWLKHGDRNTKFFHSKATQRRKRNLIEE 383
R R+ ++L+ EEI W +SR+ WL GDRNT++FHS RR RN +
Sbjct: 494 GTATPRERRQMRQDLNQAYADEEIYWQTKSRSRWLNAGDRNTRYFHSTTKTRRCRNRLLS 553
Query: 384 IQDDNGRKFVRDADIARVLTSYFQEIFSTCNPTGIE--EVATLVDGRVTDAHRRILSTPF 441
+QD +G D +IA+V +YF +++ + T + +V ++TD L P
Sbjct: 554 VQDSDGDICRGDENIAKVAINYFDDLYKSTPNTSLRYADVFQGFQQKITDEINEDLIRPV 613
Query: 442 TREDVEEALFQMHPTKAPGLDGFPALFYQKFWPIVGDDISEFCLQVLQGDISPGMVNQTL 501
T ++EE++F + P++ P DGF A FYQ+FWP + + + + + N T
Sbjct: 614 TELEIEESVFSVAPSRTPDPDGFTADFYQQFWPDIKQKVIDEVTRFFERSELDERHNHTN 673
Query: 502 IVLIPKVKKAVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVPGRLITD 561
+ LIPKV+ ++RPI+LCNV +KIISK + NR+K L I+E Q+AFVPGRLIT+
Sbjct: 674 LCLIPKVETPTTIAKFRPIALCNVSYKIISKILVNRLKKHLGGAITENQAAFVPGRLITN 733
Query: 562 NALIAFECFHYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVMN 621
NA+IA E ++ +K R + MALK D++KAYDR+EW FL + MGF W+ +M
Sbjct: 734 NAIIAHEVYYALKARKRQANSYMALKTDITKAYDRLEWDFLEETMRQMGFNTKWIERIMI 793
Query: 622 CVTTVNFAVMLNGNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHGIK 681
CVT V F+V++NG+P T P RG+R GDPLSPYLFILC E S +I+++ L GI+
Sbjct: 794 CVTMVRFSVLINGSPHGTIKPERGIRHGDPLSPYLFILCAEVLSHMIKQAEINKKLKGIR 853
Query: 682 ISRSAPVISHLLFADDSVLFARATVQEAECIKNILATYERASGQKINLEKSMLSVSRNVP 741
+S P ISHLLFADDS+ F A + IK T R
Sbjct: 854 LSTQGPFISHLLFADDSIFFTLANQRSCTAIKE-PDTKRR-------------------- 892
Query: 742 ENCFVDLQLLLGVKAVDSYDKYLGLPTIIGKSKTQIFRFVKERVWKKLKGWKEQTLSRAG 801
++ LLG+ KYLGLP K K ++F ++ E+V K +GW ++ LS+ G
Sbjct: 893 ------MRHLLGIHNEGGEGKYLGLPEQFNKKKKELFNYIIEKVKDKTQGWSKKFLSQGG 946
Query: 802 REVLIKSVAQAIPSYVMSCFVLPDGLCNEIESMISRFYWGGDASKRGIHWVKWKTMCQPK 861
+EVL+K+VA A+P Y M+ F L +C EI+S+++RF+W +G+HW WK M PK
Sbjct: 947 KEVLLKAVALAMPVYSMNIFKLTKEVCEEIDSLLARFWWSSGNETKGMHWFTWKRMSIPK 1006
Query: 862 HVGGLGFRDFKSFNLALVAKNWWRIYNYPDSLLGKVFKSVYFSSGRMECAKKGYRPSYAW 921
GGLGF++ ++FNLAL+ K W + +P+ L+ +V + YF + A +G R S+ W
Sbjct: 1007 KEGGLGFKELENFNLALLGKQTWHLLQHPNCLMARVLRGRYFPETNVMNAVQGRRASFVW 1066
Query: 922 SRILKTSAMIQKGSCWRIGEGSRVRIWEDNWL-ANGPPVNFRQ-DVVDELGLTKVADLML 979
IL +++KG +G+GS + W D WL + P ++Q D ++L + A
Sbjct: 1067 KSILHGRNLLKKGLRCCVGDGSLINAWLDPWLPLHSPRAPYKQEDAPEQLLVCSTA---- 1122
Query: 980 SGNRGWNVPLIEWTFCPATASRIMSVPLPRQPESDQLFWLGTADGLYSVKTGY---EFLQ 1036
D + W T DG+Y+VK+ Y L
Sbjct: 1123 --------------------------------RDDLIGWHYTKDGMYTVKSAYWLATHLP 1150
Query: 1037 EVVAQDSPSSSLSRGLDQTLWKRFWKAPSMPRVREVSWRVCTGALPVRTKLRQRGVDVDS 1096
P D L + WK + P+++ W++ +GA+ LR R ++ S
Sbjct: 1151 GTTGTHPPPG------DIKLKQLLWKTKTAPKIKHFCWKILSGAIATGEMLRYRHINKQS 1204
Query: 1097 SCPLCGLEDETVDHLFLGCNITRGCWFASPMGVRVDPSLKMVDFLTMV---IRDMDLEVV 1153
C C ++ET HLF C+ + W + + P+L D + + R M
Sbjct: 1205 ICKRCCRDEETSQHLFFECDYAKATWRGAGL-----PNLIFQDSIVTLEEKFRAMFTFNP 1259
Query: 1154 ACVQRF-------LFAIWEARNKHIFEEKLFCIAGVLDRAASLVSQSTLPF--------- 1197
+ + L+ +W +RN F++K + L Q L +
Sbjct: 1260 STTNYWRQLPFWILWRLWRSRNILTFQQKHI----PWEVTVQLAKQDALEWQDIEDRVQV 1315
Query: 1198 TLAASQEKDN---LKRWRRPAENVVKVNVDAAVGQDRFAGFGLVARDHAGEVLAAAAKYP 1254
S+ N RW RP K N D + + G V RD G+ +
Sbjct: 1316 INPLSRRHSNRYAANRWTRPVCGWKKCNYDGSYSTIINSKAGWVIRDEHGQFIGGGQAKG 1375
Query: 1255 IMVLSPTVAEALSLRWAMDLAIQLGFRRVQFETDCLLLQQAWKKVSGNSYLFSIIKDCHK 1314
+ + +L AM G +V FE D + + Q + +++ I+D
Sbjct: 1376 KHTNNALESALQALIIAMQSCWSHGHTKVCFEGDNIEVYQILNEGKARFDVYNWIRDIQA 1435
Query: 1315 LVVFFDHVDLTFVRREGNSCADYLAR 1340
F ++ R N AD LA+
Sbjct: 1436 WKRRFQECRFLWINRRNNKPADTLAK 1461
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 560 bits (1442), Expect = e-159
Identities = 422/1362 (30%), Positives = 644/1362 (46%), Gaps = 88/1362 (6%)
Query: 35 DIVFLMETKLFSFEMQRHRGMGGLSNIFPVQCGRNRAGGLCLLWRSEV-EVTIINASLHH 93
D++ + ET + R G N F V +GGL LLWR+ + EV++++++
Sbjct: 595 DVLAIFETHAGGDQASRICQGLGFENSFRVDAV-GHSGGLWLLWRTGIGEVSVVDSTDQF 653
Query: 94 ILFTVVHSDSVTVPMQVYALYGFPELHLKDRTWEMIRSVRPMAPIPWLCIGDFNDILSPS 153
I V+ + + +Y P + W+ + V P + GDFN I+
Sbjct: 654 IHAKDVNGKD---NVNLVVVYAAPTASRRSGLWDRLGDVIRSMDGPVVIGGDFNTIVRLD 710
Query: 154 DKLGGAPPDLARLQVATQACADCGLQRVDFSGPSFTWTNNRVGPGRVDERLDYALINQAW 213
++ GG + + D L + F G FTW R V +RLD L
Sbjct: 711 ERSGGNGRLSSDSLAFGEWINDHSLIDLGFKGNKFTWKRGREERFFVAKRLDRVLCCAHA 770
Query: 214 VNLWPVSSVSHLIRHQSDHNPVVLHCGSRRTETQRHRTRMFRFEEVWLESGEECAEIISE 273
W +SV HL SDH P+ + + R R R FRFE WL S E++
Sbjct: 771 RLKWQEASVLHLPFLASDHAPLYVQL-TPEVSGNRGR-RPFRFEAAWL-SHPGFKELLLT 827
Query: 274 GWSNTNLSILSRINLVGRSLDSWGREKYGELPKRIKEARAFLQRLQEEVQTEQVVRATRE 333
W N ++S P+ +K +Q L + Q++ +++ E
Sbjct: 828 SW-NKDIST----------------------PEALK-----VQELLDLHQSDDLLKKEEE 859
Query: 334 AEKNLDVLLKQEEIVWSQRSRANWLKHGDRNTKFFHSKATQRRKRNLIEEIQDDNGRKFV 393
K+ DV+L+QEE+VW Q+SR W HGDRNTKFFH+ RR+RN IE +QD++GR
Sbjct: 860 LLKDFDVVLEQEEVVWMQKSREKWFVHGDRNTKFFHTSTIIRRRRNQIEMLQDNDGRWLS 919
Query: 394 RDADIARVLTSYFQEIFSTCNPTGI-EEVATLVDGRVTDAHRRILSTPFTREDVEEALFQ 452
++ Y++ ++S + + E++ +++A L+ PF+ +VE A+
Sbjct: 920 NAQELETHAIDYYKRLYSLDDLDAVVEQLPQEGFTALSEADFSSLTKPFSPLEVEGAIRS 979
Query: 453 MHPTKAPGLDGFPALFYQKFWPIVGDDISEFCLQVLQGDISPGMVNQTLIVLIPKVKKAV 512
M KAPG DGF +FYQ+ W +VG+ +++F + P N L+VLI KV K
Sbjct: 980 MGKYKAPGPDGFQPVFYQQGWEVVGESVTKFVMDFFSSGSFPQETNDVLVVLIAKVLKPE 1039
Query: 513 FANQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVPGRLITDNALIAFECFHY 572
Q+RPISLCNV+FK I+K + R+K +++ LI AQ++F+PGRL TDN ++ E H
Sbjct: 1040 KITQFRPISLCNVLFKTITKVMVGRLKGVINKLIGPAQTSFIPGRLSTDNIVVVQEVVHS 1099
Query: 573 MKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVNFAVML 632
M+++ G G M LKLD+ KAYDR+ W L L G P WV +M CV + ++
Sbjct: 1100 MRRK-KGVKGWMLLKLDLEKAYDRIRWDLLEDTLKAAGLPGTWVQWIMKCVEGPSMRLLW 1158
Query: 633 NGNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHGIKISRSAPVISHL 692
NG F P RGLRQGDPLSPYLF+LC E LI+ SI+ IKIS+S P +SH+
Sbjct: 1159 NGEKTDAFKPLRGLRQGDPLSPYLFVLCIERLCHLIESSIAAKKWKPIKISQSGPRLSHI 1218
Query: 693 LFADDSVLFARATVQEAECIKNILATYERASGQKINLEKSMLSVSRNVPENCFVDLQLLL 752
FADD +LFA A++ + ++ +L + ASGQK++LEKS + S+NV + +
Sbjct: 1219 CFADDLILFAEASIDQIRVLRGVLEKFCGASGQKVSLEKSKIYFSKNVLRDLGKRISEES 1278
Query: 753 GVKAVDSYDKYLGLPTIIGKSKTQIFRFVKERVWKKLKGWKEQTLSRAGREVLIKSVAQA 812
G+KA KYLG+P + + + F V +R +L GWK + LS AGR L K+V +
Sbjct: 1279 GMKATKDLGKYLGVPILQKRINKETFGEVIKRFSSRLAGWKGRMLSFAGRLTLTKAVLTS 1338
Query: 813 IPSYVMSCFVLPDGLCNEIESMISRFYWGGDASKRGIHWVKWKTMCQPKHVGGLGFRDFK 872
I + MS LP + ++ + F WG K+ H V W +C P+ GGLG R
Sbjct: 1339 ILVHTMSTIKLPQSTLDGLDKVSRAFLWGSTLEKKKQHLVAWTRVCLPRREGGLGIRSAT 1398
Query: 873 SFNLALVAKNWWRIYNYPDSLLGKVFKSVY----FSSGRMECAKKGYRPSYAWSRI-LKT 927
+ N AL+AK WR+ N SL +V +S Y AK + S W + +
Sbjct: 1399 AMNKALIAKVGWRVLNDGSSLWAQVVRSKYKVGDVHDRNWTVAKSNW--SSTWRSVGVGL 1456
Query: 928 SAMIQKGSCWRIGEGSRVRIWEDNWLANGPPVNFRQDVVDELGLTKV----ADLMLSGNR 983
+I + W IG+G ++R W D WL+ P D + L L ++ DL G
Sbjct: 1457 RDVIWREQHWVIGDGRQIRFWTDRWLSETP---IADDSIVPLSLAQMLCTARDLWRDGT- 1512
Query: 984 GWNVPLIEWTFCPATASRIMSVPLPRQPES-DQLFWLGTADGLYSVKTGYEFLQEVVAQD 1042
GW++ I +++V + + D+L W T+DG ++VK+ + L D
Sbjct: 1513 GWDMSQIAPFVTDNKRLDLLAVIVDSVTGAHDRLAWGMTSDGRFTVKSAFAMLTN---DD 1569
Query: 1043 SPSSSLSRGLDQTLWKRFWKAPSMPRVREVSWRVCTGALPVRTKLRQRGVDVDSSCPLCG 1102
SP +S +L+ R WK + RVR W V A+ ++ ++R + C +C
Sbjct: 1570 SPRQDMS-----SLYGRVWKVQAPERVRVFLWLVVNQAIMTNSERKRRHLCDSDVCQVCR 1624
Query: 1103 LEDETVDHLFLGCNITRGCWFASPMGVRVDPSLKMVDFLTMVIRD-------MDLEVVAC 1155
E++ H+ C G W R+ P F TM + + L
Sbjct: 1625 GGIESILHVLRDCPAMSGIW------DRIVPRRLQQSFFTMSLLEWLYSNLRQGLMTEGS 1678
Query: 1156 VQRFLFAI-----WEARNKHIFEEKLFC---IAGVLDRAASLVSQSTLPFTLAASQEKDN 1207
+FA+ W+ R +IF E C + + D A + + L S + N
Sbjct: 1679 DWSTMFAMAVWWGWKWRCSNIFGENKTCRDRVRFIKDLAVEVSIAYSREVELRLSGLRVN 1738
Query: 1208 LK-RWRRPAENVVKVNVD-AAVGQDRFAGFGLVARDHAGEVLAAAAKYPIMVLSPTVAEA 1265
RW P E K+N D A+ G A G V R+ AG A I S +AE
Sbjct: 1739 KPIRWTPPMEGWYKINTDGASRGNPGLASAGGVLRNSAGAWCGGFA-VNIGRCSAPLAEL 1797
Query: 1266 LSLRWAMDLAIQLGFRRVQFETDCLLLQQAWKKVSGNSYLFS-IIKDCHKLVVFFDHVDL 1324
+ + + +A ++ E D ++ K G ++ S +++ CH + V +
Sbjct: 1798 WGVYYGLYMAWAKQLTHLELEVDSEVVVGFLKTGIGETHPLSFLVRLCHNFLSKDWTVRI 1857
Query: 1325 TFVRREGNSCADYLARNASSYPDSVWI-EEGPPGLSSLLRDD 1365
+ V RE NS AD LA +A S + + +E P L LL +D
Sbjct: 1858 SHVYREANSLADGLANHAFSLSLGLHVFDEIPISLVMLLSED 1899
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 557 bits (1436), Expect = e-158
Identities = 339/1065 (31%), Positives = 547/1065 (50%), Gaps = 21/1065 (1%)
Query: 291 RSLDSWGREKYGELPKRIKEARAFLQRLQEEVQTEQVVRATREAEKNLDVLLKQEEIVWS 350
+++ W RE RI + R L + E+ T + L ++EE W
Sbjct: 9 KAISQWKRENDFNAKSRITKLRRELDK--EKSATFPSWTQISLLQDVLGDAYREEEDFWR 66
Query: 351 QRSRANWLKHGDRNTKFFHSKATQRRKRNLIEEIQDDNGRKFVRDADIARVLTSYFQEIF 410
+SR W+ GD+N+KFF + R N + + D+NG + + + ++ ++F+++F
Sbjct: 67 LKSRDKWMVGGDKNSKFFQATVKANRVSNSLRFLVDENGNEQTVNREKGKIAVTFFEDLF 126
Query: 411 STCNPTGIEEVATLVDGRVTDAHRRILSTPFTREDVEEALFQMHPTKAPGLDGFPALFYQ 470
S+ P+ ++ V + RVT+ + L+ +++ +A+F ++ APG DGF ALF+Q
Sbjct: 127 SSSYPSSMDSVLEGFNKRVTEDMNQDLTKKVNEQEIYKAVFSINAESAPGPDGFTALFFQ 186
Query: 471 KFWPIVGDDISEFCLQVLQGDISPGMVNQTLIVLIPKVKKAVFANQYRPISLCNVIFKII 530
+ WP+V + I Q I P N T + LIPK+ K RPISLC+V++KII
Sbjct: 187 RQWPLVKNQIISDIELFFQTGILPEDWNHTHLCLIPKITKPARMADIRPISLCSVMYKII 246
Query: 531 SKTIANRMKLILHDLISEAQSAFVPGRLITDNALIAFECFHYMKKRISGRSGMMALKLDM 590
SK ++ R+K L ++S QSAFV RL++DN ++A E H ++ M K DM
Sbjct: 247 SKILSARLKKYLPVIVSPTQSAFVAERLVSDNIILAHEIVHNLRTNEKISKDFMVFKTDM 306
Query: 591 SKAYDRVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVNFAVMLNGNPQPTFAPHRGLRQGD 650
SKAYDRVEWPFL+ +L +GF W+N +M CV++V+++V++NG P PHRGLRQGD
Sbjct: 307 SKAYDRVEWPFLKGILLALGFNSTWINWMMACVSSVSYSVLINGQPFGHITPHRGLRQGD 366
Query: 651 PLSPYLFILCGEAFSALIQKSISTSALHGIKISRSAPVISHLLFADDSVLFARATVQEAE 710
PLSP+LF+LC EA ++ ++ + GI+ + + P ++HLLFADD++L +A+ E
Sbjct: 367 PLSPFLFVLCTEALIHILNQAEKIGKISGIQFNGTGPSVNHLLFADDTLLICKASQLECA 426
Query: 711 CIKNILATYERASGQKINLEKSMLSVSRNVPENCFVDLQLLLGVKAVDSYDKYLGLPTII 770
I + L+ Y SGQ IN EKS ++ V E + G++ KYLGLP
Sbjct: 427 EIMHCLSQYGHISGQMINSEKSAITFGAKVNEETKQWIMNRSGIQTEGGTGKYLGLPECF 486
Query: 771 GKSKTQIFRFVKERVWKKLKGWKEQTLSRAGREVLIKSVAQAIPSYVMSCFVLPDGLCNE 830
SK +F F+KE++ +L GW +TLS+ G+++L+KS+A A P Y M+CF L LC +
Sbjct: 487 QGSKQVLFGFIKEKLQSRLSGWYAKTLSQGGKDILLKSIAMAFPVYAMTCFRLSKTLCTK 546
Query: 831 IESMISRFYWGGDASKRGIHWVKWKTMCQPKHVGGLGFRDFKSFNLALVAKNWWRIYNYP 890
+ S++ F+W K+ IHW+ + + PK +GG GF+D + FN AL+AK R++
Sbjct: 547 LTSVMMDFWWNSVQDKKKIHWIGAQKLMLPKFLGGFGFKDLQCFNQALLAKQASRLHTDS 606
Query: 891 DSLLGKVFKSVYFSSGRMECAKKGYRPSYAWSRILKTSAMIQKGSCWRIGEGSRVRIWED 950
DSLL ++ KS Y+ + A KG RPSYAW IL ++ G IG G +W D
Sbjct: 607 DSLLSQILKSRYYMNSDFLSATKGTRPSYAWQSILYGRELLVSGLKKIIGNGENTYVWMD 666
Query: 951 NWLANGPPVNFRQ-DVVDELGLTKVADLMLSGNRGWNVPLIEWTFCPATASRIMSVPLPR 1009
NW+ + P ++ ++ L KV+ L+ +R WN+ ++ F P +I+ P
Sbjct: 667 NWIFDDKPRRPESLQIMVDIQL-KVSQLIDPFSRNWNLNMLRDLF-PWKEIQIICQQRPM 724
Query: 1010 QPESDQLFWLGTADGLYSVKTGYEFLQEVVAQDSPSSSLSRGLDQTLWKRFWKAPSMPRV 1069
D W GT GLY+VK+ Y+ V + + + L+ + W S P++
Sbjct: 725 ASRQDSFCWFGTNHGLYTVKSEYDLCSRQVHKQMFKEAEEQPSLNPLFGKIWNLNSAPKI 784
Query: 1070 REVSWRVCTGALPVRTKLRQRGVDVDSSCPLCGLEDETVDHLFLGCNITRGCWFASPMGV 1129
+ W+V GA+ V +LR RGV ++ C +C ++ET++H+ C + R W +PM
Sbjct: 785 KVFLWKVLKGAVAVEDRLRTRGVLIEDGCSMCPEKNETLNHILFQCPLARQVWALTPM-- 842
Query: 1130 RVDPSLKMVDFL----TMVIRDMDLEVVACVQRF-----LFAIWEARNKHIFEEKLFCIA 1180
P+ D + VI + ++ R+ ++ +W+ RNK +FE
Sbjct: 843 -QSPNHGFGDSIFTNVNHVIGNCHNTELSPHLRYVSPWIIWILWKNRNKRLFEGIGSVSL 901
Query: 1181 GVLDRAASLVSQSTLPFTLAASQEKDNLKRWRRPAENVVKVNVDAAVG-QDRFAGFGLVA 1239
++ +A + L S+E W P N +K N+ A + + AG V
Sbjct: 902 SIVGKALEDCKEWLKAHELICSKEPTKDLTWIPPLMNELKCNIGIAWSKKHQMAGVSWVV 961
Query: 1240 RDHAGEVLAAAAKYPIMVLSPTVAEALSLRWAMDLAIQLGFRRVQFETDCLLLQQAWKKV 1299
R+ G VL + + S A+ +A++ Q F RV F + +A K
Sbjct: 962 RNWKGRVLLHSRCSFSQISSHFDAKIKGWNYAVESMDQFKFDRVTFGASTHDIIKAMHKP 1021
Query: 1300 SGNSYLFSIIKDCHKLVVFFDHVDLTFVRREGNSCADYLARNASS 1344
+ L + I D L++ +L F+ E + C + + A S
Sbjct: 1022 NQWPALRAQIDD---LLMRAKDKELWFMAMEPHHCNEGASEIAKS 1063
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 551 bits (1421), Expect = e-157
Identities = 394/1296 (30%), Positives = 616/1296 (47%), Gaps = 113/1296 (8%)
Query: 108 MQVYALYGFPELHLKDRTWEMIRSVRPMAPIPWLCIGDFNDILSPSDKLGGAPPDLARLQ 167
M + +Y P + + W ++ V P L GDFN IL +++GG RL
Sbjct: 1 MHLIVVYAAPSVSRRSGLWGELKDVVNGLEGPLLIGGDFNTILWVDERMGGN----GRLS 56
Query: 168 VATQACAD----CGLQRVDFSGPSFTWTNNRVGPGRVDERLDYALINQAWVNLWPVSSVS 223
+ A D L + F G FTW R V +RLD + W + VS
Sbjct: 57 PDSLAFGDWINELSLIDLGFKGNKFTWRRGRQESTVVAKRLDRVFVCAHARLKWQEAVVS 116
Query: 224 HLIRHQSDHNPVVLHCGSRRTETQRHRTRMFRFEEVWLESGEECAEIISEGWSNTNLSIL 283
HL SDH P+ V LE ++
Sbjct: 117 HLPFMASDHAPLY----------------------VQLEPLQQ----------------- 137
Query: 284 SRINLVGRSLDSWGREKYGELPKRIKEARAFLQRLQE---EVQTEQVVRATREAEKNLDV 340
R L W RE +G++ R ++ A ++ +Q+ V ++ ++ K +D+
Sbjct: 138 -------RKLRKWNREVFGDIHVRKEKLVADIKEVQDLLGVVLSDDLLAKEEVLLKEMDL 190
Query: 341 LLKQEEIVWSQRSRANWLKHGDRNTKFFHSKATQRRKRNLIEEIQDDNGRKFVRDADIAR 400
+L+QEE +W Q+SR +++ GDRNT FFH+ RR+RN IE ++ D+ R ++
Sbjct: 191 VLEQEETLWFQKSREKYIELGDRNTTFFHTSTIIRRRRNRIESLKGDDDRWVTDKVELEA 250
Query: 401 VLTSYFQEIFSTCNPTGIEEVATLVD----GRVTDAHRRILSTPFTREDVEEALFQMHPT 456
+ +Y++ ++S + EV ++ +++A + L FT+ +V A+ M
Sbjct: 251 MALTYYKRLYSL---EDVSEVRNMLPTGGFASISEAEKAALLQAFTKAEVVSAVKSMGRF 307
Query: 457 KAPGLDGFPALFYQKFWPIVGDDISEFCLQVLQGDISPGMVNQTLIVLIPKVKKAVFANQ 516
KAPG DG+ +FYQ+ W VG ++ F L+ + + P N L+VLI KV K Q
Sbjct: 308 KAPGPDGYQPVFYQQCWETVGPSVTRFVLEFFETGVLPASTNDALLVLIAKVAKPERIQQ 367
Query: 517 YRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVPGRLITDNALIAFECFHYMKKR 576
+RP+SLCNV+FKII+K + R+K ++ LI AQ++F+PGRL DN ++ E H M+++
Sbjct: 368 FRPVSLCNVLFKIITKMMVTRLKNVISKLIGPAQASFIPGRLSIDNIVLVQEAVHSMRRK 427
Query: 577 ISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVNFAVMLNGNP 636
GR G M LKLD+ KAYDRV W FL+ L G W + +M VT + +V+ NG
Sbjct: 428 -KGRKGWMLLKLDLEKAYDRVRWDFLQETLEAAGLSEGWTSRIMAGVTDPSMSVLWNGER 486
Query: 637 QPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHGIKISRSAPVISHLLFAD 696
+F P RGLRQGDPLSPYLF+LC E LI+ S+ I +S +SH+ FAD
Sbjct: 487 TDSFVPARGLRQGDPLSPYLFVLCLERLCHLIEASVGKREWKPIAVSCGGSKLSHVCFAD 546
Query: 697 DSVLFARATVQEAECIKNILATYERASGQKINLEKSMLSVSRNVPENCFVDLQLLLGVKA 756
D +LFA A+V + I+ +L + ASGQK++LEKS + S NV + G+
Sbjct: 547 DLILFAEASVAQIRIIRRVLERFCEASGQKVSLEKSKIFFSHNVSREMEQLISEESGIGC 606
Query: 757 VDSYDKYLGLPTIIGKSKTQIFRFVKERVWKKLKGWKEQTLSRAGREVLIKSVAQAIPSY 816
KYLG+P + + + F V ERV +L GWK ++LS AGR L K+V +IP +
Sbjct: 607 TKELGKYLGMPILQKRMNKETFGEVLERVSARLAGWKGRSLSLAGRITLTKAVLSSIPVH 666
Query: 817 VMSCFVLPDGLCNEIESMISRFYWGGDASKRGIHWVKWKTMCQPKHVGGLGFRDFKSFNL 876
VMS +LP + ++ F WG K+ H + W+ +C+PK GG+G R + N
Sbjct: 667 VMSAILLPVSTLDTLDRYSRTFLWGSTMEKKKQHLLSWRKICKPKAEGGIGLRSARDMNK 726
Query: 877 ALVAKNWWRIYNYPDSLLGKVFKSVYFSSGRMECAKKGYRPSYAWSRILKTSA-----MI 931
ALVAK WR+ +SL +V + Y G + + +P WS ++ A ++
Sbjct: 727 ALVAKVGWRLLQDKESLWARVVRKKYKVGGVQDTS--WLKPQPRWSSTWRSVAVGLREVV 784
Query: 932 QKGSCWRIGEGSRVRIWEDNWLANGPPVNFRQDVVDELGLTKV-ADLMLSGNRGWNVPLI 990
KG W G+G +R W D WL P V D++ E KV AD L G+ GWN+ ++
Sbjct: 785 VKGVGWVPGDGCTIRFWLDRWLLQEPLVELGTDMIPEGERIKVAADYWLPGS-GWNLEIL 843
Query: 991 EWTFCPATASRIMSVPLP-RQPESDQLFWLGTADGLYSVKTGYEFLQEVVAQDSPSSSLS 1049
R++SV + D++ W GT DG ++V++ Y LQ V D P+
Sbjct: 844 GLYLPETVKRRLLSVVVQVFLGNGDEISWKGTQDGAFTVRSAYSLLQGDVG-DRPNMG-- 900
Query: 1050 RGLDQTLWKRFWKAPSMPRVREVSWRVCTGALPVRTKLRQRGVDVDSSCPLCGLEDETVD 1109
+ + R WK + RVR W V + + +R + ++ C +C +ET+
Sbjct: 901 -----SFFNRIWKLITPERVRVFIWLVSQNVIMTNVERVRRHLSENAICSVCNGAEETIL 955
Query: 1110 HLFLGCNITRGCWFASPMGVRVDPSLKMVDFLTMVIRD---MDLEVVACVQRFLFAI--- 1163
H + R C P+ R+ P + +F + + + +++ V + LF +
Sbjct: 956 H------VLRDCPAMEPIWRRLLPLRRHHEFFSQSLLEWLFTNMDPVKGIWPTLFGMGIW 1009
Query: 1164 --WEARNKHIFEEKLFCIAGVLDRAASL--VSQSTLPFTLAASQEKDN------LKRWRR 1213
W+ R +F E+ C DR + +++ + A + N + RW+
Sbjct: 1010 WAWKWRCCDVFGERKIC----RDRLKFIKDMAEEVRRVHVGAVGNRPNGVRVERMIRWQV 1065
Query: 1214 PAENVVKVNVD-AAVGQDRFAGFGLVARDHAGEVLAAAAKYPIMVLSPTVAEALSLRWAM 1272
P++ VK+ D A+ G A G R+ GE L A I + +AE + +
Sbjct: 1066 PSDGWVKITTDGASRGNHGLAAAGGAIRNGQGEWLGGFA-LNIGSCAAPLAELWGAYYGL 1124
Query: 1273 DLAIQLGFRRVQFETDCLLLQQAWKKVSGNSYLFS-IIKDCHKLVVFFDHVDLTFVRREG 1331
+A GFRRV+ + DC L+ N++ S +++ C V ++ V RE
Sbjct: 1125 LIAWDKGFRRVELDLDCKLVVGFLSTGVSNAHPLSFLVRLCQGFFTRDWLVRVSHVYREA 1184
Query: 1332 NSCADYLARNASSYPDSV-WIEEGPPGLSSLLRDDV 1366
N AD LA A + P + + P G+ LL DV
Sbjct: 1185 NRLADGLANYAFTLPLGLHCFDACPEGVRLLLLADV 1220
>At4g10830 putative protein
Length = 1294
Score = 537 bits (1383), Expect = e-152
Identities = 316/917 (34%), Positives = 486/917 (52%), Gaps = 19/917 (2%)
Query: 35 DIVFLMETKLFSFEMQRHRGMGGLSNIFPVQCGRNRAGGLCLLWRSEVEVTIINASLHHI 94
D++FL+ET + + G N+ Q + +GGL LLW+ V ++ + HI
Sbjct: 391 DVLFLIETLNKCEVISNLASVLGFPNVI-TQPPQGHSGGLALLWKDSVRLSNLYQDDRHI 449
Query: 95 LFTVVHSDSVTVPMQVYALYGFPELHLKDRTWEMIRSVRPMAPIPWLCIGDFNDILSPSD 154
VH + + +YG P + W ++ PW+ IGDFN+ILS ++
Sbjct: 450 ---DVHISINNINFYLSRVYGHPCQSERHSLWTHFENLSKTRNDPWILIGDFNEILSNNE 506
Query: 155 KLGGAPPDLARLQVATQACADCGLQRVDFSGPSFTWTNNRVGPGRVDERLDYALINQAWV 214
K+GG D + + C L+ + G F+W R V LD A IN
Sbjct: 507 KIGGPQRDEWTFRGFRNMVSTCDLKDIRSIGDRFSWVGERHSH-TVKCCLDRAFINSEGA 565
Query: 215 NLWPVSSVSHLIRHQSDHNPVVLHCGSRRTETQRHRTRMFRFEEVWLESGEECAEIISEG 274
L+P + + L SDH P+ L +TET++ R FRF++ LE + G
Sbjct: 566 FLFPFAELEFLEFTGSDHKPLFLSL--EKTETRKMRP--FRFDKRLLEV-PHFKTYVKAG 620
Query: 275 WSNT----NLSILSRINLVGRSLDSWGREKYGELPKRIKEARAFLQRLQEEV-QTEQVVR 329
W+ + ++ +++ + RI + +A L + V +TE+ R
Sbjct: 621 WNKAINGQRKHLPDQVRTCRQAMAKLKHKSNLNSRIRINQLQAALDKAMSSVNRTER--R 678
Query: 330 ATREAEKNLDVLLKQEEIVWSQRSRANWLKHGDRNTKFFHSKATQRRKRNLIEEIQDDNG 389
++ L V + EE W Q+SR W+K GDRNT+FFH+ R N + I+D+ G
Sbjct: 679 TISHIQRELTVAYRDEERYWQQKSRNQWMKEGDRNTEFFHACTKTRFSVNRLVTIKDEEG 738
Query: 390 RKFVRDADIARVLTSYFQEIF-STCNPTGIEEVATLVDGRVTDAHRRILSTPFTREDVEE 448
+ D +I +F +++ S P I + A VT+ L+ + ++
Sbjct: 739 MIYRGDKEIGVHAQEFFTKVYESNGRPVSIIDFAGFKP-IVTEQINDDLTKDLSDLEIYN 797
Query: 449 ALFQMHPTKAPGLDGFPALFYQKFWPIVGDDISEFCLQVLQGDISPGMVNQTLIVLIPKV 508
A+ + KAPG DG A FY+ W IVG D+ + + +N T I +IPK+
Sbjct: 798 AICHIGDDKAPGPDGLTARFYKSCWEIVGPDVIKEVKIFFRTSYMKQSINHTNICMIPKI 857
Query: 509 KKAVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVPGRLITDNALIAFE 568
+ YRPI+LCNV++KIISK + R+K L ++S++Q+AF+PGRL+ DN +IA E
Sbjct: 858 TNPETLSDYRPIALCNVLYKIISKCLVERLKGHLDAIVSDSQAAFIPGRLVNDNVMIAHE 917
Query: 569 CFHYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVNF 628
H +K R MA+K D+SKAYDRVEW FL + + GF W+ +M V +VN+
Sbjct: 918 MMHSLKTRKRVSQSYMAVKTDVSKAYDRVEWNFLETTMRLFGFSETWIKWIMGAVKSVNY 977
Query: 629 AVMLNGNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHGIKISRSAPV 688
+V++NG P T P RG+RQGDPLSPYLFILC + + LI+ ++ + GI+I P
Sbjct: 978 SVLVNGIPHGTIQPQRGIRQGDPLSPYLFILCADILNHLIKNRVAEGDIRGIRIGNGVPG 1037
Query: 689 ISHLLFADDSVLFARATVQEAECIKNILATYERASGQKINLEKSMLSVSRNVPENCFVDL 748
++HL FADDS+ F ++ V+ + +K++ YE SGQKIN+ KSM++ V L
Sbjct: 1038 VTHLQFADDSLFFCQSNVRNCQALKDVFDVYEYYSGQKINMSKSMITFGSRVHGTTQNRL 1097
Query: 749 QLLLGVKAVDSYDKYLGLPTIIGKSKTQIFRFVKERVWKKLKGWKEQTLSRAGREVLIKS 808
+ +LG+++ KYLGLP G+ K +F ++ ERV K+ W + LS AG+E+++KS
Sbjct: 1098 KNILGIQSHGGGGKYLGLPEQFGRKKRDMFNYIIERVKKRTSSWSAKYLSPAGKEIMLKS 1157
Query: 809 VAQAIPSYVMSCFVLPDGLCNEIESMISRFYWGGDASKRGIHWVKWKTMCQPKHVGGLGF 868
VA ++P Y MSCF LP + +EIE+++ F+W +A KR I W+ WK + K GGLGF
Sbjct: 1158 VAMSMPVYAMSCFKLPLNIVSEIEALLMNFWWEKNAKKREIPWIAWKRLQYSKKEGGLGF 1217
Query: 869 RDFKSFNLALVAKNWWRIYNYPDSLLGKVFKSVYFSSGRMECAKKGYRPSYAWSRILKTS 928
RD FN AL+AK WR+ N P+SL ++ K+ YF + AK+ SY W+ +L
Sbjct: 1218 RDLAKFNDALLAKQVWRMINNPNSLFARIMKARYFREDSILDAKRQRYQSYGWTSMLAGL 1277
Query: 929 AMIQKGSCWRIGEGSRV 945
+I+KGS + +G+G V
Sbjct: 1278 DVIKKGSRFIVGDGKTV 1294
>At3g45550 putative protein
Length = 851
Score = 518 bits (1333), Expect = e-146
Identities = 300/859 (34%), Positives = 461/859 (52%), Gaps = 42/859 (4%)
Query: 395 DADIARVLTSYFQEIFSTCNPTGIEEVATLVD-----GRVTDAHRRILSTPFTREDVEEA 449
D +I +F +IF+T GI+ + +D VT+ L+ F ++ EA
Sbjct: 3 DGEIGCHAQKFFTDIFTT---NGIQ--VSPIDFADFPSSVTNIINSELTQDFRDSEIFEA 57
Query: 450 LFQMHPTKAPGLDGFPALFYQKFWPIVGDDISEFCLQVLQGDISPGMVNQTLIVLIPKVK 509
+ Q+ KAPG DG A FY++ W IVG+D+ + + VN T I +IPK++
Sbjct: 58 ICQIGDDKAPGPDGLTARFYKQCWDIVGNDVIKEVKLFFESSHMKTSVNHTNICMIPKIQ 117
Query: 510 KAVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVPGRLITDNALIAFEC 569
+ YRPI+LCNV++K+ISK + NR+K L+ ++S++Q+AF+PGR+I DN +IA E
Sbjct: 118 NPQTLSDYRPIALCNVLYKVISKCMVNRLKAHLNSIVSDSQAAFIPGRIINDNVMIAHEI 177
Query: 570 FHYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVNFA 629
H +K R MA+K D+SKAYDRVEW FL + + GF W+ +M V +V+++
Sbjct: 178 MHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCDKWIGWIMAAVKSVHYS 237
Query: 630 VMLNGNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHGIKISRSAPVI 689
V++NG+P +P RG+RQGDPLSPYLFILCG+ S LI+ S+ + G++I AP I
Sbjct: 238 VLINGSPHGYISPTRGIRQGDPLSPYLFILCGDILSHLIKVKASSGDIRGVRIGNGAPAI 297
Query: 690 SHLLFADDSVLFARATVQEAECIKNILATYERASGQKINLEKSMLSVSRNVPENCFVDLQ 749
+HL FADDS+ F +A V+ + +K++ YE SGQKIN++KS+++ V + L+
Sbjct: 298 THLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSLITFGSRVYGSTQTRLK 357
Query: 750 LLLGVKAVDSYDKYLGLPTIIGKSKTQIFRFVKERVWKKLKGWKEQTLSRAGREVLIKSV 809
LL + KYLGLP G+ K ++F ++ +RV ++ W + LS AG+E+L+KSV
Sbjct: 358 TLLNIPNQGGGGKYLGLPEQFGRKKKEMFNYIIDRVKERTASWSAKFLSPAGKEILLKSV 417
Query: 810 AQAIPSYVMSCFVLPDGLCNEIESMISRFYWGGDASKRGIHWVKWKTMCQPKHVGGLGFR 869
A A+P Y MSCF LP G+ +EIES++ F+W ++KRGI WV WK + K GGLGFR
Sbjct: 418 ALAMPVYAMSCFKLPQGIVSEIESLLMNFWWEKASNKRGIPWVAWKRLQYSKKEGGLGFR 477
Query: 870 DFKSFNLALVAKNWWRIYNYPDSLLGKVFKSVYFSSGRMECAKKGYRPSYAWSRILKTSA 929
D FN AL+AK WRI YP+SL +V K+ YF + AK + SY WS +L A
Sbjct: 478 DLAKFNDALLAKQAWRIIQYPNSLFARVMKARYFKDNSIIDAKTRSQQSYGWSSLLSGIA 537
Query: 930 MIQKGSCWRIGEGSRVRIWEDNWLANGPPVNFRQDVVDELGLTKVADLML---SGNRGWN 986
+++KG+ + IG+G +R+ DN + + PP R + DE D + +R W+
Sbjct: 538 LLRKGTRYVIGDGKTIRLGIDNVVDSHPP---RPLLTDEQHNGLSLDNLFQHRGHSRCWD 594
Query: 987 VPLIEWTFCPATASRIMSVPLPRQPESDQLFWLGTADGLYSVKTGYEFLQEVVAQDSPSS 1046
++ + I + L + ++D+L W + G Y+V++GY + P+
Sbjct: 595 NAKLQTFVDQSDHDYIKRIYLSTRSKTDRLIWSYNSTGDYTVRSGYWLSTHDPSNTIPTM 654
Query: 1047 SLSRGLDQTLWKRFWKAPSMPRVREVSWRVCTGALPVRTKLRQRGVDVDSSCPLCGLEDE 1106
+ G L + W P MP+++ WR+ + ALP +L RG+ +D CP C E+E
Sbjct: 655 AKPHG-SVDLKTKIWNLPIMPKLKHFLWRILSKALPTTDRLTTRGMRIDPGCPRCRRENE 713
Query: 1107 TVDHLFLGCNITRGCWFASPMGVRVDPSLKMVDFLTMVIRDMDLEVVACVQR-------- 1158
+++H C W S D L L+ I D ++ +Q
Sbjct: 714 SINHALFTCPFATMAWRLS------DTPLYRSSILSNNIEDNISNILLLLQNTTITDSQK 767
Query: 1159 -----FLFAIWEARNKHIFEEKLFCIAGVLDRAAS------LVSQSTLPFTLAASQEKDN 1207
L+ IW+ARN +F + + RA + +Q+ P L
Sbjct: 768 LIPFWLLWRIWKARNNVVFNNLRESPSITVVRAKAETNEWLNATQTQGPRRLPKRTTAAG 827
Query: 1208 LKRWRRPAENVVKVNVDAA 1226
W +P +K N DA+
Sbjct: 828 NTTWVKPQMPYIKCNFDAS 846
>At2g11240 pseudogene
Length = 1044
Score = 499 bits (1284), Expect = e-141
Identities = 289/838 (34%), Positives = 444/838 (52%), Gaps = 24/838 (2%)
Query: 174 ADCGLQRVDFSGPSFTWTNNRVGPGRVDERLDYALINQAWVNLWPVSSVSHLIRHQSDHN 233
A+C L + SG +W R V RLD AL N AW +P S +L SDH
Sbjct: 2 AECDLYDLRHSGNFLSWRGKR-HDHVVHCRLDRALSNGAWAEDYPASRCIYLCFEGSDHR 60
Query: 234 PVVLHCGSRRTETQRHRTRMFRFEEVWLESGEECAEIISEGWSNTNLSILS-RINLVGRS 292
P++ H +++ + +FR++ L++ +E ++ E W+ + I+ +I+
Sbjct: 61 PLLTHFDL----SKKKKKGVFRYDRR-LKNNDEVTALVQEAWNLYDTDIVEEKISRCRLE 115
Query: 293 LDSWGREKYGELPKRIKEARAFLQRLQEEVQTEQVVRATREAEKNLDVLLKQEEIVWSQR 352
+ W R K K I+E R L+ Q + +T NL + K EE W QR
Sbjct: 116 IVKWSRAKQQSSQKLIEENRQKLEEAMSSQDHNQELLST--INTNLLLAYKAEEEYWKQR 173
Query: 353 SRANWLKHGDRNTKFFHSKATQRRKRNLIEEIQDDNGRKFVRDADIARVLTSYFQEIFST 412
SR WL GD+N+ +FH+ R N I+ ++G +A I V++ YFQ++FS
Sbjct: 174 SRQLWLALGDKNSGYFHAITRGRTVINKFSVIEKEDGVPEYEEAGILNVISEYFQKLFSA 233
Query: 413 CNPTGIEEVATLVDGRVTDAHRRILSTPFTREDVEEALFQMHPTKAPGLDGFPALFYQKF 472
AT + +A + +S E+++ A F +H KAPG DGF A F+Q
Sbjct: 234 NEGA---RAAT-----IKEAIKPFISPEQNPEEIKSACFSIHADKAPGPDGFSASFFQSN 285
Query: 473 WPIVGDDISEFCLQVLQGDISPGMVNQTLIVLIPKVKKAVFANQYRPISLCNVIFKIISK 532
W VG +I +N+T I LIPK++ YRPI+LC V +KIISK
Sbjct: 286 WMTVGPNIVLEIQSFFSSSTLQPTINKTHITLIPKIQSLKRMVDYRPIALCTVFYKIISK 345
Query: 533 TIANRMKLILHDLISEAQSAFVPGRLITDNALIAFECFHYMKKRISGRSGMMALKLDMSK 592
++ R++ IL ++ISE QSAFVP R DN LI E HY+K + + MA+K +MSK
Sbjct: 346 LLSRRLQPILQEIISENQSAFVPKRASNDNVLITHEALHYLKSLGAEKRCFMAVKTNMSK 405
Query: 593 AYDRVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVNFAVMLNGNPQPTFAPHRGLRQGDPL 652
AYDR+EW F++ V+ MGF W++ ++ C+TTV+++ +LNG+ Q P RGLRQGDPL
Sbjct: 406 AYDRIEWDFIKLVMQEMGFHQTWISWILQCITTVSYSFLLNGSAQGAVTPERGLRQGDPL 465
Query: 653 SPYLFILCGEAFSALIQKSISTSALHGIKISRSAPVISHLLFADDSVLFARATVQEAECI 712
SP+LFI+C E S L +K+ +L G+++S+ P ++HLLFADD++ F R+ ++ +
Sbjct: 466 SPFLFIICSEVLSGLCRKAQLDGSLLGLRVSKGNPRVNHLLFADDTIFFCRSDLKSCKTF 525
Query: 713 KNILATYERASGQKINLEKSMLSVSRNVPENCFVDLQLLLGVKAVDSYDKYLGLPTIIGK 772
IL YE ASGQ IN KS ++ SR P++ + Q +LG++ V KYLGLP + G+
Sbjct: 526 LCILKKYEEASGQMINKSKSAITFSRKTPDHIKTEAQQILGIQLVGGLGKYLGLPKMFGR 585
Query: 773 SKTQIFRFVKERVWKKLKGWKEQTLSRAGREVLIKSVAQAIPSYVMSCFVLPDGLCNEIE 832
K +F + +R+ ++ W + LS AG+ ++KSV ++P+Y MSCF L LC I+
Sbjct: 586 KKRDLFNQIVDRIRQRSLSWSSRFLSTAGKTTMLKSVLASMPTYTMSCFKLLVSLCKRIQ 645
Query: 833 SMISRFYWGGDASKRGIHWVKWKTMCQPKHVGGLGFRDFKSFNLALVAKNWWRIYNYPDS 892
S ++ F+W A K+ + W+ W M + K GGLGF+D +FN AL+AK WRI P
Sbjct: 646 SALTHFWWDSSADKKKMCWIAWSKMAKNKKEGGLGFKDITNFNDALLAKLSWRIVQSPSC 705
Query: 893 LLGKVFKSVYF-SSGRMECAKKGYRPSYAWSRILKTSAMIQKGSCWRIGEGSRVRIWEDN 951
+L ++ Y +S ++C+ S+ W I +I+ IG G +W +
Sbjct: 706 VLVRILLGKYCRTSSFLDCSVTA-ASSHGWRGICTGKDLIKSQLGKVIGSGLDTLVWNEP 764
Query: 952 WLANGPPVNFRQDVVDELGLTKVADLMLSGNRGWNVPLIEWTFCP----ATASRIMSV 1005
WL+ +++ VA L+ + W+ + W P ASRI S+
Sbjct: 765 WLSLSTSSTPMGPALEQFKSMTVAQLICQTTKSWDREKV-WDLSPFKTTLQASRITSM 821
Score = 67.0 bits (162), Expect = 7e-11
Identities = 60/192 (31%), Positives = 99/192 (51%), Gaps = 16/192 (8%)
Query: 1160 LFAIWEARNKHIFEEKLFCIAGVLDRAASLVSQST--LPFTLAASQEKDNLKRWRRPAE- 1216
L+ +W RNK IFE+K +D + +SQST L + AS+ K + P+E
Sbjct: 849 LWNLWNCRNKLIFEQKHI---SSMDLISQSISQSTEWLGAQIQASKSKIVIPGIS-PSEI 904
Query: 1217 --NVVKVNVDAAVGQDRF-AGFGLVARDHAGEVLAAAAKYPIMVLSPTVAEALSLRWAMD 1273
+ ++ + DA+ ++ AGFG V DH+ + + + + SP +A+A +L A+
Sbjct: 905 DLDTIQCSTDASWREETLQAGFGWVFVDHSNHLESHHKAAAMNIRSPLLAKASALSLAIQ 964
Query: 1274 LAIQLGFRRVQFETDCLLLQQAWKKVSGNSY---LFSIIKDCHKLVVFFDHVDLTFVRRE 1330
A LGF+++ +D QQ K ++G + L I+ D L + F+ +FV+RE
Sbjct: 965 HAADLGFKKLVVASDS---QQLVKVLNGEPHPMELHGIVFDISVLSLNFEENSFSFVKRE 1021
Query: 1331 GNSCADYLARNA 1342
NS AD LA+ A
Sbjct: 1022 NNSKADALAKAA 1033
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 459 bits (1181), Expect = e-129
Identities = 297/906 (32%), Positives = 447/906 (48%), Gaps = 80/906 (8%)
Query: 183 FSGPSFTWTNNRVGPGRVDERLDYALINQAWVNLWPVSSVSHLIRHQSDHNPVVLHCGSR 242
F G FTW V V +RLD L W + L C ++
Sbjct: 3 FKGNRFTWRRGLVESTFVAKRLDRVLFCAHARLKWQEA----------------LLCPAQ 46
Query: 243 RTETQRHRTRMFRFEEVWLESGEECAEIISEGWSNTNLSILSRINLVGRSLDSWGREKYG 302
+ +R R FRFE WL S E E+++ W +T LS +N + L W +E +G
Sbjct: 47 NVDARR---RPFRFEAAWL-SHEGFKELLTASW-DTGLSTPVALNRLRWQLKKWNKEVFG 101
Query: 303 ELPKRIKEARAFLQRLQ---EEVQTEQVVRATREAEKNLDVLLKQEEIVWSQRSRANWLK 359
+ R ++ + L+ +Q E VQT+ ++ K DVLL QEE +W Q+SR L
Sbjct: 102 NIHVRKEKVVSDLKAVQDLLEVVQTDDLLMKEDTLLKEFDVLLHQEETLWFQKSREKLLA 161
Query: 360 HGDRNTKFFHSKATQRRKRNLIEEIQDDNGRKFVRDADIARVLTSYFQEIFSTCNPTGIE 419
GDRNT FFH+ RR+RN IE ++D R + ++ Y+++++S +E
Sbjct: 162 LGDRNTTFFHTSTVIRRRRNRIEMLKDSEDRWVTEKEALEKLAMDYYRKLYS------LE 215
Query: 420 EVATL-----VDG--RVTDAHRRILSTPFTREDVEEALFQMHPTKAPGLDGFPALFYQKF 472
+V+ + +G R+T + L+ PFTR++V A+ M KAPG DG+ +FYQ+
Sbjct: 216 DVSVVRGTLPTEGFPRLTREEKNNLNRPFTRDEVVVAVRSMGRFKAPGPDGYQPVFYQQC 275
Query: 473 WPIVGDDISEFCLQVLQGDISPGMVNQTLIVLIPKVKKAVFANQYRPISLCNVIFKIISK 532
W VG+ +S+F ++ + + P N L+VL+ KV K Q+RP+SLCNV+FKII+K
Sbjct: 276 WETVGESVSKFVMEFFESGVLPKSTNDVLLVLLAKVAKPERITQFRPVSLCNVLFKIITK 335
Query: 533 TIANRMKLILHDLISEAQSAFVPGRLITDNALIAFECFHYMKKRISGRSGMMALKLDMSK 592
+ R+K ++ LI AQ++F+PGRL DN ++ E H M+++ GR G M LKLD+ K
Sbjct: 336 MMVIRLKNVISKLIGPAQASFIPGRLSFDNIVVVQEAVHSMRRK-KGRKGWMLLKLDLEK 394
Query: 593 AYDRVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVNFAVMLNGNPQPTFAPHRGLRQGDPL 652
AYDR+ W FL L G W+ +M CV +++ NG +F P RGLRQGDP+
Sbjct: 395 AYDRIRWDFLAETLEAAGLSEGWIKRIMECVAGPEMSLLWNGEKTDSFTPERGLRQGDPI 454
Query: 653 SPYLFILCGEAFSALIQKSISTSALHGIKISRSAPVISHLLFADDSVLFARATVQEAECI 712
SPYLF+LC E I+ ++ I IS+ P +SH+ FADD +LFA A+V
Sbjct: 455 SPYLFVLCIERLCHQIETAVGRGDWKSISISQGGPKVSHVCFADDLILFAEASV------ 508
Query: 713 KNILATYERASGQKINLEKSMLSVSRNVPENCFVDLQLLLGVKAVDSYDKYLGLPTIIGK 772
QK++LEKS + S NV + + G+ + KYLG+P + +
Sbjct: 509 -----------AQKVSLEKSKIFFSNNVSRDLEGLITAETGIGSTRELGKYLGMPVLQKR 557
Query: 773 SKTQIFRFVKERVWKKLKGWKEQTLSRAGREVLIKSVAQAIPSYVMSCFVLPDGLCNEIE 832
F V ERV +L GWK ++LS AGR L K+V +IP + MS +LP L +++
Sbjct: 558 INKDTFGEVLERVSSRLSGWKSRSLSLAGRITLTKAVLMSIPIHTMSSILLPASLLEQLD 617
Query: 833 SMISRFYWGGDASKRGIHWVKWKTMCQPKHVGGLGFRDFKSFNLALVAKNWWRIYNYPDS 892
+ F WG KR H + WK +C+PK GGLG R K N AL+AK WR+ N S
Sbjct: 618 KVSRNFLWGSTVEKRKQHLLSWKKVCRPKAAGGLGLRASKDMNRALLAKVGWRLLNDKVS 677
Query: 893 LLGKVFKSVYFSSGRMECAKKGYRPSYAWSRILKTSAMIQKGSCWR-IGEGSRVRIWEDN 951
L +V + Y + + P WS S WR IG G R +
Sbjct: 678 LWARVLRRKYKVTDVHD--SSWLVPKATWS------------STWRSIGVGLREGV-AKG 722
Query: 952 WLANGPPVNFRQDVVDELGLTKVADLMLSGNRGWNVPLIEWTFCPATASRIMSVPLPR-Q 1010
W+ + P ++ L + + GW++ + + R+ +V +
Sbjct: 723 WILHEPLCTRATCLLSPEELNARVEEFWTEGVGWDMVKLGQCLPRSVTDRLHAVVIKGVL 782
Query: 1011 PESDQLFWLGTADGLYSVKTGYEFLQEVVAQDSPSSSLSRGLDQTLWKRFWKAPSMPRVR 1070
D++ W GT+DG ++V + Y L + S+ ++ +KR W + RVR
Sbjct: 783 GLRDRISWQGTSDGDFTVGSAYVLLTQ--------EEESKPCMESFFKRIWGVIAPERVR 834
Query: 1071 EVSWRV 1076
W V
Sbjct: 835 VFLWLV 840
>At2g17610 putative non-LTR retroelement reverse transcriptase
Length = 773
Score = 447 bits (1149), Expect = e-125
Identities = 264/750 (35%), Positives = 398/750 (52%), Gaps = 36/750 (4%)
Query: 558 LITDNALIAFECFH--YMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHW 615
+ITDN LIA E H + KK + +A KLD++KA+D++EW F+ +++ MGF W
Sbjct: 1 MITDNILIAHELIHSLHTKKLVQP---FVATKLDITKAFDKIEWGFIEAIMKQMGFSEKW 57
Query: 616 VNLVMNCVTTVNFAVMLNGNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTS 675
N +M C+TT +++++NG P P RG+RQGDP+SPYL++LC E SALIQ SI
Sbjct: 58 CNWIMTCITTTTYSILINGQPVRRIIPKRGIRQGDPISPYLYLLCTEGLSALIQASIKAK 117
Query: 676 ALHGIKISRSAPVISHLLFADDSVLFARATVQEAECIKNILATYERASGQKINLEKSMLS 735
LHG K SR+ P ISHLLFA DS++F +AT++E + N+L YE+ASGQ +N +KS +
Sbjct: 118 QLHGFKASRNGPAISHLLFAHDSLVFCKATLEECMTLVNVLKLYEKASGQAVNFQKSAIL 177
Query: 736 VSRNVPENCFVDLQLLLGVKAVDSYDKYLGLPTIIGKSKTQIFRFVKERVWKKLKGWKEQ 795
+ + L LLG+ + + +YLGLP +G++KT F F+ + + +K+ W +
Sbjct: 178 FGKGLDFRTSEQLSQLLGIYKTEGFGRYLGLPEFVGRNKTNAFSFIAQTMDQKMDNWYNK 237
Query: 796 TLSRAGREVLIKSVAQAIPSYVMSCFVLPDGLCNEIESMISRFYWGGDASKRGIHWVKWK 855
LS AG+EVLIKS+ AIP+Y MSCF+LP L ++I S + F+W K I WV W
Sbjct: 238 LLSPAGKEVLIKSIVTAIPTYSMSCFLLPMRLIHQITSAMRWFWWSNTKVKHKIPWVAWS 297
Query: 856 TMCQPKHVGGLGFRDFKSFNLALVAKNWWRIYNYPDSLLGKVFKSVYFSSGRMECAKKGY 915
+ PK +GGL RD K FN+AL+AK WRI P SL+ +VFK+ YF R+ AK
Sbjct: 298 KLNDPKKMGGLAIRDLKDFNIALLAKQSWRILQQPFSLMARVFKAKYFPKERLLDAKATS 357
Query: 916 RPSYAWSRILKTSAMIQKGSCWRIGEGSRVRIWEDNWLANG---PPVNFRQDVVDELGLT 972
+ SYAW IL + +I +G + G G+ +++W+DNWL PPV + +L
Sbjct: 358 QSSYAWKSILHGTKLISRGLKYIAGNGNNIQLWKDNWLPLNPPRPPVGTCDSIYSQL--- 414
Query: 973 KVADLMLSGNRGWNVPLIEWTFCPATASRIMSVPLPRQPESDQLFWLGTADGLYSVKTGY 1032
KV+DL++ G WN L+ I ++ +D + W+ T DG YSVK+GY
Sbjct: 415 KVSDLLIEGR--WNEDLLCKLIHQNDIPHIRAIRPSITGANDAITWIYTHDGNYSVKSGY 472
Query: 1033 EFLQEVVAQDSPS-SSLSRGLDQTLWKRFWKAPSMPRVREVSWRVCTGALPVRTKLRQRG 1091
L+++ Q S S + QT++ WK + P+++ WR ALP L++R
Sbjct: 473 HLLRKLSQQQHASLPSPNEVSAQTVFTNIWKQNAPPKIKHFWWRSAHNALPTAGNLKRRR 532
Query: 1092 VDVDSSCPLCGLEDETVDHLFLGCNITRGCWFASPMGVRVDPSLKMVDFLTMV--IRDMD 1149
+ D +C CG E V+HL C +++ W + + + SL F + I+ ++
Sbjct: 533 LITDDTCQRCGEASEDVNHLLFQCRVSKEIWEQAHIKLCPGDSLMSNSFNQNLESIQKLN 592
Query: 1150 LEVVACVQRFLF---AIWEARNKHIFEEKLFCIAGVLDRAASLVSQSTLPFTLAASQEKD 1206
V F F IW+ RN IF K + I + +A L+ Q +L ++++
Sbjct: 593 QSARKDVSLFPFIGWRIWKMRNDLIFNNKRWSIPDSIQKA--LIDQQQWKESLNCNEQQQ 650
Query: 1207 NLKRWRRPAENVVKVNVDAAVGQDRFAGFGLVAR--DHAGEVLAAAAKYPIMVLSPTVAE 1264
K + + + G R A VL + P +P AE
Sbjct: 651 R------------KPQLHDSHNNKCYGGHLKTLRTLSKAQFVLRGMSSIP-PTSTPLEAE 697
Query: 1265 ALSLRWAMDLAIQLGFRRVQFETDCLLLQQ 1294
A +L+ AM +LG+ + F D L Q
Sbjct: 698 AEALKLAMIHLQRLGYEDIIFHGDVAALFQ 727
>At1g25430 hypothetical protein
Length = 1213
Score = 425 bits (1093), Expect = e-119
Identities = 325/1149 (28%), Positives = 520/1149 (44%), Gaps = 70/1149 (6%)
Query: 8 LLSWNCRGLGNLEAVRALRGLIHSQAPDIVFLMETKLFSFEMQRHRGMGGLSNIFPVQCG 67
L WN RG N+ + + + P ++ET + + ++ F
Sbjct: 5 LFCWNIRGFNNVSHRSGFKKWVKANKPIFGGVIETHVKQPKDRKFINALLPGWSFVENYA 64
Query: 68 RNRAGGLCLLWRSEVEVTIINASLHHILFTVVHSDSVTVPMQVYALYGFPELHLKDRTW- 126
+ G + ++W V+V ++ SL I V+ S + + V +Y E+ + W
Sbjct: 65 FSDLGKIWVMWDPSVQVVVVAKSLQMITCEVLLPGSPSWII-VSVVYAANEVASRKELWI 123
Query: 127 EMIRSVRP--MAPIPWLCIGDFNDILSPSDKLGGAPPDLARLQVATQACADC----GLQR 180
E++ V + PWL +GDFN +L+P + + P + + + DC L
Sbjct: 124 EIVNMVVSGIIGDRPWLVLGDFNQVLNPQEH---SNPVSLNVDINMRDFRDCLLAAELSD 180
Query: 181 VDFSGPSFTWTNNRVGPGRVDERLDYALINQAWVNLWPVSSVSHLIRHQSDHNPVVLHCG 240
+ + G +FTW N + V +++D L+N +W L+P S SDH + CG
Sbjct: 181 LRYKGNTFTWWN-KSHTTPVAKKIDRILVNDSWNALFPSSLGIFGSLDFSDH----VSCG 235
Query: 241 SRRTETQRHRTRMFRFEEVWLESGEECAEIISEGWSNTNL------SILSRINLVGRSLD 294
ET R F+F L++ + ++ + W N+ + ++ + + +
Sbjct: 236 VVLEETSIKAKRPFKFFNYLLKN-LDFLNLVRDNWFTLNVVGSSMFRVSKKLKALKKPIK 294
Query: 295 SWGREKYGELPKRIKEARAFLQRLQEEVQTEQV-VRAT--REAEKNLDVLLKQEEIVWSQ 351
+ R Y EL KR KEA FL Q+ + + A+ EAE+ +L EE + Q
Sbjct: 295 DFSRLNYSELEKRTKEAHDFLIGCQDRTLADPTPINASFELEAERKWHILTAAEESFFRQ 354
Query: 352 RSRANWLKHGDRNTKFFHSKATQRRKRNLIEEIQDDNGRKFVRDADIARVLTSYFQEIFS 411
+SR +W GD NTK+FH A R N I + D NG+ I + SYF +
Sbjct: 355 KSRISWFAEGDGNTKYFHRMADARNSSNSISALYDGNGKLVDSQEGILDLCASYFGSLLG 414
Query: 412 T-CNPTGIEE--VATLVDGRVTDAHRRILSTPFTREDVEEALFQMHPTKAPGLDGFPALF 468
+P +E+ + L+ R + A L + F+ ED+ ALF + K+ G DGF A F
Sbjct: 415 DEVDPYLMEQNDMNLLLSYRCSPAQVCELESTFSNEDIRAALFSLPRNKSCGPDGFTAEF 474
Query: 469 YQKFWPIVGDDISEFCLQVLQGDISPGMVNQTLIVLIPKVKKAVFANQYRPISLCNVIFK 528
+ W IVG ++++ + N T IVLIPK+ + +RPIS N ++K
Sbjct: 475 FIDSWSIVGAEVTDAIKEFFSSGCLLKQWNATTIVLIPKIVNPTCTSDFRPISCLNTLYK 534
Query: 529 IISKTIANRMKLILHDLISEAQSAFVPGRLITDNALIAFECFH-YMKKRISGRSGMMALK 587
+I++ + +R++ +L +IS AQSAF+PGR + +N L+A + H Y IS R GM LK
Sbjct: 535 VIARLLTDRLQRLLSGVISSAQSAFLPGRSLAENVLLATDLVHGYNWSNISPR-GM--LK 591
Query: 588 LDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVNFAVMLNGNPQPTFAPHRGLR 647
+D+ KA+D V W F+ + L + P ++N + C++T F V +NG F +GLR
Sbjct: 592 VDLKKAFDSVRWEFVIAALRALAIPEKFINWISQCISTPTFTVSINGGNGGFFKSTKGLR 651
Query: 648 QGDPLSPYLFILCGEAFSALIQKSISTSALHGIKISRSAPVISHLLFADDSVLFARATVQ 707
QGDPLSPYLF+L EAFS L+ + +H S ISHL+FADD ++F
Sbjct: 652 QGDPLSPYLFVLAMEAFSNLLHSRYESGLIH-YHPKASNLSISHLMFADDVMIFFDGGSF 710
Query: 708 EAECIKNILATYERASGQKINLEKSMLSVS--RNVPENCFVDLQLLLGVKAVDSYDKYLG 765
I L + SG K+N +KS L ++ + N +G + +YLG
Sbjct: 711 SLHGICETLDDFASWSGLKVNKDKSHLYLAGLNQLESNANAAYGFPIGTLPI----RYLG 766
Query: 766 LPTIIGKSKTQIFRFVKERVWKKLKGWKEQTLSRAGREVLIKSVAQAIPSYVMSCFVLPD 825
LP + K + + + E++ + + W + LS AGR LI SV ++ MS F+LP
Sbjct: 767 LPLMNRKLRIAEYEPLLEKITARFRSWVNKCLSFAGRIQLISSVIFGSINFWMSTFLLPK 826
Query: 826 GLCNEIESMISRFYWGGDASKRGIHWVKWKTMCQPKHVGGLGFRDFKSFNLALVAKNWWR 885
G IES+ SRF W G+ + V W +C PK GGLG R +N L + WR
Sbjct: 827 GCIKRIESLCSRFLWSGNIEQAKGIKVSWAALCLPKSEGGLGLRRLLEWNKTLSMRLIWR 886
Query: 886 IYNYPDSLLGKVFKSVYFSSGRMECAKKGYRPSYAWSRILKTSAMIQKGSCWRIGEGSRV 945
++ DSL + S G + G S+ W R+L + + ++G G +
Sbjct: 887 LFVAKDSLWADWQHLHHLSRGSFWAVEGGQSDSWTWKRLLSLRPLAHQFLVCKVGNGLKA 946
Query: 946 RIWEDNWLANGPPVNFRQDVVDELG--------LTKVADLMLSGNRGWNVPLIEWTFCPA 997
W DNW + GP FR ++ ++G L KVA GW +P+
Sbjct: 947 DYWYDNWTSLGP--LFR--IIGDIGPSSLRVPLLAKVASAF--SEDGWRLPVSRSAPAKG 1000
Query: 998 TASRIMSVPLPRQPESDQLFWLGTADGL----YSVKTGYEFLQEVVAQDSPSSSLSRGLD 1053
+ +VP+P + D + + +G +S +E ++ S +SS+
Sbjct: 1001 IHDHLCTVPVPSTAQEDVDRYEWSVNGFLCQGFSAAKTWEAIRPKATVKSWASSI----- 1055
Query: 1054 QTLWKRFWKAPSMPRVREVSWRVCTGALPVRTKLRQRGVDVDSSCPLCGLEDETVDHLFL 1113
W ++P+ W L R +L G +C LC E+ DHL L
Sbjct: 1056 -------WFKGAVPKYAFNMWVSHLNRLLTRQRLASWGHIQSDACVLCSFASESRDHLLL 1108
Query: 1114 GCNITRGCW 1122
C + W
Sbjct: 1109 ICEFSAQVW 1117
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.137 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,068,906
Number of Sequences: 26719
Number of extensions: 1337723
Number of successful extensions: 3591
Number of sequences better than 10.0: 140
Number of HSP's better than 10.0 without gapping: 109
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 2971
Number of HSP's gapped (non-prelim): 250
length of query: 1366
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1255
effective length of database: 8,352,787
effective search space: 10482747685
effective search space used: 10482747685
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0048.9


