

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
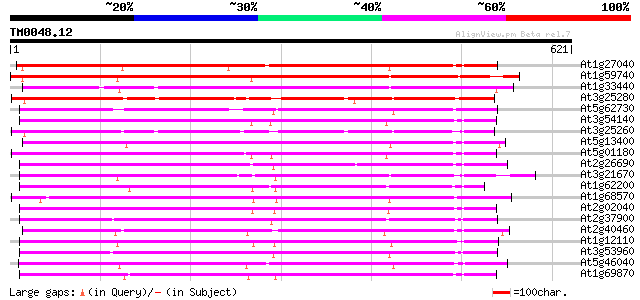
Query= TM0048.12
(621 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g27040 nitrate transporter like protein 441 e-124
At1g59740 nitrate transporter NTL1, putative 428 e-120
At1g33440 nitrate transporter NTL1, putative 409 e-114
At3g25280 nitrate transporter, putative 397 e-110
At5g62730 nitrate transporter NTL1 - like protein 390 e-108
At3g54140 peptide transport - like protein 371 e-103
At3g25260 nitrate transporter, putative 369 e-102
At5g13400 peptide transporter - like protein 361 e-100
At5g01180 oligopeptide transporter - like protein 360 e-99
At2g26690 putative nitrate transporter 358 4e-99
At3g21670 nitrate transporter 351 6e-97
At1g62200 unknown protein (At1g62200) 347 9e-96
At1g68570 peptide transporter like 342 3e-94
At2g02040 histidine transport protein (PTR2-B) 341 8e-94
At2g37900 putative peptide/amino acid transporter 340 1e-93
At2g40460 putative PTR2 family peptide transporter 336 2e-92
At1g12110 nitrate/chlorate transporter CHL1 336 2e-92
At3g53960 transporter like protein 330 2e-90
At5g46040 peptide transporter 324 8e-89
At1g69870 unknown protein 323 1e-88
>At1g27040 nitrate transporter like protein
Length = 563
Score = 441 bits (1134), Expect = e-124
Identities = 227/553 (41%), Positives = 352/553 (63%), Gaps = 27/553 (4%)
Query: 8 EWKRK-----QKGGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTN 62
+W+ K + GG A+ FV A+ L+N+ F+AN +LVLY MH LA S++ +T
Sbjct: 13 DWRNKAALRGRHGGMLAASFVLAVEILENLAFLANASNLVLYLKNFMHMSLARSSSEVTT 72
Query: 63 FMGSTFLLSLVGGFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACGKSSCI 122
FM + FLL+L+GGF++D + + L+ S+E L L++LT+QA SL P C S+ +
Sbjct: 73 FMATAFLLALLGGFLADAFFSTFVIFLISASIEFLGLILLTIQARRPSLMPPPCKSSAAL 132
Query: 123 K-----GGIAVMLYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLS 177
+ G A L+ L L++LG+GG++G++ + GA+QFDE P K +TFFN+ +
Sbjct: 133 RCEVVGGSKAAFLFVGLYLVSLGIGGIKGSLPSHGAEQFDEGTPKGRKQRSTFFNYYVFC 192
Query: 178 STLGSVVGVTGVVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRIV 237
+ G++V VT VVW+ KGW WGF + T++ + L+ LG FY+ K P SP+ I
Sbjct: 193 LSCGALVAVTFVVWIEDNKGWEWGFGVSTISIFLSILVFLLGSRFYKNKIPRGSPLTTIF 252
Query: 238 QVI----VVAFKNRNLPLPESNEQLYEAYKDATVEKIAHTNQMRFLDKATILQENFEPRP 293
+V+ +V+ ++ ++ ++ +++ T + + TN + L+KA E
Sbjct: 253 KVLLAASIVSCSSKTSSNHFTSREVQSEHEEKTPSQ-SLTNSLTCLNKAI---EGKTHHI 308
Query: 294 WKVCTVTQVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKLGSFEVPSPSI 353
W CTV QVE+VKI+ +MLPI TI++N CLAQL T+SV Q + M++K+ +F VPS S+
Sbjct: 309 WLECTVQQVEDVKIVLKMLPIFGCTIMLNCCLAQLSTYSVHQAATMNRKIVNFNVPSASL 368
Query: 354 PVIPLFFLCVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKR 413
PV P+ F+ +L P Y+ + +PFARK+T G+T LQR+GVGLVLS ++MAVA ++E+KR
Sbjct: 369 PVFPVVFMLILAPTYDHLIIPFARKVTKSEIGITHLQRIGVGLVLSIVAMAVAALVELKR 428
Query: 414 RDQAIK------DPSKPISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSF 467
+ A + + + PI+ W++ QY G AD+FTL GLLEFF+ EAP +M+SL+TS
Sbjct: 429 KQVAREAGLLDSEETLPITFLWIALQYLFLGSADLFTLAGLLEFFFTEAPSSMRSLATSL 488
Query: 468 TYLSMSLGYFLSTVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFF 527
++ S++LGY+LS+V+V ++N VTK + + WL G LN+N L+LFYW + +LS +NF
Sbjct: 489 SWASLALGYYLSSVMVPIVNRVTK--SAGQSPWL-GEKLNRNRLDLFYWLMCVLSVVNFL 545
Query: 528 NFLYWASCYKYKS 540
++L+WA YKY S
Sbjct: 546 HYLFWAKRYKYIS 558
>At1g59740 nitrate transporter NTL1, putative
Length = 591
Score = 428 bits (1100), Expect = e-120
Identities = 234/581 (40%), Positives = 353/581 (60%), Gaps = 32/581 (5%)
Query: 1 MGDKEVKEWKRK-----QKGGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLAS 55
+ ++E +W+ + + GG RA++FV L A + MG A +L+ Y I MHF L+
Sbjct: 24 LAEEESVDWRGRPSNPNKHGGMRAALFVLGLQAFEIMGIAAVGNNLITYVINEMHFPLSK 83
Query: 56 SANTLTNFMGSTFLLSLVGGFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEA 115
+AN +TNF+G+ F+ +L+GG++SD +L T ++FG +E+ ++L+VQA L L P
Sbjct: 84 AANIVTNFVGTIFIFALLGGYLSDAFLGSFWTIIIFGFVELSGFILLSVQAHLPQLKPPK 143
Query: 116 CG---KSSC--IKGGIAVMLYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATF 170
C +C KG A++ + +L L+ALG G V+ M+A GADQF + P ++K L+++
Sbjct: 144 CNPLIDQTCEEAKGFKAMIFFMALYLVALGSGCVKPNMIAHGADQFSQSHPKQSKRLSSY 203
Query: 171 FNWLLLSSTLGSVVGVTGVVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGD 230
FN + ++G ++ +T +VWV G GF + A ++G + L G ++R K P
Sbjct: 204 FNAAYFAFSMGELIALTLLVWVQTHSGMDIGFGVSAAAMTMGIISLVSGTMYFRNKRPRR 263
Query: 231 SPILRIVQVIVVAFKNRNLPLPESNEQLYEAYKDAT----VEKIAHTNQMRFLDKATI-L 285
S I VIV A R L P L+ + A + HT + RFLDKA I +
Sbjct: 264 SIFTPIAHVIVAAILKRKLASPSDPRMLHGDHHVANDVVPSSTLPHTPRFRFLDKACIKI 323
Query: 286 QE-NFEPRPWKVCTVTQVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKL- 343
Q+ N + PW++CTVTQVE+VK L ++PI ASTI+ NT LAQLQTFSV QGS M+ +L
Sbjct: 324 QDTNTKESPWRLCTVTQVEQVKTLISLVPIFASTIVFNTILAQLQTFSVQQGSSMNTRLS 383
Query: 344 GSFEVPSPSIPVIPLFFLCVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISM 403
SF +P S+ IP L L+P+Y+ VPFARK+T H SG+ L R+G+GL LS+ SM
Sbjct: 384 NSFHIPPASLQAIPYIMLIFLVPLYDSFLVPFARKLTGHNSGIPPLTRIGIGLFLSTFSM 443
Query: 404 AVAGIIEVKRRDQAIKDPSKPISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSL 463
A ++E KRRD ++ D + +S+FW++ Q+ IFG+++MFT +GL+EFFY+++ M+S
Sbjct: 444 VSAAMLEKKRRDSSVLD-GRILSIFWITPQFLIFGISEMFTAVGLIEFFYKQSAKGMESF 502
Query: 464 STSFTYLSMSLGYFLSTVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSC 523
+ TY S S G++ S+V+VSV+N +T SK GWL +DLNK+ L+LFYW LA+LS
Sbjct: 503 LMALTYCSYSFGFYFSSVLVSVVNKITSTSVDSK-GWLGENDLNKDRLDLFYWLLAVLSL 561
Query: 524 LNFFNFLYWASCYKYKSEDNSCSKLNVKAVAETTAIMVVDE 564
LNF ++L+W S+ N+K+ +V DE
Sbjct: 562 LNFLSYLFW-------------SRWNIKSSRRNNTNVVGDE 589
>At1g33440 nitrate transporter NTL1, putative
Length = 596
Score = 409 bits (1051), Expect = e-114
Identities = 227/560 (40%), Positives = 336/560 (59%), Gaps = 26/560 (4%)
Query: 15 GGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLLSLVG 74
GG RA++FV A + M A +L+ Y MHF L+ SAN +TNF+G+ FLLSL+G
Sbjct: 40 GGTRAALFVLGFQAFEMMAIAAVGNNLITYVFNEMHFPLSKSANLVTNFIGTVFLLSLLG 99
Query: 75 GFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACGKSS----CIK--GGIAV 128
GF+SD+YL T L+FG +E+ QA L L P C S C++ G A
Sbjct: 100 GFLSDSYLGSFRTMLVFGVIEISVF-----QAHLPELRPPECNMKSTTIHCVEANGYKAA 154
Query: 129 MLYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSVVGVTG 188
LYT+L L+ALG G ++ +++ GA+QF KD + L++FFN + ++G ++ +T
Sbjct: 155 TLYTALCLVALGSGCLKPNIISHGANQFQRKD---LRKLSSFFNAAYFAFSMGQLIALTL 211
Query: 189 VVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRIVQVIVVAFKNRN 248
+VWV G GF + + G + L G FYR K P S I QV V A R
Sbjct: 212 LVWVQTHSGMDVGFGVSAAVMAAGMISLVAGTSFYRNKPPSGSIFTPIAQVFVAAITKRK 271
Query: 249 LPLPESNEQLYEAYKDAT-VEKIAHTNQMRFLDKATI-LQENFEPRPWKVCTVTQVEEVK 306
P + +++ D V+ + H+N+ RFLDKA I Q PW++CT+ QV +VK
Sbjct: 272 QICPSNPNMVHQPSTDLVRVKPLLHSNKFRFLDKACIKTQGKAMESPWRLCTIEQVHQVK 331
Query: 307 ILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKLG-SFEVPSPSIPVIPLFFLCVLI 365
IL ++PI A TII NT LAQLQTFSV QGS M+ + +F++P S+ IP L +
Sbjct: 332 ILLSVIPIFACTIIFNTILAQLQTFSVQQGSSMNTHITKTFQIPPASLQAIPYIILIFFV 391
Query: 366 PIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKRRDQAIKDPSKPI 425
P+YE FVP ARK+T + SG++ LQR+G GL L++ SM A ++E KRR+ ++ + +
Sbjct: 392 PLYETFFVPLARKLTGNDSGISPLQRIGTGLFLATFSMVAAALVEKKRRESFLEQ-NVML 450
Query: 426 SLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMSLGYFLSTVVVSV 485
S+FW++ Q+ IFG+++MFT +GL+EFFY+++ +M+S T+ TY S S G++LS+V+VS
Sbjct: 451 SIFWIAPQFLIFGLSEMFTAVGLVEFFYKQSSQSMQSFLTAMTYCSYSFGFYLSSVLVST 510
Query: 486 INTVT-KRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWASCYK------- 537
+N VT + +K+GWL +DLNK+ L+ FYW LA LS +NFFN+L+W+ Y
Sbjct: 511 VNRVTSSNGSGTKEGWLGDNDLNKDRLDHFYWLLASLSFINFFNYLFWSRWYSCDPSATH 570
Query: 538 YKSEDNSCSKLNVKAVAETT 557
+ +E NS L + ++T
Sbjct: 571 HSAEVNSLEALENGEIKDST 590
>At3g25280 nitrate transporter, putative
Length = 521
Score = 397 bits (1020), Expect = e-110
Identities = 220/543 (40%), Positives = 336/543 (61%), Gaps = 36/543 (6%)
Query: 3 DKEVKEWKRKQK-----GGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSA 57
+++ ++WK K+ GG RA+ V + ++N+ F+AN + V YF+ MH+ A++A
Sbjct: 6 EEKFEDWKGKEAIPGKHGGIRAASIVCVVVMMENIVFIANGFNFVKYFMGSMHYTPATAA 65
Query: 58 NTLTNFMGSTFLLSLVGGFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACG 117
N +TNFMG++FLL+L GGFI+D+++ TT ++F +E++ L++LT QA L PE
Sbjct: 66 NMVTNFMGTSFLLTLFGGFIADSFVTHFTTFIVFCCIELMGLILLTFQAHNPKLLPEKDK 125
Query: 118 KSSCIKGGIAVMLYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLS 177
S ++ I L+T L +A+G GG++ ++ + G DQ D ++P + ++ FF+WL S
Sbjct: 126 TPSTLQSAI---LFTGLYAMAIGTGGLKASLPSHGGDQIDRRNP---RLISRFFDWLYFS 179
Query: 178 STLGSVVGVTGVVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRIV 237
G ++ VT V+W+ +KGW W F+I + I +G PFYR K P SP+ +I
Sbjct: 180 ICSGCLLAVTVVLWIEEKKGWIWSFNISVGILATALCIFTVGLPFYRFKRPNGSPLKKIA 239
Query: 238 QVIVVAFKNRNLPLPESNEQLYEAYKDATVEKIAHTNQMRFLDKATILQENFEPRPWKVC 297
VI+ A +NRN + +E++ ++ K N+++++DKAT+ + E
Sbjct: 240 IVIISAARNRN--KSDLDEEMMRGL--ISIYKNNSHNKLKWIDKATLNKNISE------- 288
Query: 298 TVTQVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKL-GSFEVPSPSIPVI 356
T+VEE + +LPI STI+M+ C+AQL TFS QG +M+KKL SFE+P PS+ I
Sbjct: 289 --TEVEETRTFLGLLPIFGSTIVMSCCVAQLSTFSAQQGMLMNKKLFHSFEIPVPSLTAI 346
Query: 357 PLFFLCVLIPIYEFIFVPFARKIT---NHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKR 413
PL F+ + IP+YEF F +KI+ N+ S L+R+G+GL LSS+SMAV+ I+E KR
Sbjct: 347 PLIFMLLSIPLYEF----FGKKISSGNNNRSSSFNLKRIGLGLALSSVSMAVSAIVEAKR 402
Query: 414 RDQAIKDPSKPISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMS 473
+ + + + + IS+ WL FQY + V+DM TL G+LEFFYREAP MKS+ST+ + S +
Sbjct: 403 KHEVVHNNFR-ISVLWLVFQYLMLSVSDMLTLGGMLEFFYREAPSNMKSISTALGWCSTA 461
Query: 474 LGYFLSTVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWA 533
LG+FLST +V V N VT R+ WL G DLNK L LFY L +L+ LN N+++WA
Sbjct: 462 LGFFLSTTLVEVTNAVTGRL---GHQWLGGEDLNKTRLELFYVLLCVLNTLNLLNYIFWA 518
Query: 534 SCY 536
Y
Sbjct: 519 KRY 521
>At5g62730 nitrate transporter NTL1 - like protein
Length = 589
Score = 390 bits (1001), Expect = e-108
Identities = 214/561 (38%), Positives = 320/561 (56%), Gaps = 59/561 (10%)
Query: 11 RKQKGGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLL 70
R + GG A+ FV + L+N+ F+AN +LVLY M F + +AN +T FMG+ F L
Sbjct: 54 RGRHGGMLAASFVLVVEVLENLAFLANASNLVLYLSTKMGFSPSGAANAVTAFMGTAFFL 113
Query: 71 SLVGGFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACGKSSCIKGGIAVML 130
+L+GGF++D + L+ ++E L L++LTVQA S P + V L
Sbjct: 114 ALLGGFLADAFFTTFHIYLVSAAIEFLGLMVLTVQAHEHSTEPWS-----------RVFL 162
Query: 131 YTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSVVGVTGVV 190
+ L L+ALG+GG++G++ GA+QFDE+ + + FFN+ + S + G+++ VT VV
Sbjct: 163 FVGLYLVALGVGGIKGSLPPHGAEQFDEETSSGRRQRSFFFNYFIFSLSCGALIAVTVVV 222
Query: 191 WVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRIVQVIVVAFKNRNLP 250
W+ KGW +GF + T A I + G YR+K P SPI + +V+ A
Sbjct: 223 WLEDNKGWSYGFGVSTAAILISVPVFLAGSRVYRLKVPSGSPITTLFKVLTAA------- 275
Query: 251 LPESNEQLYEAYKDATVEKIAHTNQMRFLDKATILQENFE-------------------- 290
LY YK +I T R ++ ++N +
Sbjct: 276 -------LYAKYKKRRTSRIVVTCHTRNDCDDSVTKQNCDGDDGFLGSFLGEVVRERESL 328
Query: 291 PRPWKVCTVTQVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKLGSFEVPS 350
PRP + CT QV++VKI+ ++LPI STI++N CLAQL TFSV Q S M+ KLGSF VP
Sbjct: 329 PRPLR-CTEEQVKDVKIVIKILPIFMSTIMLNCCLAQLSTFSVQQASTMNTKLGSFTVPP 387
Query: 351 PSIPVIPLFFLCVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIE 410
++PV P+ F+ +L P Y + +P ARK T +G+T LQR+G GLVLS ++MAVA ++E
Sbjct: 388 AALPVFPVVFMMILAPTYNHLLLPLARKSTKTETGITHLQRIGTGLVLSIVAMAVAALVE 447
Query: 411 VKRRDQAIKDPSK-----------PISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPT 459
KR+ + S PI+ W++ QY G AD+FTL G++EFF+ EAP T
Sbjct: 448 TKRKHVVVSCCSNNNSSSYSSSPLPITFLWVAIQYVFLGSADLFTLAGMMEFFFTEAPST 507
Query: 460 MKSLSTSFTYLSMSLGYFLSTVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLA 519
M+SL+TS ++ S+++GY+ S+V+VS +N VT WL G +LN+ +L FYW +
Sbjct: 508 MRSLATSLSWASLAMGYYFSSVLVSAVNFVTG--LNHHNPWLLGENLNQYHLERFYWLMC 565
Query: 520 ILSCLNFFNFLYWASCYKYKS 540
+LS +NF ++L+WAS Y Y+S
Sbjct: 566 VLSGINFLHYLFWASRYVYRS 586
>At3g54140 peptide transport - like protein
Length = 570
Score = 371 bits (952), Expect = e-103
Identities = 197/545 (36%), Positives = 316/545 (57%), Gaps = 19/545 (3%)
Query: 11 RKQKGGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLL 70
+++ G ++A F+ + + + +LV Y ++ A++AN +TN+ G+ ++
Sbjct: 22 KEKTGNWKACRFILGNECCERLAYYGMGTNLVNYLESRLNQGNATAANNVTNWSGTCYIT 81
Query: 71 SLVGGFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACGKSSCIKGGI-AVM 129
L+G FI+D YL R T F + V + +LT+ A + L P C +C +
Sbjct: 82 PLIGAFIADAYLGRYWTIATFVFIYVSGMTLLTLSASVPGLKPGNCNADTCHPNSSQTAV 141
Query: 130 LYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSVVGVTGV 189
+ +L ++ALG GG++ + +FGADQFDE D E ++FFNW S +G+++ T +
Sbjct: 142 FFVALYMIALGTGGIKPCVSSFGADQFDENDENEKIKKSSFFNWFYFSINVGALIAATVL 201
Query: 190 VWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRIVQVIVVAFKNRNL 249
VW+ M GW WGF + TVA I G FYR++ PG SP+ RI QVIV AF+ ++
Sbjct: 202 VWIQMNVGWGWGFGVPTVAMVIAVCFFFFGSRFYRLQRPGGSPLTRIFQVIVAAFRKISV 261
Query: 250 PLPESNEQLYEAYKDAT----VEKIAHTNQMRFLDKATILQE-----NFEPRPWKVCTVT 300
+PE L+E D + K+ HT+ ++F DKA + + + E PW++C+VT
Sbjct: 262 KVPEDKSLLFETADDESNIKGSRKLVHTDNLKFFDKAAVESQSDSIKDGEVNPWRLCSVT 321
Query: 301 QVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKLG-SFEVPSPSIPVIPLF 359
QVEE+K + +LP+ A+ I+ T +Q+ T V QG+ M + +G +FE+PS S+ +
Sbjct: 322 QVEELKSIITLLPVWATGIVFATVYSQMSTMFVLQGNTMDQHMGKNFEIPSASLSLFDTV 381
Query: 360 FLCVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKRRD---- 415
+ P+Y+ +P ARK T + G TQLQR+G+GLV+S +M AG++EV R D
Sbjct: 382 SVLFWTPVYDQFIIPLARKFTRNERGFTQLQRMGIGLVVSIFAMITAGVLEVVRLDYVKT 441
Query: 416 -QAIKDPSKPISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMSL 474
A +S+FW QY + G A++FT IG LEFFY +AP M+SL ++ + +++L
Sbjct: 442 HNAYDQKQIHMSIFWQIPQYLLIGCAEVFTFIGQLEFFYDQAPDAMRSLCSALSLTTVAL 501
Query: 475 GYFLSTVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWAS 534
G +LSTV+V+V+ +TK+ K GW+ +LN+ +L+ F++ LA LS LNF +L+ +
Sbjct: 502 GNYLSTVLVTVVMKITKK--NGKPGWI-PDNLNRGHLDYFFYLLATLSFLNFLVYLWISK 558
Query: 535 CYKYK 539
YKYK
Sbjct: 559 RYKYK 563
>At3g25260 nitrate transporter, putative
Length = 515
Score = 369 bits (947), Expect = e-102
Identities = 204/542 (37%), Positives = 325/542 (59%), Gaps = 40/542 (7%)
Query: 3 DKEVKEWKRKQK-----GGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSA 57
+++ ++W+ K+ GG +A+ + ++NM F+A + ++YF M++ +A
Sbjct: 6 EEKFEDWRGKEAISGKHGGIKAAFIACVVETMENMVFLACSTNFMMYFTKSMNYSTPKAA 65
Query: 58 NTLTNFMGSTFLLSLVGGFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACG 117
+TNF+G++FLL++ GGF++D++L R +LFGS+E+L L+MLT+QA + L P+
Sbjct: 66 TMVTNFVGTSFLLTIFGGFVADSFLTRFAAFVLFGSIELLGLIMLTLQAHITKLQPQGGK 125
Query: 118 KSSCIKGGIAVMLYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLS 177
K S + + +L+T L +A+G+GGV+G++ A G DQ ++ + ++ FFNW S
Sbjct: 126 KPSTPQ---STVLFTGLYAIAIGVGGVKGSLPAHGGDQIGTRN---QRLISGFFNWYFFS 179
Query: 178 STLGSVVGVTGVVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRIV 237
LG + VT +VW+ GW F+I T + + G P YR K P SP+ RIV
Sbjct: 180 VCLGGFLAVTLMVWIEENIGWSSSFTISTAVLASAIFVFVAGCPMYRFKRPAGSPLTRIV 239
Query: 238 QVIVVAFKNRNLPLPESNEQLYEAYKDATVEKIAHTNQMRFLDKATILQENFEPRPWKVC 297
V V A +NRN + ++ + +K H N+ +FL+KA + +
Sbjct: 240 NVFVSAARNRNRFVTDAE---VVTQNHNSTDKSIHHNKFKFLNKAKLNNK---------I 287
Query: 298 TVTQVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKLG-SFEVPSPSIPVI 356
+ TQVEE + +LPI STIIMN C+AQ+ TFSV QG V ++KL SFE+P S+ I
Sbjct: 288 SATQVEETRTFLALLPIFGSTIIMNCCVAQMGTFSVQQGMVTNRKLSRSFEIPVASLNAI 347
Query: 357 PLFFLCVLIPIYEFIFVPFARKITNHP--SGVTQLQRVGVGLVLSSISMAVAGIIEVKRR 414
PL + + +YE F ++I ++ S L+R+G GL L+SISMAVA I+EVKR+
Sbjct: 348 PLLCMLSSLALYEL----FGKRILSNSERSSSFNLKRIGYGLALTSISMAVAAIVEVKRK 403
Query: 415 DQAIKDPSKPISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMSL 474
+A+ + K IS+FWL Q+ + ++DM T+ G+LEFF+RE+P +M+S+ST+ + S ++
Sbjct: 404 HEAVHNNIK-ISVFWLELQFVMLSLSDMLTVGGMLEFFFRESPASMRSMSTALGWCSTAM 462
Query: 475 GYFLSTVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWAS 534
G+FLS+V+V V+N +T GWL DLN++ L LFY L +L+ LN FN+++WA
Sbjct: 463 GFFLSSVLVEVVNGIT--------GWL-RDDLNESRLELFYLVLCVLNTLNLFNYIFWAK 513
Query: 535 CY 536
Y
Sbjct: 514 RY 515
>At5g13400 peptide transporter - like protein
Length = 624
Score = 361 bits (926), Expect = e-100
Identities = 207/562 (36%), Positives = 315/562 (55%), Gaps = 31/562 (5%)
Query: 15 GGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLLSLVG 74
GG+ A+ F+F + M + V++V + VMH SS+N + NF+G + S++G
Sbjct: 66 GGWIAAFFIFGNEMAERMAYFGLSVNMVAFMFYVMHRPFESSSNAVNNFLGISQASSVLG 125
Query: 75 GFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEA--CGKSSCIKGGIA----- 127
GF++D YL R T +F ++ ++ L+ +T+ A L P+ CG+ S + G
Sbjct: 126 GFLADAYLGRYWTIAIFTTMYLVGLIGITLGASLKMFVPDQSNCGQLSLLLGNCEEAKSW 185
Query: 128 --VMLYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSVVG 185
+ LYT L + G G+R + +FGADQFDEK L FFN+ LS TLG+++
Sbjct: 186 QMLYLYTVLYITGFGAAGIRPCVSSFGADQFDEKSKDYKTHLDRFFNFFYLSVTLGAIIA 245
Query: 186 VTGVVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRIVQVIVVAFK 245
T VV+V M+ GW F + VA I + G P YR + PG SP+ R+ QV+V AF+
Sbjct: 246 FTLVVYVQMELGWGMAFGTLAVAMGISNALFFAGTPLYRHRLPGGSPLTRVAQVLVAAFR 305
Query: 246 NRNLPLPESNE-QLYEA--YKDAT--VEKIAHTNQMRFLDKATI--LQENFEPRPWKVCT 298
RN S LYE K A KI H+N +LDKA + ++ EP PWK+CT
Sbjct: 306 KRNAAFTSSEFIGLYEVPGLKSAINGSRKIPHSNDFIWLDKAALELKEDGLEPSPWKLCT 365
Query: 299 VTQVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKLGSFEVPSPSIPVIPL 358
VTQVEEVKIL R++PI TI+++ L + T SV Q ++ + ++P +PV P
Sbjct: 366 VTQVEEVKILIRLIPIPTCTIMLSLVLTEYLTLSVQQAYTLNTHIQHLKLPVTCMPVFPG 425
Query: 359 FFLCVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKRRDQAI 418
+ +++ +Y +FVP R+IT +P G +QLQRVG+GL +S IS+A AG+ E RR AI
Sbjct: 426 LSIFLILSLYYSVFVPITRRITGNPHGASQLQRVGIGLAVSIISVAWAGLFENYRRHYAI 485
Query: 419 KD--------PSKPISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYL 470
++ ++ +WL QY + G+A++F ++GLLEF Y EAP MKS+ +++ L
Sbjct: 486 QNGFEFNFLTQMPDLTAYWLLIQYCLIGIAEVFCIVGLLEFLYEEAPDAMKSIGSAYAAL 545
Query: 471 SMSLGYFLSTVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFL 530
+ LG F +T++ +++ T+ + WL ++N + YW L +LS LNF FL
Sbjct: 546 AGGLGCFAATILNNIVKAATR--DSDGKSWL-SQNINTGRFDCLYWLLTLLSFLNFCVFL 602
Query: 531 YWASCYKYKS----EDNSCSKL 548
+ A YKY++ ED S + L
Sbjct: 603 WSAHRYKYRAIESEEDKSSAVL 624
>At5g01180 oligopeptide transporter - like protein
Length = 570
Score = 360 bits (925), Expect = e-99
Identities = 192/553 (34%), Positives = 317/553 (56%), Gaps = 20/553 (3%)
Query: 3 DKEVKEWKRKQKGGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTN 62
D K + + G ++A F+ + + + +L+ Y M+ + S++ +++N
Sbjct: 15 DIHKKPANKNKTGTWKACRFILGTECCERLAYYGMSTNLINYLEKQMNMENVSASKSVSN 74
Query: 63 FMGSTFLLSLVGGFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACGKSSCI 122
+ G+ + L+G FI+D YL R T F + + + +LT+ A + L P G++
Sbjct: 75 WSGTCYATPLIGAFIADAYLGRYWTIASFVVIYIAGMTLLTISASVPGLTPTCSGETCHA 134
Query: 123 KGGIAVMLYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGS 182
G + + +L L+ALG GG++ + +FGADQFD+ D E ++ ++FFNW +G+
Sbjct: 135 TAGQTAITFIALYLIALGTGGIKPCVSSFGADQFDDTDEKEKESKSSFFNWFYFVINVGA 194
Query: 183 VVGVTGVVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRIVQVIVV 242
++ + +VW+ M GW WG + TVA +I + G FYR++ PG SP+ R++QVIV
Sbjct: 195 MIASSVLVWIQMNVGWGWGLGVPTVAMAIAVVFFFAGSNFYRLQKPGGSPLTRMLQVIVA 254
Query: 243 AFKNRNLPLPESNEQLYEAYKDAT-----VEKIAHTNQMRFLDKATILQEN-----FEPR 292
+ + + +PE LYE +DA K+ HT + F DKA + E+ +
Sbjct: 255 SCRKSKVKIPEDESLLYE-NQDAESSIIGSRKLEHTKILTFFDKAAVETESDNKGAAKSS 313
Query: 293 PWKVCTVTQVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKLG-SFEVPSP 351
WK+CTVTQVEE+K L R+LPI A+ I+ + +Q+ T V QG+ + + +G +F++PS
Sbjct: 314 SWKLCTVTQVEELKALIRLLPIWATGIVFASVYSQMGTVFVLQGNTLDQHMGPNFKIPSA 373
Query: 352 SIPVIPLFFLCVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEV 411
S+ + + P+Y+ + VPFARK T H G TQLQR+G+GLV+S SM AGI+EV
Sbjct: 374 SLSLFDTLSVLFWAPVYDKLIVPFARKYTGHERGFTQLQRIGIGLVISIFSMVSAGILEV 433
Query: 412 KRRD-----QAIKDPSKPISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTS 466
R + + + P+++FW QY + G A++FT IG LEFFY +AP M+SL ++
Sbjct: 434 ARLNYVQTHNLYNEETIPMTIFWQVPQYFLVGCAEVFTFIGQLEFFYDQAPDAMRSLCSA 493
Query: 467 FTYLSMSLGYFLSTVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNF 526
+ +++ G +LST +V+++ VT+ + + GW+ +LN +L+ F+W LA LS LNF
Sbjct: 494 LSLTAIAFGNYLSTFLVTLVTKVTR--SGGRPGWI-AKNLNNGHLDYFFWLLAGLSFLNF 550
Query: 527 FNFLYWASCYKYK 539
+L+ A Y YK
Sbjct: 551 LVYLWIAKWYTYK 563
>At2g26690 putative nitrate transporter
Length = 586
Score = 358 bits (920), Expect = 4e-99
Identities = 207/555 (37%), Positives = 321/555 (57%), Gaps = 20/555 (3%)
Query: 11 RKQKGGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLL 70
+ + GG+ + + + ++ + + V+LV Y + MH ++SAN +T+FMG++FLL
Sbjct: 31 KSKTGGWITAALILGIEVVERLSTMGIAVNLVTYLMETMHLPSSTSANIVTDFMGTSFLL 90
Query: 71 SLVGGFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACGKS-SCIKGGIAVM 129
L+GGF++D++L R T +F +++ L L V L L P C +CI M
Sbjct: 91 CLLGGFLADSFLGRFKTIGIFSTIQALGTGALAVATKLPELRPPTCHHGEACIPATAFQM 150
Query: 130 --LYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSVVGVT 187
LY SL L+ALG GG++ ++ FG+DQFD+KDP E +A FFN ++G+++ VT
Sbjct: 151 TILYVSLYLIALGTGGLKSSISGFGSDQFDDKDPKEKAHMAFFFNRFFFFISMGTLLAVT 210
Query: 188 GVVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRIVQVIVVAFKNR 247
+V++ + G W + I TV+ +I +I G YR K SP+++I QVI AF+ R
Sbjct: 211 VLVYMQDEVGRSWAYGICTVSMAIAIVIFLCGTKRYRYKKSQGSPVVQIFQVIAAAFRKR 270
Query: 248 NLPLPESNEQLYEAYKDATVEKIAHTNQMRFLDKATILQE-NFE--------PRPWKVCT 298
+ LP+S LYE + +I HT+Q LDKA I+ E +FE P PWK+ +
Sbjct: 271 KMELPQSIVYLYEDNPEGI--RIEHTDQFHLLDKAAIVAEGDFEQTLDGVAIPNPWKLSS 328
Query: 299 VTQVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKLGSFEVPSPSIPVIPL 358
VT+VEEVK++ R+LPI A+TII T AQ+ TFSV Q S M + +GSF++P+ S+ V +
Sbjct: 329 VTKVEEVKMMVRLLPIWATTIIFWTTYAQMITFSVEQASTMRRNIGSFKIPAGSLTVFFV 388
Query: 359 FFLCVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKRRDQAI 418
+ + + +Y+ +PF +K P G + LQR+ +GLVLS+ MA A ++E KR A
Sbjct: 389 AAILITLAVYDRAIMPFWKKWKGKP-GFSSLQRIAIGLVLSTAGMAAAALVEQKRLSVAK 447
Query: 419 KDPSK--PISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMSLGY 476
K PIS+F L Q+ + G + F G L+FF ++P MK++ST ++SLG+
Sbjct: 448 SSSQKTLPISVFLLVPQFFLVGAGEAFIYTGQLDFFITQSPKGMKTMSTGLFLTTLSLGF 507
Query: 477 FLSTVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWASCY 536
F+S+ +VS++ VT T + GWL ++N L+ FYW L ILS +NF ++ A +
Sbjct: 508 FVSSFLVSIVKRVTS--TSTDVGWL-ADNINHGRLDYFYWLLVILSGINFVVYIICALWF 564
Query: 537 KYKSEDNSCSKLNVK 551
K +S K N K
Sbjct: 565 KPTKGKDSVEKENGK 579
>At3g21670 nitrate transporter
Length = 590
Score = 351 bits (901), Expect = 6e-97
Identities = 209/587 (35%), Positives = 316/587 (53%), Gaps = 41/587 (6%)
Query: 11 RKQKGGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLL 70
+ + GG+ + + + + + ++LV Y + +H A SA +TNFMG+ LL
Sbjct: 27 KSKTGGWLGAGLILGSELSERICVMGISMNLVTYLVGDLHISSAKSATIVTNFMGTLNLL 86
Query: 71 SLVGGFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACGK-----SSCIK-- 123
L+GGF++D L R + S+ L +++LTV + S+ P C CI+
Sbjct: 87 GLLGGFLADAKLGRYKMVAISASVTALGVLLLTVATTISSMRPPICDDFRRLHHQCIEAN 146
Query: 124 GGIAVMLYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSV 183
G +LY +L +ALG GG++ + FG+DQFD DP E K + FFN S ++GS+
Sbjct: 147 GHQLALLYVALYTIALGGGGIKSNVSGFGSDQFDTSDPKEEKQMIFFFNRFYFSISVGSL 206
Query: 184 VGVTGVVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRIVQVIVVA 243
V +V+V G WG+ I + ++L G YR K P SP I +V +A
Sbjct: 207 FAVIALVYVQDNVGRGWGYGISAATMVVAAIVLLCGTKRYRFKKPKGSPFTTIWRVGFLA 266
Query: 244 FKNRNLPLPESNEQLYEAYKDATVEKIAHTNQMRFLDKATILQENFEPR--------PWK 295
+K R P ++ L Y + TV HT ++ LDKA I + P PW
Sbjct: 267 WKKRKESYP-AHPSLLNGYDNTTVP---HTEMLKCLDKAAISKNESSPSSKDFEEKDPWI 322
Query: 296 VCTVTQVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKLGSFEVPSPSIPV 355
V TVTQVEEVK++ +++PI A+ I+ T +Q+ TF+V Q + M +KLGSF VP+ S
Sbjct: 323 VSTVTQVEEVKLVMKLVPIWATNILFWTIYSQMTTFTVEQATFMDRKLGSFTVPAGSYSA 382
Query: 356 IPLFFLCVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKRRD 415
+ + + + E +FVP R++T P G+T LQR+GVGLV S +MAVA +IE RR+
Sbjct: 383 FLILTILLFTSLNERVFVPLTRRLTKKPQGITSLQRIGVGLVFSMAAMAVAAVIENARRE 442
Query: 416 QAIKDPSKPISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMSLG 475
A+ + K IS FWL QY + G + F +G LEFF REAP MKS+ST ++S+G
Sbjct: 443 AAVNN-DKKISAFWLVPQYFLVGAGEAFAYVGQLEFFIREAPERMKSMSTGLFLSTISMG 501
Query: 476 YFLSTVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWASC 535
+F+S+++VS+++ VT + WL S+LNK LN FYW L +L LNF F+ +A
Sbjct: 502 FFVSSLLVSLVDRVTDK------SWL-RSNLNKARLNYFYWLLVVLGALNFLIFIVFAMK 554
Query: 536 YKYKSEDNSCSKLNVKAVAETTAIMVVDEKKHDKEDMKDHGSTQDLR 582
++YK A+ ++V D+ +KE K S +L+
Sbjct: 555 HQYK--------------ADVITVVVTDDDSVEKEVTKKESSEFELK 587
>At1g62200 unknown protein (At1g62200)
Length = 590
Score = 347 bits (891), Expect = 9e-96
Identities = 193/530 (36%), Positives = 298/530 (55%), Gaps = 21/530 (3%)
Query: 11 RKQKGGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLL 70
+K+ G ++A F+ + + + +L+ Y+ +H S+A+ + + G+ ++
Sbjct: 50 KKKTGNWKACPFILGNECCERLAYYGIAKNLITYYTSELHESNVSAASDVMIWQGTCYIT 109
Query: 71 SLVGGFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEAC-GKSSCIKGGIAVM 129
L+G I+D+Y R T F ++ + + +LT+ A L L P AC G ++ + +
Sbjct: 110 PLIGAVIADSYWGRYWTIASFSAIYFIGMALLTLSASLPVLKPAACAGVAAALCSPATTV 169
Query: 130 LY----TSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSVVG 185
Y T L L+ALG GG++ + +FGADQFD+ DP E A+FFNW S +GS +
Sbjct: 170 QYAVFFTGLYLIALGTGGIKPCVSSFGADQFDDTDPRERVRKASFFNWFYFSINIGSFIS 229
Query: 186 VTGVVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRIVQVIVVAFK 245
T +VWV GW GF I TV + +G P YR + PG SPI R+ QV+V A++
Sbjct: 230 STLLVWVQENVGWGLGFLIPTVFMGVSIASFFIGTPLYRFQKPGGSPITRVCQVLVAAYR 289
Query: 246 NRNLPLPESNEQLYEAYKDATV----EKIAHTNQMRFLDKATILQENFEPR------PWK 295
L LPE LYE + ++ KI HT+ +FLDKA ++ E +E + PWK
Sbjct: 290 KLKLNLPEDISFLYETREKNSMIAGSRKIQHTDGYKFLDKAAVISE-YESKSGAFSNPWK 348
Query: 296 VCTVTQVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKLGSFEVPSPSIPV 355
+CTVTQVEEVK L RM PI AS I+ + +Q+ T V QG M++ + SFE+P S V
Sbjct: 349 LCTVTQVEEVKTLIRMFPIWASGIVYSVLYSQISTLFVQQGRSMNRIIRSFEIPPASFGV 408
Query: 356 IPLFFLCVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKRRD 415
+ + IPIY+ VPF R+ T P G+T LQR+G+GL LS +S+A A I+E R
Sbjct: 409 FDTLIVLISIPIYDRFLVPFVRRFTGIPKGLTDLQRMGIGLFLSVLSIAAAAIVETVRLQ 468
Query: 416 QAIKDPSKPISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMSLG 475
A +S+FW QY + G+A++F IG +EFFY E+P M+S+ ++ L+ ++G
Sbjct: 469 LA--QDFVAMSIFWQIPQYILMGIAEVFFFIGRVEFFYDESPDAMRSVCSALALLNTAVG 526
Query: 476 YFLSTVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLN 525
+LS+++++++ T K GW+ DLNK +L+ F+W L L +N
Sbjct: 527 SYLSSLILTLVAYFT--ALGGKDGWV-PDDLNKGHLDYFFWLLVSLGLVN 573
>At1g68570 peptide transporter like
Length = 596
Score = 342 bits (878), Expect = 3e-94
Identities = 209/577 (36%), Positives = 310/577 (53%), Gaps = 29/577 (5%)
Query: 3 DKEVKEWKRKQKGGFRASMFVFALSALDNM---GFVANMVSLVLYFIMVMHFDLASSANT 59
+K++ + KGG F+FA + + GF ANM+S Y +H L +ANT
Sbjct: 13 EKQLHGRPNRPKGGLITMPFIFANEICEKLAVVGFHANMIS---YLTTQLHLPLTKAANT 69
Query: 60 LTNFMGSTFLLSLVGGFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEAC-GK 118
LTNF G++ L L+G FI+D++ R T + + + +LT+ A + +L P C G+
Sbjct: 70 LTNFAGTSSLTPLLGAFIADSFAGRFWTITFASIIYQIGMTLLTISAIIPTLRPPPCKGE 129
Query: 119 SSCIKGGIAVM--LYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLL 176
C+ A + LY +L L ALG GG+R +VAFGADQFDE DP + +FNW
Sbjct: 130 EVCVVADTAQLSILYVALLLGALGSGGIRPCVVAFGADQFDESDPNQTTKTWNYFNWYYF 189
Query: 177 SSTLGSVVGVTGVVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRI 236
++ VT +VW+ GW G I TVA + + G YR P SP R+
Sbjct: 190 CMGAAVLLAVTVLVWIQDNVGWGLGLGIPTVAMFLSVIAFVGGFQLYRHLVPAGSPFTRL 249
Query: 237 VQVIVVAFKNRNLPLPESNEQLY-EAYKDATVE---KIAHTNQMRFLDKATILQENFEPR 292
+QV V AF+ R L + LY DA + K+ HT M FLDKA I+ E +
Sbjct: 250 IQVGVAAFRKRKLRMVSDPSLLYFNDEIDAPISLGGKLTHTKHMSFLDKAAIVTEEDNLK 309
Query: 293 P------WKVCTVTQVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKL-GS 345
P W++ TV +VEE+K + RM PI AS I++ T AQ TFS+ Q M++ L S
Sbjct: 310 PGQIPNHWRLSTVHRVEELKSVIRMGPIGASGILLITAYAQQGTFSLQQAKTMNRHLTNS 369
Query: 346 FEVPSPSIPVIPLFFLCVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAV 405
F++P+ S+ V + I Y+ +FV ARK T G+T L R+G+G V+S I+ V
Sbjct: 370 FQIPAGSMSVFTTVAMLTTIIFYDRVFVKVARKFTGLERGITFLHRMGIGFVISIIATLV 429
Query: 406 AGIIEVKRRDQAIK-------DPSKPISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPP 458
AG +EVKR+ AI+ PIS WL QY + GVA+ F IG LEFFY +AP
Sbjct: 430 AGFVEVKRKSVAIEHGLLDKPHTIVPISFLWLIPQYGLHGVAEAFMSIGHLEFFYDQAPE 489
Query: 459 TMKSLSTSFTYLSMSLGYFLSTVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFL 518
+M+S +T+ ++++S+G ++ST++V++++ + + P WL ++LN+ L FYW +
Sbjct: 490 SMRSTATALFWMAISIGNYVSTLLVTLVHKFSAK--PDGSNWLPDNNLNRGRLEYFYWLI 547
Query: 519 AILSCLNFFNFLYWASCYKYKSEDNSCSKLNVKAVAE 555
+L +N +L+ A Y YK SK + V E
Sbjct: 548 TVLQAVNLVYYLWCAKIYTYKPVQVHHSKEDSSPVKE 584
>At2g02040 histidine transport protein (PTR2-B)
Length = 585
Score = 341 bits (874), Expect = 8e-94
Identities = 189/546 (34%), Positives = 299/546 (54%), Gaps = 20/546 (3%)
Query: 11 RKQKGGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLL 70
+++ G ++A F+ + + + +L+ Y +H S+A +T + G+ +L
Sbjct: 39 KEKTGNWKACPFILGNECCERLAYYGIAGNLITYLTTKLHQGNVSAATNVTTWQGTCYLT 98
Query: 71 SLVGGFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACGKSSCIKGGIA--V 128
L+G ++D Y R T F + + + LT+ A + +L P C C A
Sbjct: 99 PLIGAVLADAYWGRYWTIACFSGIYFIGMSALTLSASVPALKPAECIGDFCPSATPAQYA 158
Query: 129 MLYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSVVGVTG 188
M + L L+ALG GG++ + +FGADQFD+ D E A+FFNW S +G++V +
Sbjct: 159 MFFGGLYLIALGTGGIKPCVSSFGADQFDDTDSRERVRKASFFNWFYFSINIGALVSSSL 218
Query: 189 VVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRIVQVIVVAFKNRN 248
+VW+ +GW GF I TV + G P YR + PG SPI RI QV+V +F+ +
Sbjct: 219 LVWIQENRGWGLGFGIPTVFMGLAIASFFFGTPLYRFQKPGGSPITRISQVVVASFRKSS 278
Query: 249 LPLPESNEQLYEAY-KDATV---EKIAHTNQMRFLDKATILQENFEP-----RPWKVCTV 299
+ +PE LYE K++ + KI HT+ ++LDKA ++ E W++CTV
Sbjct: 279 VKVPEDATLLYETQDKNSAIAGSRKIEHTDDCQYLDKAAVISEEESKSGDYSNSWRLCTV 338
Query: 300 TQVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKLGSFEVPSPSIPVIPLF 359
TQVEE+KIL RM PI AS II + AQ+ T V QG M+ K+GSF++P ++
Sbjct: 339 TQVEELKILIRMFPIWASGIIFSAVYAQMSTMFVQQGRAMNCKIGSFQLPPAALGTFDTA 398
Query: 360 FLCVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKRRDQA-- 417
+ + +P+Y+ VP ARK T G T++QR+G+GL +S + MA A I+E+ R A
Sbjct: 399 SVIIWVPLYDRFIVPLARKFTGVDKGFTEIQRMGIGLFVSVLCMAAAAIVEIIRLHMAND 458
Query: 418 ----IKDPSKPISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMS 473
PIS+ W QY I G A++F IG LEFFY ++P M+SL ++ L+ +
Sbjct: 459 LGLVESGAPVPISVLWQIPQYFILGAAEVFYFIGQLEFFYDQSPDAMRSLCSALALLTNA 518
Query: 474 LGYFLSTVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWA 533
LG +LS+++++++ T R ++GW+ +LN +L+ F+W LA LS +N + + A
Sbjct: 519 LGNYLSSLILTLVTYFTTR--NGQEGWI-SDNLNSGHLDYFFWLLAGLSLVNMAVYFFSA 575
Query: 534 SCYKYK 539
+ YK K
Sbjct: 576 ARYKQK 581
>At2g37900 putative peptide/amino acid transporter
Length = 575
Score = 340 bits (872), Expect = 1e-93
Identities = 182/539 (33%), Positives = 304/539 (55%), Gaps = 16/539 (2%)
Query: 11 RKQKGGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLL 70
R + G +RA++F+ A+ + + + +LV+Y +++ DL + + + G T L+
Sbjct: 36 RARTGAWRAALFIIAIEFSERLSYFGLATNLVVYLTTILNQDLKMAIRNVNYWSGVTTLM 95
Query: 71 SLVGGFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACGKSSCIKGGIA--V 128
L+GGFI+D YL R T L+ ++ ++ LV+LT+ + L P C + C++ A V
Sbjct: 96 PLLGGFIADAYLGRYATVLVATTIYLMGLVLLTMSWFIPGLKP--CHQEVCVEPRKAHEV 153
Query: 129 MLYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSVVGVTG 188
+ ++ L+++G GG + ++ +FGADQFD+ E K +FFNW +S G + VT
Sbjct: 154 AFFIAIYLISIGTGGHKPSLESFGADQFDDDHVEERKMKMSFFNWWNVSLCAGILTAVTA 213
Query: 189 VVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRIVQVIVVAFKNRN 248
V ++ + GW I+TV +I +I +GKPFYR + P SP+ I+QV V A RN
Sbjct: 214 VAYIEDRVGWGVAGIILTVVMAISLIIFFIGKPFYRYRTPSGSPLTPILQVFVAAIAKRN 273
Query: 249 LPLPESNEQLYEAYKD--ATVEKIAHTNQMRFLDKATILQEN-----FEPRPWKVCTVTQ 301
LP P L+E K + + HT ++FLDKA I+++ + PW++ T+T+
Sbjct: 274 LPYPSDPSLLHEVSKTEFTSGRLLCHTEHLKFLDKAAIIEDKNPLALEKQSPWRLLTLTK 333
Query: 302 VEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKLGSFEVPSPSIPVIPLFFL 361
VEE K++ ++PI ST+ C Q TF + Q M + +G F VP S+ + L
Sbjct: 334 VEETKLIINVIPIWFSTLAFGICATQASTFFIKQAITMDRHIGGFTVPPASMFTLTALTL 393
Query: 362 CVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKRRDQAIKDP 421
+ + +YE + VP R IT + G+ LQR+G G++ S I+M +A ++E +R D+ +
Sbjct: 394 IISLTVYEKLLVPLLRSITRNQRGINILQRIGTGMIFSLITMIIAALVEKQRLDRT--NN 451
Query: 422 SKPISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMSLGYFLSTV 481
+KP+S+ WL+ Q+ + G AD FTL+GL E+FY + P +M+SL +F + FL+ +
Sbjct: 452 NKPMSVIWLAPQFMVIGFADAFTLVGLQEYFYHQVPDSMRSLGIAFYLSVIGAASFLNNL 511
Query: 482 VVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWASCYKYKS 540
+++ ++T+ + S + W +G DLN + L+ FYWFLA + N F+ A YKS
Sbjct: 512 LITAVDTLAENF--SGKSW-FGKDLNSSRLDRFYWFLAGVIAANICVFVIVAKRCPYKS 567
>At2g40460 putative PTR2 family peptide transporter
Length = 583
Score = 336 bits (862), Expect = 2e-92
Identities = 197/562 (35%), Positives = 316/562 (56%), Gaps = 34/562 (6%)
Query: 15 GGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLLSLVG 74
G +RA F+ A + M F +LV Y +H D SS + N+ G+ ++ + G
Sbjct: 26 GRWRACSFLLGYEAFERMAFYGIASNLVNYLTKRLHEDTISSVRNVNNWSGAVWITPIAG 85
Query: 75 GFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEA----CGKSSCIKGGIAVML 130
+I+D+Y+ R T + VL +++LT+ + SL P C K+S ++
Sbjct: 86 AYIADSYIGRFWTFTASSLIYVLGMILLTMAVTVKSLRPTCENGVCNKASSLQ---VTFF 142
Query: 131 YTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSVVGVTGVV 190
Y SL +A+G GG + + FGADQFD E K +FFNW + SS LG++ G+V
Sbjct: 143 YISLYTIAIGAGGTKPNISTFGADQFDSYSIEEKKQKVSFFNWWMFSSFLGALFATLGLV 202
Query: 191 WVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKA-PGDSPILRIVQVIVVAFKNRNL 249
++ GW G+ I TV + ++ +G PFYR K D+ +VQV + AFKNR L
Sbjct: 203 YIQENLGWGLGYGIPTVGLLVSLVVFYIGTPFYRHKVIKTDNLAKDLVQVPIAAFKNRKL 262
Query: 250 PLPESNEQLYEA----YKDATVEKIAHTNQMRFLDKATILQENFEPRPWKVCTVTQVEEV 305
P+ + +LYE YK ++ HT RFLDKA I + P CTVT+VE
Sbjct: 263 QCPDDHLELYELDSHYYKSNGKHQVHHTPVFRFLDKAAIKTSSRVP-----CTVTKVEVA 317
Query: 306 KILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKLGS-FEVPSPSIPVIPLFFLCVL 364
K + ++ I T+I +T AQ+ T V QG+ + +K+GS F++P+ S+ + +
Sbjct: 318 KRVLGLIFIWLVTLIPSTLWAQVNTLFVKQGTTLDRKIGSNFQIPAASLGSFVTLSMLLS 377
Query: 365 IPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKR----RDQAIKD 420
+P+Y+ FVPF RK T +P G+T LQR+GVG + +++A+A +EVKR ++ I
Sbjct: 378 VPMYDQSFVPFMRKKTGNPRGITLLQRLGVGFAIQIVAIAIASAVEVKRMRVIKEFHITS 437
Query: 421 PSK--PISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMSLGYFL 478
P++ P+S+FWL QY++ G+ D+F IGLLEFFY ++P M+SL T+F + LG FL
Sbjct: 438 PTQVVPMSIFWLLPQYSLLGIGDVFNAIGLLEFFYDQSPEEMQSLGTTFFTSGIGLGNFL 497
Query: 479 STVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWASCYKY 538
++ +V++I+ +T + + W+ G++LN + L+ +Y FL ++S +N F++ AS Y Y
Sbjct: 498 NSFLVTMIDKITSK--GGGKSWI-GNNLNDSRLDYYYGFLVVISIVNMGLFVWAASKYVY 554
Query: 539 KSEDNS-------CSKLNVKAV 553
KS+D++ C ++ KA+
Sbjct: 555 KSDDDTKEFSGGGCVQMEAKAL 576
>At1g12110 nitrate/chlorate transporter CHL1
Length = 590
Score = 336 bits (862), Expect = 2e-92
Identities = 197/552 (35%), Positives = 303/552 (54%), Gaps = 26/552 (4%)
Query: 11 RKQKGGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLL 70
R + GG+ ++ + + A++ + + V+LV Y MH A++ANT+TNF+G++F+L
Sbjct: 25 RSKTGGWASAAMILCIEAVERLTTLGIGVNLVTYLTGTMHLGNATAANTVTNFLGTSFML 84
Query: 71 SLVGGFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACG---KSSCIKG-GI 126
L+GGFI+DT+L R T +F +++ + +LT+ + L P C S C + GI
Sbjct: 85 CLLGGFIADTFLGRYLTIAIFAAIQATGVSILTLSTIIPGLRPPRCNPTTSSHCEQASGI 144
Query: 127 AV-MLYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSVVG 185
+ +LY +L L ALG GGV+ ++ FG+DQFDE +P E + FFN +GS++
Sbjct: 145 QLTVLYLALYLTALGTGGVKASVSGFGSDQFDETEPKERSKMTYFFNRFFFCINVGSLLA 204
Query: 186 VTGVVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRIVQVIVVAFK 245
VT +V+V G WG+ I A + + G YR K SP+ ++ VIV A++
Sbjct: 205 VTVLVYVQDDVGRKWGYGICAFAIVLALSVFLAGTNRYRFKKLIGSPMTQVAAVIVAAWR 264
Query: 246 NRNLPLPESNEQLYEAYKDATVE-------KIAHTNQMRFLDKATILQENFEP-----RP 293
NR L LP LY+ E K+ HT Q R LDKA I +
Sbjct: 265 NRKLELPADPSYLYDVDDIIAAEGSMKGKQKLPHTEQFRSLDKAAIRDQEAGVTSNVFNK 324
Query: 294 WKVCTVTQVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKLGSFEVPSPSI 353
W + T+T VEEVK + RMLPI A+ I+ T AQL T SV+Q + + +GSFE+P S+
Sbjct: 325 WTLSTLTDVEEVKQIVRMLPIWATCILFWTVHAQLTTLSVAQSETLDRSIGSFEIPPASM 384
Query: 354 PVIPLFFLCVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKR 413
V + L + +Y+ + + +K+ N+P G+ LQR+G+GL S++MAVA ++E+KR
Sbjct: 385 AVFYVGGLLLTTAVYDRVAIRLCKKLFNYPHGLRPLQRIGLGLFFGSMAMAVAALVELKR 444
Query: 414 -RDQAIKDP---SKPISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTY 469
R P + P+ + L QY I G+ + G L+FF RE P MK +ST
Sbjct: 445 LRTAHAHGPTVKTLPLGFYLLIPQYLIVGIGEALIYTGQLDFFLRECPKGMKGMSTGLLL 504
Query: 470 LSMSLGYFLSTVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNF 529
+++LG+F S+V+V+++ T + P W+ DLNK L FYW +A+L LNF F
Sbjct: 505 STLALGFFFSSVLVTIVEKFTGKAHP----WI-ADDLNKGRLYNFYWLVAVLVALNFLIF 559
Query: 530 LYWASCYKYKSE 541
L ++ Y YK +
Sbjct: 560 LVFSKWYVYKEK 571
>At3g53960 transporter like protein
Length = 620
Score = 330 bits (845), Expect = 2e-90
Identities = 180/545 (33%), Positives = 306/545 (56%), Gaps = 20/545 (3%)
Query: 11 RKQKGGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLL 70
R Q G +RA++F+ + + + + +LV+Y ++H DL + + G T L+
Sbjct: 35 RAQTGAWRAALFIIGIEFSERLSYFGISTNLVVYLTTILHQDLKMAVKNTNYWSGVTTLM 94
Query: 71 SLVGGFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACGKSSCIKGGIA--V 128
L+GGF++D YL R T LL ++ ++ L++LT+ + L +AC + C++ A +
Sbjct: 95 PLLGGFVADAYLGRYGTVLLATTIYLMGLILLTLSWFIPGL--KACHEDMCVEPRKAHEI 152
Query: 129 MLYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSVVGVTG 188
+ ++ L+++G GG + ++ +FGADQF++ P E K ++FNW G + VT
Sbjct: 153 AFFIAIYLISIGTGGHKPSLESFGADQFEDGHPEERKMKMSYFNWWNAGLCAGILTAVTV 212
Query: 189 VVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRIVQVIVVAFKNRN 248
+V++ + GW I+T+ + F I +GKPFYR +AP SP+ ++QV V A RN
Sbjct: 213 IVYIEDRIGWGVASIILTIVMATSFFIFRIGKPFYRYRAPSGSPLTPMLQVFVAAIAKRN 272
Query: 249 LPLPESNEQLYEAYKDATVE--KIAHTNQMRFLDKATILQENFE------PRPWKVCTVT 300
LP P + L+E + + ++ + ++FLDKA ++++ E PW++ TVT
Sbjct: 273 LPCPSDSSLLHELTNEEYTKGRLLSSSKNLKFLDKAAVIEDRNENTKAEKQSPWRLATVT 332
Query: 301 QVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKL--GSFEVPSPSIPVIPL 358
+VEEVK+L M+PI T+ C Q T + Q +M + + SF VP S+ +
Sbjct: 333 KVEEVKLLINMIPIWFFTLAFGVCATQSSTLFIKQAIIMDRHITGTSFIVPPASLFSLIA 392
Query: 359 FFLCVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKRRDQAI 418
+ + + IYE + VP R+ T + G++ LQR+GVG+V S +M +A +IE KR D A
Sbjct: 393 LSIIITVTIYEKLLVPLLRRATGNERGISILQRIGVGMVFSLFAMIIAALIEKKRLDYAK 452
Query: 419 K---DPSKPISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMSLG 475
+ + + +S WL+ Q+ + GVAD FTL+GL E+FY + P +M+SL +F +
Sbjct: 453 EHHMNKTMTLSAIWLAPQFLVLGVADAFTLVGLQEYFYDQVPDSMRSLGIAFYLSVLGAA 512
Query: 476 YFLSTVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWASC 535
F++ ++++V + + + I S +GW +G DLN + L+ FYW LA L+ N F+ A
Sbjct: 513 SFVNNLLITVSDHLAEEI--SGKGW-FGKDLNSSRLDRFYWMLAALTAANICCFVIVAMR 569
Query: 536 YKYKS 540
Y YK+
Sbjct: 570 YTYKT 574
>At5g46040 peptide transporter
Length = 586
Score = 324 bits (831), Expect = 8e-89
Identities = 180/559 (32%), Positives = 308/559 (54%), Gaps = 23/559 (4%)
Query: 10 KRKQKGGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFL 69
+R Q G ++A FV + M + +LV+Y +H S+N +TN++G+++L
Sbjct: 24 RRSQTGRWKACSFVVVYEVFERMAYYGISSNLVIYMTTKLHQGTVKSSNNVTNWVGTSWL 83
Query: 70 LSLVGGFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACGKSS---CIKGGI 126
++G +++D + R T ++ ++ +L + +LT+ L L P C ++ C K +
Sbjct: 84 TPILGAYVADAHFGRYITFVISSAIYLLGMALLTLSVSLPGLKPPKCSTANVENCEKASV 143
Query: 127 A--VMLYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSVV 184
+ + +L LA+G GG + + GADQFDE DP + +FFNW + S G+
Sbjct: 144 IQLAVFFGALYTLAIGTGGTKPNISTIGADQFDEFDPKDKIHKHSFFNWWMFSIFFGTFF 203
Query: 185 GVTGVVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRIVQVIVVAF 244
T +V+V GW G+ + T+ + I LG YR K P SP ++ +VIV +
Sbjct: 204 ATTVLVYVQDNVGWAIGYGLSTLGLAFSIFIFLLGTRLYRHKLPMGSPFTKMARVIVASL 263
Query: 245 KNRNLPLPESNEQLYE----AYKDATVEKIAHTNQMRFLDKATILQENFEPRPWKVCTVT 300
+ P+ + + YE Y I T+ +RFL++A++ + W++CT+T
Sbjct: 264 RKAREPMSSDSTRFYELPPMEYASKRAFPIHSTSSLRFLNRASL--KTGSTHKWRLCTIT 321
Query: 301 QVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKL-GSFEVPSPSIPVIPLF 359
+VEE K + +MLP+L T + + LAQ+ T + QG+ + ++L +F +P S+ F
Sbjct: 322 EVEETKQMLKMLPVLFVTFVPSMMLAQIMTLFIKQGTTLDRRLTNNFSIPPASLLGFTTF 381
Query: 360 FLCVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKRRDQAIK 419
+ V I IY+ +FV F RK+T +P G+T LQR+G+G++L + M +A I E R A +
Sbjct: 382 SMLVSIVIYDRVFVKFMRKLTGNPRGITLLQRMGIGMILHILIMIIASITERYRLKVAAE 441
Query: 420 DPSK-------PISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSM 472
P+S+F L QY + G+AD F I LEFFY +AP +MKSL TS+T SM
Sbjct: 442 HGLTHQTAVPIPLSIFTLLPQYVLMGLADAFIEIAKLEFFYDQAPESMKSLGTSYTSTSM 501
Query: 473 SLGYFLSTVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYW 532
++GYF+S++++S ++ +TK+ +GW+ ++LN++ L+ +Y F A+L+ LNF FL
Sbjct: 502 AVGYFMSSILLSSVSQITKK---QGRGWIQ-NNLNESRLDNYYMFFAVLNLLNFILFLVV 557
Query: 533 ASCYKYKSEDNSCSKLNVK 551
Y+Y+++ + + K
Sbjct: 558 IRFYEYRADVTQSANVEQK 576
>At1g69870 unknown protein
Length = 620
Score = 323 bits (829), Expect = 1e-88
Identities = 194/549 (35%), Positives = 300/549 (54%), Gaps = 25/549 (4%)
Query: 12 KQKGGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLLS 71
K+ GG+RA F+ L+ +G + + + ++Y V H + +AN + + G T L
Sbjct: 50 KKPGGWRAVSFILGNETLERLGSIGLLANFMVYLTKVFHLEQVDAANVINIWSGFTNLTP 109
Query: 72 LVGGFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACGKSSCIKGG------ 125
LVG +ISDTY+ R T +L L+ +T+ A LHP +C + G
Sbjct: 110 LVGAYISDTYVGRFKTIAFASFATLLGLITITLTASFPQLHPASCNSQDPLSCGGPNKLQ 169
Query: 126 IAVMLYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSVVG 185
I V+L L L++G GG+R + FG DQFD++ K +A+FFNW ++ T+ ++
Sbjct: 170 IGVLLL-GLCFLSVGSGGIRPCSIPFGVDQFDQRTEEGVKGVASFFNWYYMTFTVVLIIT 228
Query: 186 VTGVVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPILRIVQVIVVAFK 245
T VV++ Q W GFSI T ++ ++ G Y P S I QVIV A K
Sbjct: 229 QTVVVYIQDQVSWIIGFSIPTGLMALAVVMFFAGMKRYVYVKPEGSIFSGIAQVIVAARK 288
Query: 246 NRNLPLPESNEQLYEAY----KDATVEKIAHTNQMRFLDKATILQE-NFEPR-----PWK 295
R L LP ++ Y K + + K+ +NQ R LDKA ++ E + P W+
Sbjct: 289 KRKLKLPAEDDGTVTYYDPAIKSSVLSKLHRSNQFRCLDKAAVVIEGDLTPEGPPADKWR 348
Query: 296 VCTVTQVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKLG-SFEVPSPSIP 354
+C+V +VEEVK L R++PI ++ II + TF+VSQ M + LG FE+P+ S+
Sbjct: 349 LCSVQEVEEVKCLIRIVPIWSAGIISLAAMTTQGTFTVSQALKMDRNLGPKFEIPAGSLS 408
Query: 355 VIPLFFLCVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKRR 414
VI L + + +P Y+ +FVPF R+IT H SG+T LQR+G G+V + SM VAGI+E RR
Sbjct: 409 VISLLTIGIFLPFYDRVFVPFMRRITGHKSGITLLQRIGTGIVFAIFSMIVAGIVERMRR 468
Query: 415 DQAIK--DPS--KPISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYL 470
++I DP+ P+S+FWLS Q + G+ + F +IG +EFF + P M+S++ S L
Sbjct: 469 IRSINAGDPTGMTPMSVFWLSPQLILMGLCEAFNIIGQIEFFNSQFPEHMRSIANSLFSL 528
Query: 471 SMSLGYFLSTVVVSVINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFL 530
S + +LS+ +V+V++ + + WL +LN L+ FY+ +A+L +N F
Sbjct: 529 SFAGSSYLSSFLVTVVHKFSG--GHDRPDWL-NKNLNAGKLDYFYYLIAVLGVVNLVYFW 585
Query: 531 YWASCYKYK 539
Y A Y+YK
Sbjct: 586 YCARGYRYK 594
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,246,112
Number of Sequences: 26719
Number of extensions: 547937
Number of successful extensions: 2378
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 64
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 1977
Number of HSP's gapped (non-prelim): 145
length of query: 621
length of database: 11,318,596
effective HSP length: 105
effective length of query: 516
effective length of database: 8,513,101
effective search space: 4392760116
effective search space used: 4392760116
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0048.12


