

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0048.6
(1124 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
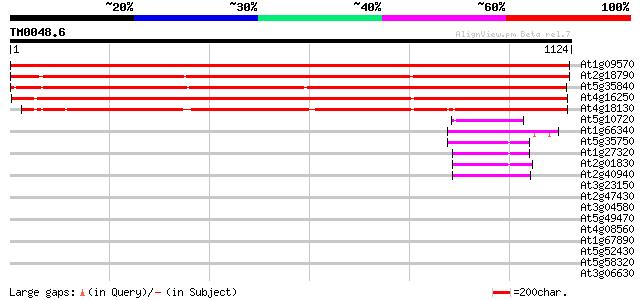
Score E
Sequences producing significant alignments: (bits) Value
At1g09570 putative phytochrome A 1808 0.0
At2g18790 phytochrome B 1157 0.0
At5g35840 phytochrome C (sp|P14714) 1155 0.0
At4g16250 phytochrome D 1150 0.0
At4g18130 phytochrome E 982 0.0
At5g10720 histidine kinase - like protein 65 3e-10
At1g66340 ethylene-response protein, ETR1 61 3e-09
At5g35750 histidine kinase (AHK2) 59 2e-08
At1g27320 histidine kinase (AHK3) 56 1e-07
At2g01830 cytokinin receptor CRE1b (CRE1) 52 1e-06
At2g40940 ethylene response sensor (ERS) 49 2e-05
At3g23150 thylene receptor like protein 40 0.006
At2g47430 putative histidine kinase 36 0.11
At3g04580 putative ethylene receptor 36 0.14
At5g49470 protein kinase like protein 35 0.18
At4g08560 putative self-incompatability RNA-binding protein 33 0.91
At1g67890 putative protein kinase (At1g67890) 32 1.6
At5g52430 putative protein 32 2.0
At5g58320 contains similarity to unknown protein (gb|AAB63087.1) 31 3.5
At3g06630 protein kinase, putative 31 3.5
>At1g09570 putative phytochrome A
Length = 1122
Score = 1808 bits (4682), Expect = 0.0
Identities = 886/1122 (78%), Positives = 1010/1122 (89%), Gaps = 1/1122 (0%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MS SRP+QSS S RSRHSAR+IAQTTVDAK+HA+FEESGSSFDYS+SVR +G +
Sbjct: 1 MSGSRPTQSSEGSRRSRHSARIIAQTTVDAKLHADFEESGSSFDYSTSVRVTGPVVENQP 60
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
P+S+KVTT YLHHIQ+GKLIQPFGCLLALDEKT KVIAYSENA E+LTM SHAVPSVGEH
Sbjct: 61 PRSDKVTTTYLHHIQKGKLIQPFGCLLALDEKTFKVIAYSENASELLTMASHAVPSVGEH 120
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
P LGI TDIR++FTAPSASALQKALGF +V+LLNPILVHC+TS KPFYAIIHRVTGS+II
Sbjct: 121 PVLGIGTDIRSLFTAPSASALQKALGFGDVSLLNPILVHCRTSAKPFYAIIHRVTGSIII 180
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM
Sbjct: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
AYKFHEDDHGEV++E+TKPGLEPYLGLHYPATDIPQA+RFLFMKNKVRMIVDC+AK +V
Sbjct: 241 AYKFHEDDHGEVVSEVTKPGLEPYLGLHYPATDIPQAARFLFMKNKVRMIVDCNAKHARV 300
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSV-QPQ 359
L DEKL FDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVN+ D +GD D+ QPQ
Sbjct: 301 LQDEKLSFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNEEDGEGDAPDATTQPQ 360
Query: 360 KRKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLL 419
KRKRLWGLVVCHNT+PRFVPFPLRYACEFLAQVFAIHVNKE+EL+ Q++EKNILRTQTLL
Sbjct: 361 KRKRLWGLVVCHNTTPRFVPFPLRYACEFLAQVFAIHVNKEVELDNQMVEKNILRTQTLL 420
Query: 420 CDMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHT 479
CDMLMRDAPLGI++QSPN+MDLVKCDGAALLYK+K+W LG TPSE H+++IASWL +YH
Sbjct: 421 CDMLMRDAPLGIVSQSPNIMDLVKCDGAALLYKDKIWKLGTTPSEFHLQEIASWLCEYHM 480
Query: 480 DSTGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPG 539
DSTGLSTDSL DAGFP ALSLGD VCGMAAVRI+ KD++FWFRSHTA E+RWGGAKH+P
Sbjct: 481 DSTGLSTDSLHDAGFPRALSLGDSVCGMAAVRISSKDMIFWFRSHTAGEVRWGGAKHDPD 540
Query: 540 EQDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTAI 599
++DD ++MHPRSSFKAFLEVV+ RS PWKDYEMDAIHSLQLILRNAFKD+++ D+NT I
Sbjct: 541 DRDDARRMHPRSSFKAFLEVVKTRSLPWKDYEMDAIHSLQLILRNAFKDSETTDVNTKVI 600
Query: 600 DTRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEA 659
++L+DLKI+G+QELEAVTSEMVRLIETATVPILAVD DGLVNGWN KIAELTGL V EA
Sbjct: 601 YSKLNDLKIDGIQELEAVTSEMVRLIETATVPILAVDSDGLVNGWNTKIAELTGLSVDEA 660
Query: 660 IGKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRD 719
IGKH LTLVED S + VK+ML+ AL G EE+NVQFEIKTH S+ ++GPISLVVNACASRD
Sbjct: 661 IGKHFLTLVEDSSVEIVKRMLENALEGTEEQNVQFEIKTHLSRADAGPISLVVNACASRD 720
Query: 720 LRENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNP 779
L ENVVGVCFVA D+T QKTVMDKFTRIEGDYKAI+QNPNPLIPPIFGTDEFGWC EWNP
Sbjct: 721 LHENVVGVCFVAHDLTGQKTVMDKFTRIEGDYKAIIQNPNPLIPPIFGTDEFGWCTEWNP 780
Query: 780 AMTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFG 839
AM+KLTG KREEV+DKMLLGEVFGT + CRLKNQEAFVN GIVLN A+T + EKV F
Sbjct: 781 AMSKLTGLKREEVIDKMLLGEVFGTQKSCCRLKNQEAFVNLGIVLNNAVTSQDPEKVSFA 840
Query: 840 FFARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKAL 899
FF R GKYVECLL VSKKLD EG+VTGVFCFLQLAS ELQQALH+Q L+E+TA+KRLKAL
Sbjct: 841 FFTRGGKYVECLLCVSKKLDREGVVTGVFCFLQLASHELQQALHVQRLAERTAVKRLKAL 900
Query: 900 TYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGY 959
Y+KRQIRNPLSGI+F+RK +EGT+LG EQ+R++ TSA CQ+QLSKILDDSDL+SI++G
Sbjct: 901 AYIKRQIRNPLSGIMFTRKMIEGTELGPEQRRILQTSALCQKQLSKILDDSDLESIIEGC 960
Query: 960 LDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFL 1019
LDLEM EFTL +VL S SQ+M +S+ + +RI N+ EE+M + LYGDS+RLQQVLADF+
Sbjct: 961 LDLEMKEFTLNEVLTASTSQVMMKSNGKSVRITNETGEEVMSDTLYGDSIRLQQVLADFM 1020
Query: 1020 LISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGLES 1079
L+++N TP+GGQ+ V+ASL K+QLG+SVHLANLE+ +TH G+G+PE LLNQMFG + S
Sbjct: 1021 LMAVNFTPSGGQLTVSASLRKDQLGRSVHLANLEIRLTHTGAGIPEFLLNQMFGTEEDVS 1080
Query: 1080 EEGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHK 1121
EEG+SL++SRKL+KLM+GDV+YLR+AGKSSFI++ ELAAA+K
Sbjct: 1081 EEGLSLMVSRKLVKLMNGDVQYLRQAGKSSFIITAELAAANK 1122
>At2g18790 phytochrome B
Length = 1172
Score = 1157 bits (2993), Expect = 0.0
Identities = 587/1130 (51%), Positives = 795/1130 (69%), Gaps = 22/1130 (1%)
Query: 2 SSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFE---ESGSSFDYSSSVRAS--GTAD 56
+ S ++S + ++ I Q TVDA++HA FE ESG SFDYS S++ + G++
Sbjct: 37 AQSSGTKSLRPRSNTESMSKAIQQYTVDARLHAVFEQSGESGKSFDYSQSLKTTTYGSSV 96
Query: 57 ADHQPKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPS 116
+ Q TAYL IQRG IQPFGC++A+DE + ++I YSENA EML ++ +VP+
Sbjct: 97 PEQQ------ITAYLSRIQRGGYIQPFGCMIAVDESSFRIIGYSENAREMLGIMPQSVPT 150
Query: 117 VGEHPALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTG 176
+ + L + TD+R++FT+ S+ L++A E+TLLNP+ +H K +GKPFYAI+HR+
Sbjct: 151 LEKPEILAMGTDVRSLFTSSSSILLERAFVAREITLLNPVWIHSKNTGKPFYAILHRIDV 210
Query: 177 SLIIDFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGY 236
++ID EP + + ++ AGA+QS KLA +AI++LQ+LP G ++ LCDT+V+ V +LTGY
Sbjct: 211 GVVIDLEPARTEDPALSIAGAVQSQKLAVRAISQLQALPGGDIKLLCDTVVESVRDLTGY 270
Query: 237 DRVMAYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAK 296
DRVM YKFHED+HGEV+AE + LEPY+GLHYPATDIPQASRFLF +N+VRMIVDC+A
Sbjct: 271 DRVMVYKFHEDEHGEVVAESKRDDLEPYIGLHYPATDIPQASRFLFKQNRVRMIVDCNAT 330
Query: 297 QVKVLIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSV 356
V V+ D++L + L GSTLRAPH CH QYMANM SIASL MAV++N N++DG S+
Sbjct: 331 PVLVVQDDRLTQSMCLVGSTLRAPHGCHSQYMANMGSIASLAMAVIINGNEDDG--SNVA 388
Query: 357 QPQKRKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQ 416
+ RLWGLVVCH+TS R +PFPLRYACEFL Q F + +N E++L Q+ EK +LRTQ
Sbjct: 389 SGRSSMRLWGLVVCHHTSSRCIPFPLRYACEFLMQAFGLQLNMELQLALQMSEKRVLRTQ 448
Query: 417 TLLCDMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSK 476
TLLCDML+RD+P GI+TQSP++MDLVKCDGAA LY K + LGV PSE+ I+D+ WL
Sbjct: 449 TLLCDMLLRDSPAGIVTQSPSIMDLVKCDGAAFLYHGKYYPLGVAPSEVQIKDVVEWLLA 508
Query: 477 YHTDSTGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKH 536
H DSTGLSTDSL DAG+PGA +LGD VCGMA IT +D +FWFRSHTA EI+WGGAKH
Sbjct: 509 NHADSTGLSTDSLGDAGYPGAAALGDAVCGMAVAYITKRDFLFWFRSHTAKEIKWGGAKH 568
Query: 537 EPGEQDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINT 596
P ++DDG++MHPRSSF+AFLEVV++RS PW+ EMDAIHSLQLILR++FK++++ +N+
Sbjct: 569 HPEDKDDGQRMHPRSSFQAFLEVVKSRSQPWETAEMDAIHSLQLILRDSFKESEAA-MNS 627
Query: 597 TAIDTRLSDLK----IEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELT 652
+D + + +G+ EL AV EMVRLIETATVPI AVD G +NGWN KIAELT
Sbjct: 628 KVVDGVVQPCRDMAGEQGIDELGAVAREMVRLIETATVPIFAVDAGGCINGWNAKIAELT 687
Query: 653 GLPVGEAIGKHLLT-LVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLV 711
GL V EA+GK L++ L+ + V K+L AL G+EEKNV+ ++KT +++ + +V
Sbjct: 688 GLSVEEAMGKSLVSDLIYKENEATVNKLLSRALRGDEEKNVEVKLKTFSPELQGKAVFVV 747
Query: 712 VNACASRDLRENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEF 771
VNAC+S+D N+VGVCFV QD+T+QK VMDKF I+GDYKAIV +PNPLIPPIF DE
Sbjct: 748 VNACSSKDYLNNIVGVCFVGQDVTSQKIVMDKFINIQGDYKAIVHSPNPLIPPIFAADEN 807
Query: 772 GWCCEWNPAMTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGS 831
C EWN AM KLTGW R EV+ KM++GEVFG + C LK +A F IVL+ A+ G
Sbjct: 808 TCCLEWNMAMEKLTGWSRSEVIGKMIVGEVFG---SCCMLKGPDALTKFMIVLHNAIGGQ 864
Query: 832 ETEKVGFGFFARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQT 891
+T+K F FF R+GK+V+ LL+ +K++ +EG V G FCFLQ+ SPELQQAL +Q +
Sbjct: 865 DTDKFPFPFFDRNGKFVQALLTANKRVSLEGKVIGAFCFLQIPSPELQQALAVQRRQDTE 924
Query: 892 ALKRLKALTYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSD 951
+ K L Y+ + I+NPLSG+ F+ LE TDL +QK+L+ TS C++Q+S+I+ D D
Sbjct: 925 CFTKAKELAYICQVIKNPLSGMRFANSLLEATDLNEDQKQLLETSVSCEKQISRIVGDMD 984
Query: 952 LDSIMDGYLDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRL 1011
L+SI DG L+ EF L V+ +SQ M RG++++ D+ EEI ++GD +R+
Sbjct: 985 LESIEDGSFVLKREEFFLGSVINAIVSQAMFLLRDRGLQLIRDIPEEIKSIEVFGDQIRI 1044
Query: 1012 QQVLADFLLISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQM 1071
QQ+LA+FLL I P+ V + S +Q+ E + G G+P L+ M
Sbjct: 1045 QQLLAEFLLSIIRYAPSQEWVEIHLSQLSKQMADGFAAIRTEFRMACPGEGLPPELVRDM 1104
Query: 1072 FGNDGLESEEGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHK 1121
F + S EG+ L + RK+LKLM+G+V+Y+RE+ +S F++ +EL K
Sbjct: 1105 FHSSRWTSPEGLGLSVCRKILKLMNGEVQYIRESERSYFLIILELPVPRK 1154
>At5g35840 phytochrome C (sp|P14714)
Length = 1111
Score = 1155 bits (2988), Expect = 0.0
Identities = 592/1119 (52%), Positives = 788/1119 (69%), Gaps = 19/1119 (1%)
Query: 3 SSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQPK 62
SS S+S S RSR ++RV +Q VDAK+H NFEES FDYS+S+ + + + P
Sbjct: 2 SSNTSRSC--STRSRQNSRVSSQVLVDAKLHGNFEESERLFDYSASINLNMPSSSCEIPS 59
Query: 63 SNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEHPA 122
S + YL IQRG LIQPFGCL+ +DEK KVIA+SEN EML ++ H VPS+ + A
Sbjct: 60 S--AVSTYLQKIQRGMLIQPFGCLIVVDEKNLKVIAFSENTQEMLGLIPHTVPSMEQREA 117
Query: 123 LGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLIIDF 182
L I TD++++F +P SAL+KA+ F E+++LNPI +HC++S KPFYAI+HR+ L+ID
Sbjct: 118 LTIGTDVKSLFLSPGCSALEKAVDFGEISILNPITLHCRSSSKPFYAILHRIEEGLVIDL 177
Query: 183 EPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVMAY 242
EPV P EVP+TAAGAL+SYKLAAK+I+RLQ+LPSG+M LCD +V+EV ELTGYDRVM Y
Sbjct: 178 EPVSPDEVPVTAAGALRSYKLAAKSISRLQALPSGNMLLLCDALVKEVSELTGYDRVMVY 237
Query: 243 KFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKVLI 302
KFHED HGEVIAE + +EPYLGLHY ATDIPQASRFLFM+NKVRMI DC A VKV+
Sbjct: 238 KFHEDGHGEVIAECCREDMEPYLGLHYSATDIPQASRFLFMRNKVRMICDCSAVPVKVVQ 297
Query: 303 DEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSVQPQKRK 362
D+ L ++L GSTLRAPH CH QYM+NM S+ASLVM+V +N +D D D Q +
Sbjct: 298 DKSLSQPISLSGSTLRAPHGCHAQYMSNMGSVASLVMSVTINGSDSDEMNRDL---QTGR 354
Query: 363 RLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLLCDM 422
LWGLVVCH+ SPRFVPFPLRYACEFL QVF + +NKE E + EK IL+TQ++LCDM
Sbjct: 355 HLWGLVVCHHASPRFVPFPLRYACEFLTQVFGVQINKEAESAVLLKEKRILQTQSVLCDM 414
Query: 423 LMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHTDST 482
L R+AP+GI+TQSPN+MDLVKCDGAAL Y++ +W LGVTP+E IRD+ W+ K H +T
Sbjct: 415 LFRNAPIGIVTQSPNIMDLVKCDGAALYYRDNLWSLGVTPTETQIRDLIDWVLKSHGGNT 474
Query: 483 GLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPGEQD 542
G +T+SL ++G+P A LG+ +CGMAAV I+ KD +FWFRS TA +I+WGGA+H+P ++
Sbjct: 475 GFTTESLMESGYPDASVLGESICGMAAVYISEKDFLFWFRSSTAKQIKWGGARHDPNDR- 533
Query: 543 DGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTAIDTR 602
DGK+MHPRSSFKAF+E+VR +S PW D EMDAI+SLQLI++ + ++ S T +D
Sbjct: 534 DGKRMHPRSSFKAFMEIVRWKSVPWDDMEMDAINSLQLIIKGSLQEEHS----KTVVDVP 589
Query: 603 LSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEAIGK 662
L D +++ + EL + +EMVRLI+TA VPI AVD G++NGWN K AE+TGL V +AIGK
Sbjct: 590 LVDNRVQKVDELCVIVNEMVRLIDTAAVPIFAVDASGVINGWNSKAAEVTGLAVEQAIGK 649
Query: 663 HLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRDLRE 722
+ LVED S + VK ML LAL G EE+ + I+ G K +S P+ LVVN C SRD+
Sbjct: 650 PVSDLVEDDSVETVKNMLALALEGSEERGAEIRIRAFGPKRKSSPVELVVNTCCSRDMTN 709
Query: 723 NVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNPAMT 782
NV+GVCF+ QD+T QKT+ + ++R++GDY I+ +P+ LIPPIF T+E G C EWN AM
Sbjct: 710 NVLGVCFIGQDVTGQKTLTENYSRVKGDYARIMWSPSTLIPPIFITNENGVCSEWNNAMQ 769
Query: 783 KLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSET-EKVGFGFF 841
KL+G KREEV++K+LLGEVF T C LK+ + I N ++G + EK+ FGF+
Sbjct: 770 KLSGIKREEVVNKILLGEVFTTDDYGCCLKDHDTLTKLRIGFNAVISGQKNIEKLLFGFY 829
Query: 842 ARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKALTY 901
R G ++E LLS +K+ D+EG VTGV CFLQ+ SPELQ AL +Q +SE L L Y
Sbjct: 830 HRDGSFIEALLSANKRTDIEGKVTGVLCFLQVPSPELQYALQVQQISEHAIACALNKLAY 889
Query: 902 MKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYLD 961
++ ++++P I F + L + L +QKRL+ TS C+ QL+K++ DSD++ I +GY++
Sbjct: 890 LRHEVKDPEKAISFLQDLLHSSGLSEDQKRLLRTSVLCREQLAKVISDSDIEGIEEGYVE 949
Query: 962 LEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFLLI 1021
L+ +EF LQ+ L + Q+M S R ++I D +E+ LYGD+LRLQQ+L++ LL
Sbjct: 950 LDCSEFGLQESLEAVVKQVMELSIERKVQISCDYPQEVSSMRLYGDNLRLQQILSETLLS 1009
Query: 1022 SINCTP--NGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFG--NDGL 1077
SI TP G V E +GK + LE I H G+PE L+ +MF G
Sbjct: 1010 SIRFTPALRGLCVSFKVIARIEAIGKRMKRVELEFRIIHPAPGLPEDLVREMFQPLRKG- 1068
Query: 1078 ESEEGISLLISRKLLKLMS-GDVRYLREAGKSSFILSVE 1115
S EG+ L I++KL+KLM G +RYLRE+ S+F++ E
Sbjct: 1069 TSREGLGLHITQKLVKLMERGTLRYLRESEMSAFVILTE 1107
>At4g16250 phytochrome D
Length = 1164
Score = 1150 bits (2976), Expect = 0.0
Identities = 585/1121 (52%), Positives = 782/1121 (69%), Gaps = 16/1121 (1%)
Query: 4 SRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFE---ESGSSFDYSSSVRASGTADADHQ 60
S+ Q N+ G + + + I Q TVDA++HA FE ESG SFDYS S++ TA D
Sbjct: 41 SQNQQPQNHGGGTESTNKAIQQYTVDARLHAVFEQSGESGKSFDYSQSLK---TAPYDSS 97
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
++T AYL IQRG QPFGCL+A++E T +I YSENA EML ++S +VPS+ +
Sbjct: 98 VPEQQIT-AYLSRIQRGGYTQPFGCLIAVEESTFTIIGYSENAREMLGLMSQSVPSIEDK 156
Query: 121 P-ALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLI 179
L I TD+R++F + S L++A E+TLLNPI +H +GKPFYAI+HRV ++
Sbjct: 157 SEVLTIGTDLRSLFKSSSYLLLERAFVAREITLLNPIWIHSNNTGKPFYAILHRVDVGIL 216
Query: 180 IDFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRV 239
ID EP + + ++ AGA+QS KLA +AI+ LQSLPSG ++ LCDT+V+ V +LTGYDRV
Sbjct: 217 IDLEPARTEDPALSIAGAVQSQKLAVRAISHLQSLPSGDIKLLCDTVVESVRDLTGYDRV 276
Query: 240 MAYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVK 299
M YKFHED+HGEV+AE + LEPY+GLHYPATDIPQASRFLF +N+VRMIVDC+A V+
Sbjct: 277 MVYKFHEDEHGEVVAESKRNDLEPYIGLHYPATDIPQASRFLFKQNRVRMIVDCYASPVR 336
Query: 300 VLIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSVQPQ 359
V+ D++L + L GSTLRAPH CH QYM NM SIASL MAV++N N+EDG+G ++ +
Sbjct: 337 VVQDDRLTQFICLVGSTLRAPHGCHAQYMTNMGSIASLAMAVIINGNEEDGNGVNT-GGR 395
Query: 360 KRKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLL 419
RLWGLVVCH+TS R +PFPLRYACEFL Q F + +N E++L Q+ EK +LR QTLL
Sbjct: 396 NSMRLWGLVVCHHTSARCIPFPLRYACEFLMQAFGLQLNMELQLALQVSEKRVLRMQTLL 455
Query: 420 CDMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHT 479
CDML+RD+P GI+TQ P++MDLVKC+GAA LY+ K + LGVTP++ I DI WL H+
Sbjct: 456 CDMLLRDSPAGIVTQRPSIMDLVKCNGAAFLYQGKYYPLGVTPTDSQINDIVEWLVANHS 515
Query: 480 DSTGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPG 539
DSTGLSTDSL DAG+P A +LGD VCGMA IT +D +FWFRSHT EI+WGGAKH P
Sbjct: 516 DSTGLSTDSLGDAGYPRAAALGDAVCGMAVACITKRDFLFWFRSHTEKEIKWGGAKHHPE 575
Query: 540 EQDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTT-- 597
++DDG++M+PRSSF+ FLEVV++R PW+ EMDAIHSLQLILR++FK++++MD
Sbjct: 576 DKDDGQRMNPRSSFQTFLEVVKSRCQPWETAEMDAIHSLQLILRDSFKESEAMDSKAAAA 635
Query: 598 -AIDTRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPV 656
A+ D+ +GMQE+ AV EMVRLIETATVPI AVDIDG +NGWN KIAELTGL V
Sbjct: 636 GAVQPHGDDMVQQGMQEIGAVAREMVRLIETATVPIFAVDIDGCINGWNAKIAELTGLSV 695
Query: 657 GEAIGKHLL-TLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNAC 715
+A+GK L+ L+ + V ++L AL G+E KNV+ ++KT GS+++ + +VVNAC
Sbjct: 696 EDAMGKSLVRELIYKEYKETVDRLLSCALKGDEGKNVEVKLKTFGSELQGKAMFVVVNAC 755
Query: 716 ASRDLRENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCC 775
+S+D N+VGVCFV QD+T K VMDKF I+GDYKAI+ +PNPLIPPIF DE C
Sbjct: 756 SSKDYLNNIVGVCFVGQDVTGHKIVMDKFINIQGDYKAIIHSPNPLIPPIFAADENTCCL 815
Query: 776 EWNPAMTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEK 835
EWN AM KLTGW R EV+ K+L+ EVFG++ CRLK +A F IVL+ A+ G +T+K
Sbjct: 816 EWNTAMEKLTGWPRSEVIGKLLVREVFGSY---CRLKGPDALTKFMIVLHNAIGGQDTDK 872
Query: 836 VGFGFFARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKR 895
F FF R G++++ LL+++K++ ++G + G FCFLQ+ SPELQQAL +Q E R
Sbjct: 873 FPFPFFDRKGEFIQALLTLNKRVSIDGKIIGAFCFLQIPSPELQQALEVQRRQESEYFSR 932
Query: 896 LKALTYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSI 955
K L Y+ + I+NPLSG+ F+ LE DL +QK+L+ TS C++Q+SKI+ D D+ SI
Sbjct: 933 RKELAYIFQVIKNPLSGLRFTNSLLEDMDLNEDQKQLLETSVSCEKQISKIVGDMDVKSI 992
Query: 956 MDGYLDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVL 1015
DG LE EF + +V +SQ+M R ++++ ++ E+ +YGD +RLQQVL
Sbjct: 993 DDGSFLLERTEFFIGNVTNAVVSQVMLVVRERNLQLIRNIPTEVKSMAVYGDQIRLQQVL 1052
Query: 1016 ADFLLISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGND 1075
A+FLL + P G V + T Q+ LE + G GVP + MF +
Sbjct: 1053 AEFLLSIVRYAPMEGSVELHLCPTLNQMADGFSAVRLEFRMACAGEGVPPEKVQDMFHSS 1112
Query: 1076 GLESEEGISLLISRKLLKLMSGDVRYLREAGKSSFILSVEL 1116
S EG+ L + RK+LKLM+G V+Y+RE +S F++ +EL
Sbjct: 1113 RWTSPEGLGLSVCRKILKLMNGGVQYIREFERSYFLIVIEL 1153
>At4g18130 phytochrome E
Length = 1112
Score = 982 bits (2538), Expect = 0.0
Identities = 529/1110 (47%), Positives = 731/1110 (65%), Gaps = 55/1110 (4%)
Query: 24 AQTTVDAKIHANFEES---GSSFDYSSSVRASGTADADHQPKSNKVTTAYLHHIQRGKLI 80
AQ +VDA + A+F +S G SF+YS SV + +H P + TAYL +IQRG L+
Sbjct: 23 AQYSVDAALFADFAQSIYTGKSFNYSKSV----ISPPNHVPDEH--ITAYLSNIQRGGLV 76
Query: 81 QPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSV---GEHPALG--IDTDIRTIFTA 135
QPFGCL+A++E + +++ S+N+ + L ++S +PS GE + I D RT+FT
Sbjct: 77 QPFGCLIAVEEPSFRILGLSDNSSDFLGLLS--LPSTSHSGEFDKVKGLIGIDARTLFTP 134
Query: 136 PSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLIIDFEPVKPYEVPMTAA 195
S ++L KA F E++LLNP+LVH +T+ KPFYAI+HR+ +++D EP K + +T A
Sbjct: 135 SSGASLSKAASFTEISLLNPVLVHSRTTQKPFYAILHRIDAGIVMDLEPAKSGDPALTLA 194
Query: 196 GALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVMAYKFHEDDHGEVIAE 255
GA+QS KLA +AI+RLQSLP G + LCDT+V++V LTGYDRVM Y+FHEDDHGEV++E
Sbjct: 195 GAVQSQKLAVRAISRLQSLPGGDIGALCDTVVEDVQRLTGYDRVMVYQFHEDDHGEVVSE 254
Query: 256 ITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKVLIDEKLPFDLTLCGS 315
I + LEPYLGLHYPATDIPQA+RFLF +N+VRMI DC+A VKV+ E+L L L S
Sbjct: 255 IRRSDLEPYLGLHYPATDIPQAARFLFKQNRVRMICDCNATPVKVVQSEELKRPLCLVNS 314
Query: 316 TLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSVQPQKRKRLWGLVVCHNTSP 375
TLRAPH CH QYMANM S+ASL +A+VV D +LWGLVV H+ SP
Sbjct: 315 TLRAPHGCHTQYMANMGSVASLALAIVVKGKDS-------------SKLWGLVVGHHCSP 361
Query: 376 RFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLLCDMLMRDAPLGILTQS 435
R+VPFPLRYACEFL Q F + + E++L Q+ EK +RTQTLLCDML+RD I+TQS
Sbjct: 362 RYVPFPLRYACEFLMQAFGLQLQMELQLASQLAEKKAMRTQTLLCDMLLRDTVSAIVTQS 421
Query: 436 PNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYH-TDSTGLSTDSLSDAGF 494
P +MDLVKCDGAAL YK K W++GVTP+E ++D+ +WL + H DSTGL+TDSL DAG+
Sbjct: 422 PGIMDLVKCDGAALYYKGKCWLVGVTPNESQVKDLVNWLVENHGDDSTGLTTDSLVDAGY 481
Query: 495 PGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPGEQDDGKKMHPRSSFK 554
PGA+SLGD VCG+AA + KD + WFRS+TA+ I+WGGAKH P ++DD +MHPRSSF
Sbjct: 482 PGAISLGDAVCGVAAAEFSSKDYLLWFRSNTASAIKWGGAKHHPKDKDDAGRMHPRSSFT 541
Query: 555 AFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDT-DSMDINTTAIDTRLSDLKIEGMQE 613
AFLEV ++RS PW+ E+DAIHSL+LI+R +F + + N A D E
Sbjct: 542 AFLEVAKSRSLPWEISEIDAIHSLRLIMRESFTSSRPVLSGNGVARDA----------NE 591
Query: 614 LEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEAIGKHLL-TLVEDCS 672
L + EMVR+IETAT PI VD G +NGWN K AE+TGL EA+GK L +V++ S
Sbjct: 592 LTSFVCEMVRVIETATAPIFGVDSSGCINGWNKKTAEMTGLLASEAMGKSLADEIVQEES 651
Query: 673 TDRVKKMLDLALSGEEEKNVQFEIKTHGSKME---SGPISLVVNACASRDLRENVVGVCF 729
++ +L AL GEEEK+V +++ G S + ++VN+C SRD EN++GVCF
Sbjct: 652 RAALESLLCKALQGEEEKSVMLKLRKFGQNNHPDYSSDVCVLVNSCTSRDYTENIIGVCF 711
Query: 730 VAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNPAMTKLTGWKR 789
V QDIT++K + D+F R++GDYK IVQ+ NPLIPPIF +DE C EWN AM KLTGW +
Sbjct: 712 VGQDITSEKAITDRFIRLQGDYKTIVQSLNPLIPPIFASDENACCSEWNAAMEKLTGWSK 771
Query: 790 EEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSET-EKVGFGFFARSGKYV 848
EV+ KML GEVFG C++K Q++ F I L + + G E FF + GKY+
Sbjct: 772 HEVIGKMLPGEVFGVF---CKVKCQDSLTKFLISLYQGIAGDNVPESSLVEFFNKEGKYI 828
Query: 849 ECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKALTYMKRQIRN 908
E L+ +K ++EG V F FLQ+ + E L L E + + L LTY++++I+N
Sbjct: 829 EASLTANKSTNIEGKVIRCFFFLQIINKE--SGLSCPELKE--SAQSLNELTYVRQEIKN 884
Query: 909 PLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYLDLEMAEFT 968
PL+GI F+ K LE +++ Q++ + TS C++Q++ I++ +DL SI +G L LE EF
Sbjct: 885 PLNGIRFAHKLLESSEISASQRQFLETSDACEKQITTIIESTDLKSIEEGKLQLETEEFR 944
Query: 969 LQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFLLISINCTP- 1027
L+++L T +SQ+M R ++ +VAEEI L GD ++LQ +LAD L +N P
Sbjct: 945 LENILDTIISQVMIILRERNSQLRVEVAEEIKTLPLNGDRVKLQLILADLLRNIVNHAPF 1004
Query: 1028 NGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMF-GNDGLESEEGISLL 1086
V ++ S +E + +L+ + H G G+P +L+ MF DG + +G+ L
Sbjct: 1005 PNSWVGISISPGQELSRDNGRYIHLQFRMIHPGKGLPSEMLSDMFETRDGWVTPDGLGLK 1064
Query: 1087 ISRKLLKLMSGDVRYLREAGKSSFILSVEL 1116
+SRKLL+ M+G V Y+RE + F + +++
Sbjct: 1065 LSRKLLEQMNGRVSYVREDERCFFQVDLQV 1094
>At5g10720 histidine kinase - like protein
Length = 950
Score = 64.7 bits (156), Expect = 3e-10
Identities = 40/145 (27%), Positives = 83/145 (56%), Gaps = 4/145 (2%)
Query: 886 HLSEQTALKRLKALTYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSK 945
H++E+T ++ + L M +IR+PLSG+V + L T L EQ++L++ + +
Sbjct: 355 HITEET-MRAKQMLATMSHEIRSPLSGVVGMAEILSTTKLDKEQRQLLNVMISSGDLVLQ 413
Query: 946 ILDD-SDLDSIMDGYLDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEIL 1004
+++D DL + G + LE +F ++V + + Q A S + + + ++A+++ +E++
Sbjct: 414 LINDILDLSKVESGVMRLEATKFRPREV-VKHVLQTAAASLKKSLTLEGNIADDVPIEVV 472
Query: 1005 YGDSLRLQQVLADFLLISINCTPNG 1029
GD LR++Q+L + + +I T G
Sbjct: 473 -GDVLRIRQILTNLISNAIKFTHEG 496
>At1g66340 ethylene-response protein, ETR1
Length = 738
Score = 61.2 bits (147), Expect = 3e-09
Identities = 55/240 (22%), Positives = 107/240 (43%), Gaps = 19/240 (7%)
Query: 877 ELQQALHIQHLSEQTALK-RLKALTYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHT 935
E AL + +TA++ R L M ++R P+ I+ L+ T+L EQ+ +V T
Sbjct: 325 EQNVALDLARREAETAIRARNDFLAVMNHEMRTPMHAIIALSSLLQETELTPEQRLMVET 384
Query: 936 SAQCQRQLSKILDD-SDLDSIMDGYLDLEMAEFTLQDVLITSLSQIMARSSARGIRIVND 994
+ L+ +++D DL + DG L LE+ F L + L+ I + + + I +
Sbjct: 385 ILKSSNLLATLMNDVLDLSRLEDGSLQLELGTFNLHTLFREVLNLIKPIAVVKKLPITLN 444
Query: 995 VAEEIMVEILYGDSLRLQQVLADFLLISINCTPNGGQVVVAASLTKEQLGKSVHLA---- 1050
+A + + E + GD RL Q++ + + ++ + G + V A +TK +
Sbjct: 445 LAPD-LPEFVVGDEKRLMQIILNIVGNAVKFSKQ-GSISVTALVTKSDTRAADFFVVPTG 502
Query: 1051 ---NLELSITHGGSGVPEALLNQMFGNDGLESE--------EGISLLISRKLLKLMSGDV 1099
L + + G+G+ + ++F G+ L IS++ + LM G++
Sbjct: 503 SHFYLRVKVKDSGAGINPQDIPKIFTKFAQTQSLATRSSGGSGLGLAISKRFVNLMEGNI 562
>At5g35750 histidine kinase (AHK2)
Length = 1176
Score = 58.5 bits (140), Expect = 2e-08
Identities = 37/166 (22%), Positives = 83/166 (49%), Gaps = 2/166 (1%)
Query: 877 ELQQALHIQHLSEQTALKRLKALTYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTS 936
+ Q+ ++ +E + + + L + +IR P++G++ K L TDL +Q T+
Sbjct: 570 DCQKMRELKARAEAADIAKSQFLATVSHEIRTPMNGVLGMLKMLMDTDLDAKQMDYAQTA 629
Query: 937 AQCQRQLSKILDD-SDLDSIMDGYLDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDV 995
+ L+ ++++ D I G L+LE F ++ +L S + +++ +GI + V
Sbjct: 630 HGSGKDLTSLINEVLDQAKIESGRLELENVPFDMRFILDNVSSLLSGKANEKGIELAVYV 689
Query: 996 AEEIMVEILYGDSLRLQQVLADFLLISINCTPNGGQVVVAASLTKE 1041
+ ++ +++ GD R +Q++ + + SI T G + ++ L E
Sbjct: 690 SSQV-PDVVVGDPSRFRQIITNLVGNSIKFTQERGHIFISVHLADE 734
>At1g27320 histidine kinase (AHK3)
Length = 1036
Score = 55.8 bits (133), Expect = 1e-07
Identities = 39/155 (25%), Positives = 77/155 (49%), Gaps = 3/155 (1%)
Query: 888 SEQTALKRLKALTYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKIL 947
+E + + + L + +IR P++G++ L T+L + Q+ V T+ + L ++
Sbjct: 444 AEAADVAKSQFLATVSHEIRTPMNGVLGMLHMLMDTELDVTQQDYVRTAQASGKALVSLI 503
Query: 948 DD-SDLDSIMDGYLDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYG 1006
++ D I G L+LE F L+ +L LS ++S +G+ + +++ + ++L G
Sbjct: 504 NEVLDQAKIESGKLELEEVRFDLRGILDDVLSLFSSKSQQKGVELAVYISDRV-PDMLIG 562
Query: 1007 DSLRLQQVLADFLLISINCTPNGGQVVVAASLTKE 1041
D R +Q+L + + SI T G + V L E
Sbjct: 563 DPGRFRQILTNLMGNSIKFTEK-GHIFVTVHLVDE 596
>At2g01830 cytokinin receptor CRE1b (CRE1)
Length = 844
Score = 52.4 bits (124), Expect = 1e-06
Identities = 40/160 (25%), Positives = 78/160 (48%), Gaps = 3/160 (1%)
Query: 888 SEQTALKRLKALTYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKIL 947
+E + + + L + +IR P++GI+ L T+L Q+ T+ C + L ++
Sbjct: 230 AEAADVAKSQFLATVSHEIRTPMNGILGMLAMLLDTELSSTQRDYAQTAQVCGKALIALI 289
Query: 948 DD-SDLDSIMDGYLDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYG 1006
++ D I G L+LE F ++ +L LS S + I + V++++ EI+ G
Sbjct: 290 NEVLDRAKIEAGKLELESVPFDIRSILDDVLSLFSEESRNKSIELAVFVSDKV-PEIVKG 348
Query: 1007 DSLRLQQVLADFLLISINCTPNGGQVVVAASLTKEQLGKS 1046
DS R +Q++ + + S+ T G + V L ++ +S
Sbjct: 349 DSGRFRQIIINLVGNSVKFTEK-GHIFVKVHLAEQSKDES 387
>At2g40940 ethylene response sensor (ERS)
Length = 613
Score = 48.9 bits (115), Expect = 2e-05
Identities = 38/157 (24%), Positives = 70/157 (44%), Gaps = 2/157 (1%)
Query: 888 SEQTALKRLKALTYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKIL 947
+E R L M ++R P+ I+ L T+L EQ+ ++ T + ++ ++
Sbjct: 337 AEMAVHARNDFLAVMNHEMRTPMHAIISLSSLLLETELSPEQRVMIETILKSSNLVATLI 396
Query: 948 DD-SDLDSIMDGYLDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYG 1006
D DL + DG L LE F+LQ + +S I +S + + N + + G
Sbjct: 397 SDVLDLSRLEDGSLLLENEPFSLQAIFEEVISLIKPIASVKKLS-TNLILSADLPTYAIG 455
Query: 1007 DSLRLQQVLADFLLISINCTPNGGQVVVAASLTKEQL 1043
D RL Q + + + ++ T G ++A+ + E L
Sbjct: 456 DEKRLMQTILNIMGNAVKFTKEGYISIIASIMKPESL 492
>At3g23150 thylene receptor like protein
Length = 773
Score = 40.4 bits (93), Expect = 0.006
Identities = 36/148 (24%), Positives = 65/148 (43%), Gaps = 7/148 (4%)
Query: 872 QLASPELQQALHIQHLSEQTALKRLKALTY----MKRQIRNPLSGIVFSRKTLEGTDLGI 927
QL +L + ++++ AL+ +A M +R P+ I+ ++ L
Sbjct: 341 QLMREKLAEQNRALQMAKRDALRASQARNAFQKTMSEGMRRPMHSILGLLSMIQDEKLSD 400
Query: 928 EQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYLDLEMAEFTLQDVLITSLSQIMARSSAR 987
EQK +V T + +S ++ DS + DG EM F+L + +
Sbjct: 401 EQKMIVDTMVKTGNVMSNLVGDS--MDVPDGRFGTEMKPFSLHRTIHEAACMARCLCLCN 458
Query: 988 GIRIVNDVAEEIMVEILYGDSLRLQQVL 1015
GIR + D AE+ + + + GD R+ QV+
Sbjct: 459 GIRFLVD-AEKSLPDNVVGDERRVFQVI 485
>At2g47430 putative histidine kinase
Length = 1122
Score = 36.2 bits (82), Expect = 0.11
Identities = 56/267 (20%), Positives = 103/267 (37%), Gaps = 49/267 (18%)
Query: 888 SEQTALKRLKALTYMKRQIRNPLSGIV-FSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKI 946
+E+ ++ + +A IR L+G+ +G G + ++ C + L +
Sbjct: 389 AERKSMNKSQAFANASHDIRGALAGMKGLIDICRDGVKPGSDVDTTLNQVNVCAKDLVAL 448
Query: 947 LDDS-DLDSIMDGYLDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEI-- 1003
L+ D+ I G + L +F L +L + + +G+ +V D + + +
Sbjct: 449 LNSVLDMSKIESGKMQLVEEDFNLSKLLEDVIDFYHPVAMKKGVDVVLDPHDGSVFKFSN 508
Query: 1004 LYGDSLRLQQVLADFLLISINCT-------------PNGGQVVVAASLTK--EQLGKSVH 1048
+ GDS RL+Q+L + + ++ T P VV AS K + KS+
Sbjct: 509 VRGDSGRLKQILNNLVSNAVKFTVDGHIAVRAWAQRPGSNSSVVLASYPKGVSKFVKSMF 568
Query: 1049 LANLELSITHG-----------------------GSGVPEALLNQMFGNDGLESEE---- 1081
N E S T+ G G+P + +F N E
Sbjct: 569 CKNKEESSTYETEISNSIRNNANTMEFVFEVDDTGKGIPMEMRKSVFENYVQVRETAQGH 628
Query: 1082 ---GISLLISRKLLKLMSGDVRYLREA 1105
G+ L I + L++LM G++R +A
Sbjct: 629 QGTGLGLGIVQSLVRLMGGEIRITDKA 655
>At3g04580 putative ethylene receptor
Length = 766
Score = 35.8 bits (81), Expect = 0.14
Identities = 44/222 (19%), Positives = 91/222 (40%), Gaps = 27/222 (12%)
Query: 902 MKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDS-DLDSIMDGYL 960
M +R P+ I+ + + ++QK +V + LS +++D D+ +G
Sbjct: 375 MSHGMRRPMHTILGLLSMFQSESMSLDQKIIVDALMKTSTVLSALINDVIDISPKDNGKS 434
Query: 961 DLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFLL 1020
LE+ F L ++ + S +G DV + ++ GD R Q++ L
Sbjct: 435 ALEVKRFQLHSLIREAACVAKCLSVYKGYGFEMDVQTRLP-NLVVGDEKRTFQLVMYMLG 493
Query: 1021 ISINCTPNGGQVV---------VAASLTKEQLGK-SVHLANLELSITH------------ 1058
++ T G V + +K + G H+++ L +
Sbjct: 494 YILDMTDGGKTVTFRVICEGTGTSQDKSKRETGMWKSHMSDDSLGVKFEVEINEIQNPPL 553
Query: 1059 -GGSGVPEALLNQMFGNDGLESEEGISLLISRKLLKLMSGDV 1099
G + + N+ + ++G++ EG+SL + RKL ++M G++
Sbjct: 554 DGSAMAMRHIPNRRYHSNGIK--EGLSLGMCRKLAQMMQGNI 593
>At5g49470 protein kinase like protein
Length = 770
Score = 35.4 bits (80), Expect = 0.18
Identities = 24/125 (19%), Positives = 50/125 (39%), Gaps = 4/125 (3%)
Query: 609 EGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEAIGKHLLTLV 668
EG + + + ++++ + D++G + WN +L G EA+GK + ++
Sbjct: 90 EGQSAGKFTDKQYLNILQSMAQAVHVFDLNGQIIFWNSMAEKLYGFSASEALGKDPIDIL 149
Query: 669 EDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRDLRENVVGVC 728
D V + + S E +F +K + S+V S D ++G+
Sbjct: 150 VDVQDASVAQNITRRCSSGESWTGEFPVKNKAGER----FSVVTTMSPSYDDDGCLIGII 205
Query: 729 FVAQD 733
+ D
Sbjct: 206 CITND 210
>At4g08560 putative self-incompatability RNA-binding protein
Length = 477
Score = 33.1 bits (74), Expect = 0.91
Identities = 23/100 (23%), Positives = 51/100 (51%), Gaps = 13/100 (13%)
Query: 667 LVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRDLRENVVG 726
L+E C+ +++ ++LD+ L E + V+ + THG+ G L+ + C+ + +
Sbjct: 207 LMEKCTDEQITRVLDIVLE-EPFEFVRLCVHTHGTHAIQG---LMRSLCSEEQISRFMET 262
Query: 727 VCFVA----QDITAQKTVMDKFTRIEGDY-----KAIVQN 757
+C+V+ +D+ A + ++ F + + + IVQN
Sbjct: 263 LCYVSLLLTKDVIAHRVILFCFNQFSPSHTRYLLEVIVQN 302
>At1g67890 putative protein kinase (At1g67890)
Length = 765
Score = 32.3 bits (72), Expect = 1.6
Identities = 17/78 (21%), Positives = 35/78 (44%)
Query: 620 EMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEAIGKHLLTLVEDCSTDRVKKM 679
+ + ++++ + D++G + WN +L G EA+GK + ++ D V K
Sbjct: 102 QYLNILQSMAQAVHVFDLNGQIIFWNSMAEKLYGFSAAEALGKDSINILVDGQDAAVAKN 161
Query: 680 LDLALSGEEEKNVQFEIK 697
+ S E +F +K
Sbjct: 162 IFQRCSSGESWTGEFPVK 179
>At5g52430 putative protein
Length = 438
Score = 32.0 bits (71), Expect = 2.0
Identities = 29/92 (31%), Positives = 40/92 (42%), Gaps = 14/92 (15%)
Query: 1020 LISINCTPNG------GQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQ--- 1070
++S N TPN Q+ ASL G V +A+ +S G V L ++
Sbjct: 297 IVSGNLTPNNTTWPLQNQISEVASLANSDHGSEVMVADHRVSFELTGEDVARCLASKLNR 356
Query: 1071 ----MFGNDGLESEEGISLLISRKLLKLMSGD 1098
M ND +E+EE S I R + K SGD
Sbjct: 357 SHDRMNNNDRIETEESSSTDIRRNIEK-RSGD 387
>At5g58320 contains similarity to unknown protein (gb|AAB63087.1)
Length = 589
Score = 31.2 bits (69), Expect = 3.5
Identities = 26/94 (27%), Positives = 39/94 (40%), Gaps = 15/94 (15%)
Query: 423 LMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSK------ 476
L + +PL + +Q L D+ D +AL N+V LG PS +L
Sbjct: 132 LRKGSPLELQSQGSGLSDISASDLSALWTSNEVNRLGRPPSGRRAPGFEYFLGNGGLPSD 191
Query: 477 -YHTD--------STGLSTDSLSDAGFPGALSLG 501
YH D + L +D S +PG +S+G
Sbjct: 192 LYHKDGDDSASITDSELESDDSSVTNYPGYVSIG 225
>At3g06630 protein kinase, putative
Length = 671
Score = 31.2 bits (69), Expect = 3.5
Identities = 23/116 (19%), Positives = 49/116 (41%), Gaps = 6/116 (5%)
Query: 620 EMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEAIGKHLLTLVEDCSTDRVKKM 679
+ + ++++ + +D++ + WN +L G E +G++ + ++ D
Sbjct: 65 QYLNILQSLAQSVHVLDLNTRIIFWNAMSEKLYGYSAAEVVGRNPVHVIVDDQNAAFALN 124
Query: 680 LDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACAS-RDLRENVVGVCFVAQDI 734
+ + E +F +KT +SG I V C+ D VVG+ + DI
Sbjct: 125 VARRCANGESWTGEFPVKT-----KSGKIFSAVTTCSPFYDDNGTVVGIISITSDI 175
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,806,315
Number of Sequences: 26719
Number of extensions: 1011642
Number of successful extensions: 2603
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 2542
Number of HSP's gapped (non-prelim): 25
length of query: 1124
length of database: 11,318,596
effective HSP length: 110
effective length of query: 1014
effective length of database: 8,379,506
effective search space: 8496819084
effective search space used: 8496819084
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0048.6


