

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0046a.3
(160 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
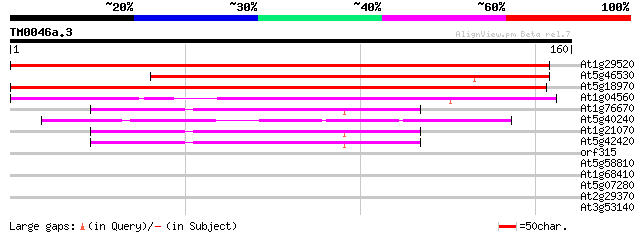
Score E
Sequences producing significant alignments: (bits) Value
At1g29520 plasma membrane associated protein, putative 253 3e-68
At5g46530 putative protein 166 7e-42
At5g18970 plasma membrane associated protein -like 134 2e-32
At1g04560 unknown protein 112 9e-26
At1g76670 unknown protein 43 6e-05
At5g40240 unknown protein 43 8e-05
At1g21070 unknown protein 42 1e-04
At5g42420 unknown protein 41 3e-04
orf315 -mitochondrial genome- 31 0.33
At5g58810 cucumisin precursor - like 29 0.95
At1g68410 putative protein phosphatase 28 2.8
At5g07280 receptor-like protein kinase-like protein 27 3.6
At2g29370 putative tropinone reductase 27 4.7
At3g53140 caffeic acid O-methyltransferase - like protein 26 8.1
>At1g29520 plasma membrane associated protein, putative
Length = 158
Score = 253 bits (646), Expect = 3e-68
Identities = 121/154 (78%), Positives = 140/154 (90%)
Query: 1 MANEQMKPIATLLLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMG 60
MA EQ+KP+A+ LL LNFCMYVIVLGIGGWAMNRAID GF +GP L+LPAHFSPI+FPMG
Sbjct: 1 MAGEQIKPVASGLLVLNFCMYVIVLGIGGWAMNRAIDHGFEVGPNLELPAHFSPIYFPMG 60
Query: 61 NASTGFFVTFALIAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWK 120
NA+TGFFV FAL+AGVVGAAS ISG++HIRSWT SLP+AA+ AT+AWTLT+LAMGFAWK
Sbjct: 61 NAATGFFVIFALLAGVVGAASTISGLSHIRSWTVGSLPAAATAATIAWTLTVLAMGFAWK 120
Query: 121 EVELRIRNARLKTMEAFLIILSATQLFYIAAIHG 154
E+EL+ RNA+L+TMEAFLIILS TQL YIAA+HG
Sbjct: 121 EIELQGRNAKLRTMEAFLIILSVTQLIYIAAVHG 154
>At5g46530 putative protein
Length = 138
Score = 166 bits (419), Expect = 7e-42
Identities = 85/120 (70%), Positives = 98/120 (80%), Gaps = 6/120 (5%)
Query: 41 IIGPELQLPAHFSPIFFPMGNASTGFFVTFALIAGVVGAASAISGINHIRSWTAESLPSA 100
I G + LPAHFSPI FPMGNA+TGFF+ FALIAGV GAAS ISGI+H++SWT SLP+A
Sbjct: 16 IQGADYSLPAHFSPIHFPMGNAATGFFIMFALIAGVAGAASVISGISHLQSWTTTSLPAA 75
Query: 101 ASVATMAWTLTLLAMGFAWKEVELRIRNARL------KTMEAFLIILSATQLFYIAAIHG 154
S AT+AW+LTLLAMGF KE+EL +RNARL +TMEAFLIILSATQL YIAAI+G
Sbjct: 76 VSAATIAWSLTLLAMGFGCKEIELGMRNARLVSKPLMRTMEAFLIILSATQLLYIAAIYG 135
>At5g18970 plasma membrane associated protein -like
Length = 171
Score = 134 bits (337), Expect = 2e-32
Identities = 70/153 (45%), Positives = 99/153 (63%)
Query: 1 MANEQMKPIATLLLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMG 60
MA+ K A +LL LN +Y ++ I WA+N I++ L LPA PI+FP+G
Sbjct: 1 MASGGSKSAAFMLLMLNLGLYFVITIIASWAVNHGIERTRESASTLSLPAKIFPIYFPVG 60
Query: 61 NASTGFFVTFALIAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWK 120
N +TGFFV F LIAGVVG A++++GI ++ W + +L SAA+ + ++W+LTLLAMG A K
Sbjct: 61 NMATGFFVIFTLIAGVVGMATSLTGIINVLQWDSPNLHSAAASSLISWSLTLLAMGLACK 120
Query: 121 EVELRIRNARLKTMEAFLIILSATQLFYIAAIH 153
E+ + A L+T+E II+SATQL AIH
Sbjct: 121 EINIGWTEANLRTLEVLTIIVSATQLLCTGAIH 153
>At1g04560 unknown protein
Length = 186
Score = 112 bits (280), Expect = 9e-26
Identities = 66/157 (42%), Positives = 92/157 (58%), Gaps = 14/157 (8%)
Query: 1 MANEQMKPIATLLLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMG 60
MA + IA LL LN MY+IVLG W +N+ I+ G P G
Sbjct: 1 MATTVGRNIAAPLLFLNLVMYLIVLGFASWCLNKYIN-GQTNHPSFG------------G 47
Query: 61 NASTGFFVTFALIAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWK 120
N +T FF+TF+++A VVG AS ++G NHIR W +SL +A + + +AW +T LAMG A K
Sbjct: 48 NGATPFFLTFSILAAVVGVASKLAGANHIRFWRNDSLAAAGASSIVAWAITALAMGLACK 107
Query: 121 EVEL-RIRNARLKTMEAFLIILSATQLFYIAAIHGAA 156
++ + R RLK +EAF+IIL+ TQL Y+ IH +
Sbjct: 108 QINIGGWRGWRLKMIEAFIIILTFTQLLYLMLIHAGS 144
>At1g76670 unknown protein
Length = 347
Score = 43.1 bits (100), Expect = 6e-05
Identities = 28/96 (29%), Positives = 45/96 (46%), Gaps = 4/96 (4%)
Query: 24 VLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMGNASTGFFVTFALIAGVVGAASAI 83
V +G WAMN G I+ + + + S F TGF F + G+V A+ +
Sbjct: 12 VSDVGAWAMNVISSVGIIMANKQLMSS--SGFGFGFATTLTGFHFAFTALVGMVSNATGL 69
Query: 84 SGINHIRSWTA--ESLPSAASVATMAWTLTLLAMGF 117
S H+ W S+ + S+A M ++L L ++GF
Sbjct: 70 SASKHVPLWELLWFSIVANISIAAMNFSLMLNSVGF 105
>At5g40240 unknown protein
Length = 368
Score = 42.7 bits (99), Expect = 8e-05
Identities = 42/134 (31%), Positives = 64/134 (47%), Gaps = 16/134 (11%)
Query: 10 ATLLLGLNFCMYVIVLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMGNASTGFFVT 69
A L GL+F ++V I + + I G +LPA SP+FF +
Sbjct: 40 AATLRGLSFYVFVFYSYIVSTLL--LLPLSVIFGRSRRLPAAKSPLFFKI---------- 87
Query: 70 FALIAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTLTLLAMGFAWKEVELRIRNA 129
+ G+VG S I+G I ++++ +L SA S T A+T T LA+ F ++V LR
Sbjct: 88 --FLLGLVGFMSQIAGCKGI-AYSSPTLASAISNLTPAFTFT-LAVIFRMEQVRLRSSAT 143
Query: 130 RLKTMEAFLIILSA 143
+ K + A L I A
Sbjct: 144 QAKIIGAILSISGA 157
>At1g21070 unknown protein
Length = 348
Score = 42.4 bits (98), Expect = 1e-04
Identities = 28/96 (29%), Positives = 44/96 (45%), Gaps = 4/96 (4%)
Query: 24 VLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMGNASTGFFVTFALIAGVVGAASAI 83
V +G WAMN G I+ + + + S F TGF + G+V A+ +
Sbjct: 13 VSDVGAWAMNVTSSVGIIMANKQLMSS--SGFGFSFATTLTGFHFALTALVGMVSNATGL 70
Query: 84 SGINHIRSWTA--ESLPSAASVATMAWTLTLLAMGF 117
S H+ W SL + S+A M ++L L ++GF
Sbjct: 71 SASKHVPLWELLWFSLVANISIAAMNFSLMLNSVGF 106
>At5g42420 unknown protein
Length = 350
Score = 40.8 bits (94), Expect = 3e-04
Identities = 27/96 (28%), Positives = 43/96 (44%), Gaps = 4/96 (4%)
Query: 24 VLGIGGWAMNRAIDQGFIIGPELQLPAHFSPIFFPMGNASTGFFVTFALIAGVVGAASAI 83
V +G WAMN G I+ + + + S F TGF + G+V A+
Sbjct: 13 VSDMGAWAMNVISSVGIIMANKQLMSS--SGFAFSFATTLTGFHFALTALVGMVSNATGF 70
Query: 84 SGINHIRSWTA--ESLPSAASVATMAWTLTLLAMGF 117
S H+ W S+ + S+A M ++L L ++GF
Sbjct: 71 SASKHVPMWELIWFSIVANVSIAAMNFSLMLNSVGF 106
>orf315 -mitochondrial genome-
Length = 315
Score = 30.8 bits (68), Expect = 0.33
Identities = 26/85 (30%), Positives = 38/85 (44%), Gaps = 5/85 (5%)
Query: 74 AGVVGAASAISGINHIRSWTAESL---PSAASVATMAWTLTLLAMGFAWKEVELRIRNAR 130
A + +A A GI ++ S S+ PS A+ + TLTLL A+ R++
Sbjct: 23 AATIASAGAAIGIGNVFSSLIHSVARNPSLATTTVLVVTLTLLGGVAAFYLHSFRLKGPL 82
Query: 131 LKTMEAFLIILSAT--QLFYIAAIH 153
K + FL+ A L I AIH
Sbjct: 83 KKIIYLFLVFFIAVGISLIRIKAIH 107
>At5g58810 cucumisin precursor - like
Length = 693
Score = 29.3 bits (64), Expect = 0.95
Identities = 18/73 (24%), Positives = 36/73 (48%), Gaps = 3/73 (4%)
Query: 41 IIGPELQLPAHFSPIFFPMGNASTGFFVTFALIAGVVGAASAISGI-NHIRSWTAESLPS 99
I P +++ A +SP+ P V + + +G A +SG+ +I+++ E PS
Sbjct: 439 ITAPGVEILAAYSPLVSPSATTLDNRRVNYTITSGTSMACPHVSGVAAYIKTFHPEWYPS 498
Query: 100 --AASVATMAWTL 110
+++ T AW +
Sbjct: 499 MIQSAIMTTAWPM 511
>At1g68410 putative protein phosphatase
Length = 436
Score = 27.7 bits (60), Expect = 2.8
Identities = 24/81 (29%), Positives = 36/81 (43%), Gaps = 2/81 (2%)
Query: 61 NASTGFFVTFALIAGVVGAASAISGINHIRSWTAESLPSAASVATMAWTL-TLLAMGFAW 119
N+ST F V FA+ G G A+A+ ++ + +LPS S L L GF
Sbjct: 66 NSSTAFSV-FAVFDGHNGKAAAVYTRENLLNHVISALPSGLSRDEWLHALPRALVSGFVK 124
Query: 120 KEVELRIRNARLKTMEAFLII 140
+ E + R T F+I+
Sbjct: 125 TDKEFQSRGETSGTTATFVIV 145
>At5g07280 receptor-like protein kinase-like protein
Length = 1192
Score = 27.3 bits (59), Expect = 3.6
Identities = 11/30 (36%), Positives = 17/30 (56%)
Query: 9 IATLLLGLNFCMYVIVLGIGGWAMNRAIDQ 38
IA L+LG ++V V + WAM + + Q
Sbjct: 830 IAGLMLGFTIIVFVFVFSLRRWAMTKRVKQ 859
>At2g29370 putative tropinone reductase
Length = 268
Score = 26.9 bits (58), Expect = 4.7
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 1/39 (2%)
Query: 40 FIIGPELQLPAHFSPIFFPMGNAS-TGFFVTFALIAGVV 77
FII L+ HFS + P+ AS +G V + +AGVV
Sbjct: 122 FIIATNLESTFHFSQLAHPLLKASGSGNIVLMSSVAGVV 160
>At3g53140 caffeic acid O-methyltransferase - like protein
Length = 359
Score = 26.2 bits (56), Expect = 8.1
Identities = 14/36 (38%), Positives = 20/36 (54%), Gaps = 1/36 (2%)
Query: 76 VVGAASAISGINHIRSWTAESLPSAASVATMAWTLT 111
VV A I G+ H+ +S+PSA ++ M W LT
Sbjct: 230 VVAKAPNIPGVTHVGGDMFQSVPSADAI-FMKWVLT 264
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.138 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,074,361
Number of Sequences: 26719
Number of extensions: 109455
Number of successful extensions: 339
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 326
Number of HSP's gapped (non-prelim): 15
length of query: 160
length of database: 11,318,596
effective HSP length: 91
effective length of query: 69
effective length of database: 8,887,167
effective search space: 613214523
effective search space used: 613214523
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0046a.3


