

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0048.13
(98 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
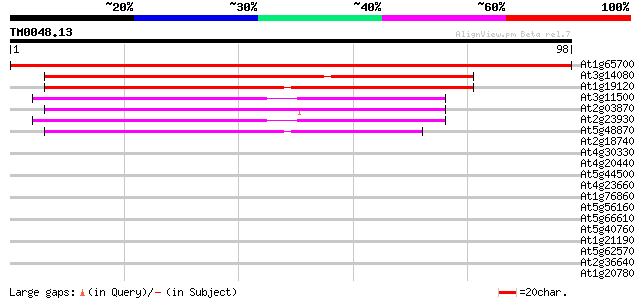
At1g65700 small nuclear ribonucleoprotein, putative 157 1e-39
At3g14080 Sm protein, putative 65 6e-12
At1g19120 unknown protein 64 2e-11
At3g11500 putative small nuclear ribonucleoprotein polypeptide G 47 2e-06
At2g03870 putative snRNP splicing factor 47 2e-06
At2g23930 putative small nuclear ribonucleoprotein E 45 5e-06
At5g48870 Sm-like protein (SAD1) 39 6e-04
At2g18740 small nuclear ribonucleoprotein E like protein 36 0.004
At4g30330 small nuclear ribonucleoprotein homolog 35 0.006
At4g20440 putative snRNP protein 33 0.018
At5g44500 unknown protein 33 0.024
At4g23660 para-hydroxy bezoate polyprenyl diphosphate transferas... 29 0.46
At1g76860 unknown protein 29 0.46
At5g56160 putative protein 28 0.78
At5g66610 putative protein 28 1.0
At5g40760 glucose-6-phosphate dehydrogenase 27 1.3
At1g21190 putative protein 27 1.3
At5g62570 unknown protein 26 3.0
At2g36640 late embryogenesis abundant protein (AtECP63) 26 3.0
At1g20780 unknown protein (At1g20780) 26 3.0
>At1g65700 small nuclear ribonucleoprotein, putative
Length = 98
Score = 157 bits (396), Expect = 1e-39
Identities = 79/98 (80%), Positives = 86/98 (87%)
Query: 1 MSTGPGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALG 60
M+ GLE+LVDQ ISVITNDGRNIVG LKGFDQATNIILDESHERVFSTKEGVQ LG
Sbjct: 1 MAATTGLETLVDQIISVITNDGRNIVGVLKGFDQATNIILDESHERVFSTKEGVQQHVLG 60
Query: 61 LYIIRGDNISVVGELDEDLDKNLDKSKIRAQPLKPVIH 98
LYIIRGDNI V+GELDE+LD +LD SK+RA PLKPV+H
Sbjct: 61 LYIIRGDNIGVIGELDEELDASLDFSKLRAHPLKPVVH 98
>At3g14080 Sm protein, putative
Length = 128
Score = 65.1 bits (157), Expect = 6e-12
Identities = 31/75 (41%), Positives = 52/75 (69%), Gaps = 1/75 (1%)
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRG 66
L S +D+++ V+ DGR ++GTL+ FDQ N +L+ + ERV ++ I LGLY+IRG
Sbjct: 15 LASYLDRKLLVLLRDGRKLMGTLRSFDQFANAVLEGACERVIVGEQYCD-IPLGLYVIRG 73
Query: 67 DNISVVGELDEDLDK 81
+N+ ++GELD + ++
Sbjct: 74 ENVVLIGELDTEREE 88
>At1g19120 unknown protein
Length = 128
Score = 63.5 bits (153), Expect = 2e-11
Identities = 31/75 (41%), Positives = 52/75 (69%), Gaps = 1/75 (1%)
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRG 66
L + +D+++ V+ DGR ++G L+ FDQ N +L+E++ERV + I LGLYIIRG
Sbjct: 15 LAAYLDKKLLVLLRDGRKLMGLLRSFDQFANAVLEEAYERVI-VGDLYCDIPLGLYIIRG 73
Query: 67 DNISVVGELDEDLDK 81
+N+ ++GELD + ++
Sbjct: 74 ENVVLIGELDVEKEE 88
>At3g11500 putative small nuclear ribonucleoprotein polypeptide G
Length = 79
Score = 47.0 bits (110), Expect = 2e-06
Identities = 24/72 (33%), Positives = 42/72 (58%), Gaps = 5/72 (6%)
Query: 5 PGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYII 64
P L+ +D+++ + N R +VGTL+GFDQ N+++D + E G +G+ +I
Sbjct: 8 PDLKKYMDKKLQIKLNANRMVVGTLRGFDQFMNLVVDNTVE-----VNGDDKTDIGMVVI 62
Query: 65 RGDNISVVGELD 76
RG++I V L+
Sbjct: 63 RGNSIVTVEALE 74
>At2g03870 putative snRNP splicing factor
Length = 99
Score = 47.0 bits (110), Expect = 2e-06
Identities = 28/74 (37%), Positives = 38/74 (50%), Gaps = 4/74 (5%)
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFS----TKEGVQLIALGLY 62
L VD+ + V GR + GTLKG+DQ N++LDE+ E V K Q LGL
Sbjct: 11 LAKFVDKGVQVKLTGGRQVTGTLKGYDQLLNLVLDEAVEFVRDHDDPLKTTDQTRRLGLI 70
Query: 63 IIRGDNISVVGELD 76
+ RG + +V D
Sbjct: 71 VCRGTAVMLVSPTD 84
>At2g23930 putative small nuclear ribonucleoprotein E
Length = 80
Score = 45.4 bits (106), Expect = 5e-06
Identities = 23/72 (31%), Positives = 41/72 (56%), Gaps = 5/72 (6%)
Query: 5 PGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYII 64
P L+ +D+++ + N R + GTL+GFDQ N+++D + E G +G+ +I
Sbjct: 8 PDLKKYMDKKLQIKLNANRMVTGTLRGFDQFMNLVVDNTVE-----VNGNDKTDIGMVVI 62
Query: 65 RGDNISVVGELD 76
RG++I V L+
Sbjct: 63 RGNSIVTVEALE 74
>At5g48870 Sm-like protein (SAD1)
Length = 88
Score = 38.5 bits (88), Expect = 6e-04
Identities = 20/66 (30%), Positives = 38/66 (57%), Gaps = 1/66 (1%)
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRG 66
++ + +I VI + +VG LKGFD N++L++ E T EG ++ L ++ G
Sbjct: 14 IDRCIGSKIWVIMKGDKELVGILKGFDVYVNMVLEDVTEYEI-TAEGRRVTKLDQILLNG 72
Query: 67 DNISVV 72
+NI+++
Sbjct: 73 NNIAIL 78
>At2g18740 small nuclear ribonucleoprotein E like protein
Length = 88
Score = 35.8 bits (81), Expect = 0.004
Identities = 22/66 (33%), Positives = 37/66 (55%), Gaps = 3/66 (4%)
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRG 66
L+S QI + I G + GFD+ N++LDE+ E S K+ + LG +++G
Sbjct: 21 LQSKARIQIWLFEQKDLRIEGRITGFDEYMNLVLDEAEE--VSIKKNTRK-PLGRILLKG 77
Query: 67 DNISVV 72
DNI+++
Sbjct: 78 DNITLM 83
>At4g30330 small nuclear ribonucleoprotein homolog
Length = 88
Score = 35.0 bits (79), Expect = 0.006
Identities = 21/66 (31%), Positives = 34/66 (50%), Gaps = 3/66 (4%)
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRG 66
L+S QI + I G + GFD+ N++LDE+ E K LG +++G
Sbjct: 21 LQSKARIQIWLFEQKDLRIEGRITGFDEYMNLVLDEAEEVSIKKKTRK---PLGRILLKG 77
Query: 67 DNISVV 72
DNI+++
Sbjct: 78 DNITLM 83
>At4g20440 putative snRNP protein
Length = 257
Score = 33.5 bits (75), Expect = 0.018
Identities = 20/77 (25%), Positives = 37/77 (47%), Gaps = 8/77 (10%)
Query: 1 MSTGPGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHE-RVFSTKEGVQL--- 56
MS + ++ ++ V DGR +VG FD+ N++L + E R +G ++
Sbjct: 3 MSKSSKMLQFINYRMRVTIQDGRQLVGKFMAFDRHMNLVLGDCEEFRKLPPAKGKKINEE 62
Query: 57 ----IALGLYIIRGDNI 69
LGL ++RG+ +
Sbjct: 63 REDRRTLGLVLLRGEEV 79
>At5g44500 unknown protein
Length = 254
Score = 33.1 bits (74), Expect = 0.024
Identities = 18/78 (23%), Positives = 36/78 (46%), Gaps = 9/78 (11%)
Query: 1 MSTGPGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHE---------RVFSTK 51
MS + ++ ++ V DGR ++G FD+ N++L + E + +
Sbjct: 3 MSKSSKMLQFINYRMRVTIQDGRQLIGKFMAFDRHMNLVLGDCEEFRKLPPAKGNKKTNE 62
Query: 52 EGVQLIALGLYIIRGDNI 69
E + LGL ++RG+ +
Sbjct: 63 EREERRTLGLVLLRGEEV 80
>At4g23660 para-hydroxy bezoate polyprenyl diphosphate transferase
(AtPPT1)
Length = 407
Score = 28.9 bits (63), Expect = 0.46
Identities = 11/34 (32%), Positives = 21/34 (61%)
Query: 56 LIALGLYIIRGDNISVVGELDEDLDKNLDKSKIR 89
L G ++RG ++ LD+D+D +D++K+R
Sbjct: 167 LFGCGALLLRGAGCTINDLLDQDIDTKVDRTKLR 200
>At1g76860 unknown protein
Length = 98
Score = 28.9 bits (63), Expect = 0.46
Identities = 21/76 (27%), Positives = 36/76 (46%), Gaps = 19/76 (25%)
Query: 11 VDQQISVITNDGRNIVGTLKGFDQATNIIL-------------DESHERVF-STKEGVQL 56
+D++I V R + G L FDQ N+IL DE++E + +TK ++
Sbjct: 20 LDERIYVKLRSDRELRGKLHAFDQHLNMILGDVEETITTVEIDDETYEEIVRTTKRTIEF 79
Query: 57 IALGLYIIRGDNISVV 72
+ +RGD + +V
Sbjct: 80 L-----FVRGDGVILV 90
>At5g56160 putative protein
Length = 592
Score = 28.1 bits (61), Expect = 0.78
Identities = 14/45 (31%), Positives = 24/45 (53%), Gaps = 3/45 (6%)
Query: 19 TNDGRNIVGTLKGFDQATNIILDESHERVF---STKEGVQLIALG 60
T+DG + T++ + A + +LD E F S +EG Q++ G
Sbjct: 414 TSDGDKFITTVESIESAQSQLLDADTENTFANTSVREGGQILRFG 458
>At5g66610 putative protein
Length = 529
Score = 27.7 bits (60), Expect = 1.0
Identities = 16/67 (23%), Positives = 30/67 (43%), Gaps = 3/67 (4%)
Query: 12 DQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRGDNISV 71
DQQ+S I + G K F+ ++ ++ +E + ++ L + NIS
Sbjct: 94 DQQLSKIVEESLKEKGKSKQFEDDQ---VENDEQQALMVQESLYMVELSAQLEEDKNIST 150
Query: 72 VGELDED 78
+ L+ED
Sbjct: 151 IPPLNED 157
>At5g40760 glucose-6-phosphate dehydrogenase
Length = 515
Score = 27.3 bits (59), Expect = 1.3
Identities = 16/59 (27%), Positives = 31/59 (52%), Gaps = 6/59 (10%)
Query: 38 IILDESHER-VFSTKEGVQLIALGLYIIRGDNISVVGELDEDLDKNLDKSKIRAQPLKP 95
+ + E++ER + T +G Q + +R D + V E+ L +DK ++++ P KP
Sbjct: 432 VAIPEAYERLILDTIKGDQQ-----HFVRRDELKVAWEIFTPLLHRIDKGEVKSIPYKP 485
>At1g21190 putative protein
Length = 97
Score = 27.3 bits (59), Expect = 1.3
Identities = 21/76 (27%), Positives = 35/76 (45%), Gaps = 19/76 (25%)
Query: 11 VDQQISVITNDGRNIVGTLKGFDQATNIIL-------------DESHERVF-STKEGVQL 56
++++I V R + G L FDQ N+IL DE++E + +TK V
Sbjct: 20 IEERIYVKLRSDRELRGKLHAFDQHLNMILGDVEEVITTIEIDDETYEEIVRTTKRTVPF 79
Query: 57 IALGLYIIRGDNISVV 72
+ +RGD + +V
Sbjct: 80 L-----FVRGDGVILV 90
>At5g62570 unknown protein
Length = 494
Score = 26.2 bits (56), Expect = 3.0
Identities = 15/55 (27%), Positives = 29/55 (52%), Gaps = 1/55 (1%)
Query: 39 ILDESHERVFSTKEGVQLIALGLYIIRGDNISVVGELDEDLDKNLDKSKIRAQPL 93
++D S ++FS+ L ++++ GD SV DED+ N+ + + +PL
Sbjct: 101 LIDPSTGQIFSSGPASSA-KLEVFVVEGDFNSVSDWTDEDIRNNIVREREGKKPL 154
>At2g36640 late embryogenesis abundant protein (AtECP63)
Length = 448
Score = 26.2 bits (56), Expect = 3.0
Identities = 14/40 (35%), Positives = 24/40 (60%), Gaps = 3/40 (7%)
Query: 17 VITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQL 56
VI + R + GT ++ A + ++ ++HE STKEG Q+
Sbjct: 54 VIGSVFRAVQGT---YEHARDAVVGKTHEAAESTKEGAQI 90
>At1g20780 unknown protein (At1g20780)
Length = 383
Score = 26.2 bits (56), Expect = 3.0
Identities = 15/62 (24%), Positives = 32/62 (51%), Gaps = 5/62 (8%)
Query: 30 KGFDQATNIILDESHERVFSTK----EGVQLIALGLYIIRGDNISVVGELDEDLDKNLDK 85
KG + A +++ + S K G ++ +GL +N+S+V + D L+ N+++
Sbjct: 201 KGREAAVSLLFELSKSEALCEKIGSIHGALILLVGLTSSNSENVSIVEKADRTLE-NMER 259
Query: 86 SK 87
S+
Sbjct: 260 SE 261
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.138 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,123,007
Number of Sequences: 26719
Number of extensions: 80135
Number of successful extensions: 184
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 158
Number of HSP's gapped (non-prelim): 31
length of query: 98
length of database: 11,318,596
effective HSP length: 74
effective length of query: 24
effective length of database: 9,341,390
effective search space: 224193360
effective search space used: 224193360
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0048.13


