

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0042.15
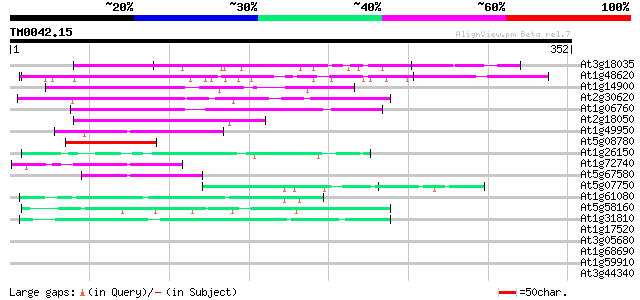
(352 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g18035 unknown protein 139 3e-33
At1g48620 unknown protein 137 9e-33
At1g14900 linker histone protein, putative 101 7e-22
At2g30620 putative histone H1 protein 88 6e-18
At1g06760 histone H1 like protein 79 5e-15
At2g18050 histone H1 59 3e-09
At1g49950 telomere repeat binding factor 1 (TRB1)/DNA-binding pr... 59 5e-09
At5g08780 putative protein 55 8e-08
At1g26150 Pto kinase interactor, putative 51 1e-06
At1g72740 DNA-binding like protein 49 5e-06
At5g67580 telomere repeat binding factor 2 (TRB2) 44 1e-04
At5g07750 putative protein 44 1e-04
At1g61080 hypothetical protein 44 1e-04
At5g58160 strong similarity to unknown protein (gb|AAD23008.1) 43 3e-04
At1g31810 hypothetical protein 41 9e-04
At1g17520 myb-related DNA-binding like protein 40 0.003
At3g05680 unknown protein 39 0.003
At1g68690 protein kinase, putative 39 0.003
At1g59910 hypothetical protein 39 0.006
At3g44340 putative protein 38 0.007
>At3g18035 unknown protein
Length = 427
Score = 139 bits (349), Expect = 3e-33
Identities = 102/244 (41%), Positives = 130/244 (52%), Gaps = 40/244 (16%)
Query: 41 HPPYAEMIYRAIEALKEKDGSSKTAIAKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIM 100
HPPY+EMI AI AL E DGSSK AI++YIE YT L AH+ LLTHHLK LK +G+L M
Sbjct: 12 HPPYSEMICAAIAALNEPDGSSKMAISRYIERCYTGLTSAHAALLTHHLKTLKTSGVLSM 71
Query: 101 LKKSYKL--PSSLPSDITQNAAQPVRRGRPPRSGI------------------PKRRGRP 140
+KKSYK+ S+ P+ + AA + PRS I + RGRP
Sbjct: 72 VKKSYKIAGSSTPPASVAVAAAAAAQGLDVPRSEILHSSNNDPMASGSASQPLKRGRGRP 131
Query: 141 PKPK--SLSNGLKRRPPKHQLQATVIPFADPSLAQPPVPVGSP------RPRGRPRKNAA 192
PKPK S L++ PP +Q+QA P + Q PVPV +P R GRPRKN +
Sbjct: 132 PKPKPESQPQPLQQLPPTNQVQANGQPIWEQQQVQSPVPVPTPVTESAKRGPGRPRKNGS 191
Query: 193 ALPPPHAVGGVDSSLMVPGRPQKLAVRGRPKNPAGRPVGR---PKGTTS-ANIAQRIANE 248
A P + V +S+M + RGRP P R GR PK +S A++ +AN
Sbjct: 192 AAPATAPI--VQASVMA----GIMKRRGRP--PGRRAAGRQRKPKSVSSTASVYPYVANG 243
Query: 249 NLRR 252
RR
Sbjct: 244 ARRR 247
Score = 70.5 bits (171), Expect = 1e-12
Identities = 67/258 (25%), Positives = 116/258 (43%), Gaps = 48/258 (18%)
Query: 91 RLKDTGLLIMLKKSYKLPSSLPSDITQNAAQPVRRGRPPRS---------------GIPK 135
+++ G I ++ + P +P+ +T++A + R R S GI K
Sbjct: 151 QVQANGQPIWEQQQVQSPVPVPTPVTESAKRGPGRPRKNGSAAPATAPIVQASVMAGIMK 210
Query: 136 RRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSLAQPPVPVGSPRPRGRPRK--NAAA 193
RRGRPP ++ G +R+P A+V P+ R RGRPR+ + ++
Sbjct: 211 RRGRPPGRRAA--GRQRKPKSVSSTASVYPYV----------ANGARRRGRPRRVVDPSS 258
Query: 194 LPPPHAVGGVDSSLMVPG------RPQKLA-----VRGRPKNPAGRPVGRPKGTTSANIA 242
+ VGG + + + PG RP K+ + +PK GRPVGRP+ ++
Sbjct: 259 IVSVAPVGGENVAAVAPGMKRGRGRPPKIGGVISRLIMKPKRGRGRPVGRPRKIGTSVTT 318
Query: 243 QRIANENLRRKLEYFQSKVKESLDVLKPYFDHASPVPAIAAIQELEIVATSDLKAPLRDV 302
+ L++K + FQ KVKE + VLK + + AI++LE + ++ P
Sbjct: 319 GTQDSGELKKKFDIFQEKVKEIVKVLKDGVTSENQA-VVQAIKDLEALTVTETVEP---- 373
Query: 303 PSQKIPEQLPLQEHLLPQ 320
++ E++ +E PQ
Sbjct: 374 ---QVMEEVQPEETAAPQ 388
>At1g48620 unknown protein
Length = 479
Score = 137 bits (345), Expect = 9e-33
Identities = 112/313 (35%), Positives = 149/313 (46%), Gaps = 84/313 (26%)
Query: 7 PPPPPPPTAVPFTQE----AINH---------APDSNIPINSSAPA-------------- 39
PPPP + PFT + NH AP +NI + +AP
Sbjct: 14 PPPPQFTSFPPFTNTNPFASPNHPFFTGPTAVAPPNNIHLYQAAPPQQPQTSPVPPHPSI 73
Query: 40 NHPPYAEMIYRAIEALKEKDGSSKTAIAKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLI 99
+HPPY++MI AI AL E DGSSK AI++YIE YT +P AH LLTHHLK LK +G+L+
Sbjct: 74 SHPPYSDMICTAIAALNEPDGSSKQAISRYIERIYTGIPTAHGALLTHHLKTLKTSGILV 133
Query: 100 MLKKSYKLPSSLP---------------SDITQNAAQ----PVRRGRPPRSGIPKRRGRP 140
M+KKSYKL S+ P SD N Q PV P++ I + RGRP
Sbjct: 134 MVKKSYKLASTPPPPPPTSVAPSLEPPRSDFIVNENQPLPDPVLASSTPQT-IKRGRGRP 192
Query: 141 PK-------PKSLSNGL---------KRRPPKHQLQATVIPFADPSLAQPPVPVGSPRPR 184
PK P+ L+NG RP + Q+Q +P QP PV RP
Sbjct: 193 PKAKPDVVQPQPLTNGKLTWEQSELPVSRPEEIQIQPPQLPL------QPQQPV--KRPP 244
Query: 185 GRPRKNAAALPPPHAVGGVDSSLMVPGRPQKLAVRGRPKNPAGRPVGRPKG----TTSAN 240
GRPRK+ + V + V G + + RGRP P+GR GR + + A+
Sbjct: 245 GRPRKDGTS-------PTVKPAASVSGGVETVKRRGRP--PSGRAAGRERKPIVVSAPAS 295
Query: 241 IAQRIANENLRRK 253
+ +AN +RR+
Sbjct: 296 VFPYVANGGVRRR 308
Score = 83.2 bits (204), Expect = 2e-16
Identities = 96/371 (25%), Positives = 154/371 (40%), Gaps = 77/371 (20%)
Query: 8 PPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDGSSKTAIA 67
PPPPPPT+V + E P S+ +N + P P A + I+ + + +K +
Sbjct: 145 PPPPPPTSVAPSLEP----PRSDFIVNENQPLPDPVLASSTPQTIKRGRGRPPKAKPDVV 200
Query: 68 KYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNAAQPVRR-- 125
+ LT+ + + L + + ++ P + QPV+R
Sbjct: 201 Q-------------PQPLTNGKLTWEQSELPVSRPEEIQIQ---PPQLPLQPQQPVKRPP 244
Query: 126 GRPPRSGIP----------------KRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADP 169
GRP + G KRRGRPP ++ G +R+P A+V P+
Sbjct: 245 GRPRKDGTSPTVKPAASVSGGVETVKRRGRPPSGRAA--GRERKPIVVSAPASVFPY--- 299
Query: 170 SLAQPPVPVGSPRPRGRPRK------NAAALPPPHAV----GGVDSSLMVPGRPQKLAVR 219
V G R RGRP++ ++ A PPP GG + ++ GR + +
Sbjct: 300 ------VANGGVRRRGRPKRVDAGGASSVAPPPPPPTNVESGGEEVAVKKRGRGRPPKIG 353
Query: 220 G---RPKNP------AGRPVGRP-KGTTSANIAQRIANE--NLRRKLEYFQSKVKESLDV 267
G +P P G+PVGRP K S + R + L++K E FQ++ K+ + V
Sbjct: 354 GVIRKPMKPMRSFARTGKPVGRPRKNAVSVGASGRQDGDYGELKKKFELFQARAKDIVIV 413
Query: 268 LKPYFDHASPVPAIAAIQELEIVATSDLKAPLRDVPSQKIPEQLPLQEHLLPQPLPEPQI 327
LK + + AIQ+LE +A + + P QLP +EHL +P E Q
Sbjct: 414 LKSEIGGSGNQAVVQAIQDLEGIAET------TNEPKHMEEVQLPDEEHLETEPEAEGQG 467
Query: 328 TYQQQHLQTQL 338
+ + +Q L
Sbjct: 468 QTEAEAMQEAL 478
>At1g14900 linker histone protein, putative
Length = 204
Score = 101 bits (251), Expect = 7e-22
Identities = 74/220 (33%), Positives = 99/220 (44%), Gaps = 56/220 (25%)
Query: 23 INHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDGSSKTAIAKYIEEAYTDLPPAHS 82
++H S+ + + PPY +MI AIE+L +K+G +KT IAK+IE LPP+H
Sbjct: 5 LHHGSASDTHSSELPSFSLPPYPQMIMEAIESLNDKNGCNKTTIAKHIESTQQTLPPSHM 64
Query: 83 TLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNAAQPVR-RGRPPR----------- 130
TLL++HL ++K TG LIM+K +Y P A P R RGRPP+
Sbjct: 65 TLLSYHLNQMKKTGQLIMVKNNYMKPDP--------DAPPKRGRGRPPKQKTQAESDAAA 116
Query: 131 ----------SGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSLAQPPVPVGS 180
+ P+ RGRPPKPK S PP Q V GS
Sbjct: 117 AAVVAATVVSTDPPRSRGRPPKPKDPS-----EPP-----------------QEKVITGS 154
Query: 181 PRPRG----RPRKNAAALPPPHAVGGVDSSLMVPGRPQKL 216
RPRG RPR ++ + P GRP K+
Sbjct: 155 GRPRGRPPKRPRTDSETVAAPEPAAQATGERRGRGRPPKV 194
>At2g30620 putative histone H1 protein
Length = 273
Score = 88.2 bits (217), Expect = 6e-18
Identities = 73/246 (29%), Positives = 112/246 (44%), Gaps = 26/246 (10%)
Query: 6 VPPPPPPPTAVPFTQEAINHAPDSNIPINSSAP------ANHPPYAEMIYRAIEALKEKD 59
V P P A + A + P+ ++AP ++HP Y EMI AI LKE+
Sbjct: 21 VKSPEKKPAAKGGKSKKTTTAKATKKPVKAAAPTKKKTTSSHPTYEEMIKDAIVTLKERT 80
Query: 60 GSSKTAIAKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNA 119
GSS+ AI K+IEE + LPP LL +LKRL + L+ +K S+K+PS+ S T
Sbjct: 81 GSSQYAIQKFIEEKHKSLPPTFRKLLLVNLKRLVASEKLVKVKASFKIPSA-RSAATPKP 139
Query: 120 AQPVRRGRPPRSGIPKRRGR------PPKPKSLSNGLKRRPPKHQLQATVIPFADPSLAQ 173
A PV++ + + K +G+ P K K+ + G K+ K +A V A+
Sbjct: 140 AAPVKK---KATVVAKPKGKVAAAVAPAKAKAAAKGTKKPAAKVVAKAKV-------TAK 189
Query: 174 PPVPVGSPRPRGRPRKNAAALPPPHAVGGVDSSLMVPGRPQKLAVRGRPKNPAGRPVGRP 233
P V + +P+ K+ AA+ AV + P + + + R P P +
Sbjct: 190 PKAKVTAAKPKS---KSVAAVSKTKAVAAKPKAKERPAKASRTSTRTSPGKKVAAPAKKV 246
Query: 234 KGTTSA 239
T A
Sbjct: 247 AVTKKA 252
>At1g06760 histone H1 like protein
Length = 274
Score = 78.6 bits (192), Expect = 5e-15
Identities = 60/196 (30%), Positives = 86/196 (43%), Gaps = 16/196 (8%)
Query: 39 ANHPPYAEMIYRAIEALKEKDGSSKTAIAKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLL 98
++HP Y EMI AI LKE+ GSS+ AI K+IEE +LPP LL +LKRL +G L
Sbjct: 60 SSHPTYEEMIKDAIVTLKERTGSSQYAIQKFIEEKRKELPPTFRKLLLLNLKRLVASGKL 119
Query: 99 IMLKKSYKLPSSLPSDITQNAAQPVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQ 158
+ +K S+KLPS+ + P++ K KP +++ +R
Sbjct: 120 VKVKASFKLPSA------------SAKASSPKAAAEKSAPAKKKPATVAVTKAKRKVAAA 167
Query: 159 LQATVIPFADPSLAQPPVPVGSPRPRGRPRKNAAALPPPHAVGGVDSSLMVPGRPQKLAV 218
+A P A + + PR AAA VD+ RP K A
Sbjct: 168 SKAKKTIAVKPKTAAAKKVTAKAKAKPVPRATAAATKRK----AVDAKPKAKARPAKAAK 223
Query: 219 RGRPKNPAGRPVGRPK 234
+ +PA + V K
Sbjct: 224 TAKVTSPAKKAVAATK 239
>At2g18050 histone H1
Length = 167
Score = 59.3 bits (142), Expect = 3e-09
Identities = 44/125 (35%), Positives = 60/125 (47%), Gaps = 5/125 (4%)
Query: 41 HPPYAEMIYRAIEALKEKDGSSKTAIAKYIEEAYTD-LPPAHSTLLTHHLKRLKDTGLLI 99
HPPY +MI A+ LKEK+GSS AIAK IEE + LP + L+ LK G L+
Sbjct: 24 HPPYFQMIKEALMVLKEKNGSSPYAIAKKIEEKHKSLLPESFRKTLSLQLKNSVAKGKLV 83
Query: 100 MLKKSYKLPSSLPSDITQNAAQPVRRGRPPRSGIPKR----RGRPPKPKSLSNGLKRRPP 155
++ SYKL + Q + + + I KR RP K S++ K+R
Sbjct: 84 KIRASYKLSDTTKMITRQQDKKNKKNMKQEDKEITKRTRSSSTRPKKTVSVNKQEKKRKV 143
Query: 156 KHQLQ 160
K Q
Sbjct: 144 KKARQ 148
>At1g49950 telomere repeat binding factor 1 (TRB1)/DNA-binding
protein PcMYB1, putative
Length = 300
Score = 58.5 bits (140), Expect = 5e-09
Identities = 36/108 (33%), Positives = 53/108 (48%), Gaps = 3/108 (2%)
Query: 29 SNIPINSSAPANHPPYA--EMIYRAIEALKEKDGSSKTAIAKYIEEAYTDLPPAHSTLLT 86
S + ++S+ P P +I AI LKE G +KT I YIE+ Y PP LL+
Sbjct: 104 SGLQVSSNPPPRRPNVRLDSLIMEAIATLKEPGGCNKTTIGAYIEDQY-HAPPDFKRLLS 162
Query: 87 HHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNAAQPVRRGRPPRSGIP 134
LK L G L+ +K+ Y++P+S P + V G+ S +P
Sbjct: 163 TKLKYLTSCGKLVKVKRKYRIPNSTPLSSHRRKGLGVFGGKQRTSSLP 210
>At5g08780 putative protein
Length = 463
Score = 54.7 bits (130), Expect = 8e-08
Identities = 25/57 (43%), Positives = 40/57 (69%)
Query: 36 SAPANHPPYAEMIYRAIEALKEKDGSSKTAIAKYIEEAYTDLPPAHSTLLTHHLKRL 92
S +HP Y+ MI+ AI L ++ G+S+ AI+++I+ Y +LP AH+ LL+HHL +L
Sbjct: 50 SRTPDHPTYSAMIFIAIMDLNKEGGASEDAISEFIKSKYKNLPFAHTNLLSHHLAKL 106
>At1g26150 Pto kinase interactor, putative
Length = 760
Score = 50.8 bits (120), Expect = 1e-06
Identities = 60/225 (26%), Positives = 83/225 (36%), Gaps = 45/225 (20%)
Query: 8 PPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDGSSKTAIA 67
PPPPPPT P T + +P +N P P PP +L D S
Sbjct: 124 PPPPPPTEAPPTTPITSPSPPTNPP----PPPESPP----------SLPAPDPPSNPLPP 169
Query: 68 KYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNAAQPVRRGR 127
+ +PP+HS + L + LPS S+
Sbjct: 170 PKL------VPPSHSPP-----RHLPSPPASEIPPPPRHLPSPPASERPSTPPSDSEHPS 218
Query: 128 PPRSGIPKRRGRPPKPKSLSNGLKR---RPPKHQLQATVIPFADPSLAQPPVPVGSPRPR 184
PP G PKRR +PP P G KR PP + + PS + +P P P
Sbjct: 219 PPPPGHPKRREQPPPP-----GSKRPTPSPPSPSDSKRPVHPSPPSPPEETLPPPKPSPD 273
Query: 185 GRPRKNAA---ALPPPHAVGGVDSSLMVPGRPQKLAVRGRPKNPA 226
P +++ LPP SS++ P P + +V G P NP+
Sbjct: 274 PLPSNSSSPPTLLPP--------SSVVSPPSPPRKSVSG-PDNPS 309
>At1g72740 DNA-binding like protein
Length = 291
Score = 48.5 bits (114), Expect = 5e-06
Identities = 34/111 (30%), Positives = 54/111 (48%), Gaps = 15/111 (13%)
Query: 2 DSIWVPPP----PPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKE 57
D++ +P P PPPP E I PD N N P Y +I+ A+ AL +
Sbjct: 87 DAVTIPRPIPTIPPPPGRRTLPSELI---PDENTK-------NAPRYDGVIFEALSALAD 136
Query: 58 KDGSSKTAIAKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLP 108
+GS ++I +IE + ++PP +L+ L+RL L ++ YK+P
Sbjct: 137 GNGSDVSSIYHFIEPRH-EVPPNFRRILSTRLRRLAAQSKLEKIQNFYKIP 186
>At5g67580 telomere repeat binding factor 2 (TRB2)
Length = 299
Score = 43.9 bits (102), Expect = 1e-04
Identities = 27/77 (35%), Positives = 41/77 (53%), Gaps = 2/77 (2%)
Query: 46 EMIYRAIEALKEKDGSSKTAIAKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSY 105
++I+ AI L+E GS +T+I YIEE + PP + LK L G L+ +K Y
Sbjct: 127 KIIFEAITNLRELRGSDRTSIFLYIEENF-KTPPNMKRHVAVRLKHLSSNGTLVKIKHKY 185
Query: 106 KLPSS-LPSDITQNAAQ 121
+ S+ +P+ Q A Q
Sbjct: 186 RFSSNFIPAGARQKAPQ 202
>At5g07750 putative protein
Length = 1289
Score = 43.9 bits (102), Expect = 1e-04
Identities = 51/194 (26%), Positives = 73/194 (37%), Gaps = 29/194 (14%)
Query: 122 PVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSL---AQPPVP- 177
P+R G PP P R G PP P G PP + P P + A PP P
Sbjct: 1080 PMRGGAPPPPPPPMRGGAPPPPPPPMRGGAPPPPPPPMHGGAPPPPPPPMRGGAPPPPPP 1139
Query: 178 -----VGSPRPRGRPRKNAAALPP---PHAVGGVDSSLMVPGRPQKLAVRGRPKNPAGRP 229
G+P P P A PP P GG P P L RG +P G
Sbjct: 1140 PGGRGPGAPPPPPPPGGRAPGPPPPPGPRPPGG------GPPPPPMLGARGAAVDPRG-- 1191
Query: 230 VGRPKGTTSANIAQRIANENLRRKLEYFQSKVKESL-----DVLKPYFDHASPVPAIAAI 284
GR +G ++ + L + KV +L D L+ + + S P+ +
Sbjct: 1192 AGRGRGLPRPGFGSAAQKKSSLKPLHWV--KVTRALQGSLWDELQRHGE--SQTPSEFDV 1247
Query: 285 QELEIVATSDLKAP 298
E+E + ++ ++ P
Sbjct: 1248 SEIETLFSATVQKP 1261
Score = 41.6 bits (96), Expect = 7e-04
Identities = 33/110 (30%), Positives = 39/110 (35%), Gaps = 6/110 (5%)
Query: 122 PVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSLAQPPVPVGSP 181
P+ G P P R G PP P G PP ++ P PP+ G+P
Sbjct: 1068 PMFGGAQPPPPPPMRGGAPPPPPPPMRGGAPPPPPPPMRG-----GAPPPPPPPMHGGAP 1122
Query: 182 RPRGRPRKNAAALPPPHAVGGVDSSLMVPGRPQKLAVRGRPKNPAGRPVG 231
P P + A PPP GG P P G P P RP G
Sbjct: 1123 PPPPPPMRGGAP-PPPPPPGGRGPGAPPPPPPPGGRAPGPPPPPGPRPPG 1171
Score = 38.9 bits (89), Expect = 0.004
Identities = 30/106 (28%), Positives = 39/106 (36%), Gaps = 7/106 (6%)
Query: 128 PPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSLAQPPVPVGSPRPRGRP 187
PP P G PP P G + PP ++ P PP+ G+P P P
Sbjct: 1050 PPPPPPPMHGGAPPPPPPPMFGGAQPPPPPPMRG-----GAPPPPPPPMRGGAPPPPPPP 1104
Query: 188 RKNAAALPPPHAVGGVDSSLMVPGRPQKLAVRGRPKNPAGRPVGRP 233
+ A PPP + G + P P + P P GR G P
Sbjct: 1105 MRGGAPPPPPPPMHG--GAPPPPPPPMRGGAPPPPPPPGGRGPGAP 1148
Score = 38.1 bits (87), Expect = 0.007
Identities = 42/202 (20%), Positives = 77/202 (37%), Gaps = 17/202 (8%)
Query: 129 PRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSLAQPPVPVGSPRPRGRPR 188
P++ + K+ G KP + + LK + + + Q + A P+ +P +G +
Sbjct: 485 PQATLRKQVGANAKPAAAGDSLKPKSKQQETQGPNVRMAKPNAVSRWIPSN----KGSYK 540
Query: 189 KNAAALPPPHAVGGVDSSLMVPGRPQKLAVRGRPKNPAGRPVGRPKGTTSANIAQRIANE 248
+ PP + +S+ + K R +P G PK + + +++
Sbjct: 541 DSMHVAYPPTRINSAPASITTSLKDGK-----RATSPDGVI---PKDAKTKYLRASVSSP 592
Query: 249 NLRRKLEYFQS---KVKESLDVLKPYFDHASPVPAIAAIQELEIVATSD--LKAPLRDVP 303
++R + S KE+ L P H +P P + E + V S + +P P
Sbjct: 593 DMRSRAPICSSPDSSPKETPSSLPPASPHQAPPPLPSLTSEAKTVLHSSQAVASPPPPPP 652
Query: 304 SQKIPEQLPLQEHLLPQPLPEP 325
+P Q LP P P P
Sbjct: 653 PPPLPTYSHYQTSQLPPPPPPP 674
Score = 37.7 bits (86), Expect = 0.010
Identities = 58/230 (25%), Positives = 75/230 (32%), Gaps = 27/230 (11%)
Query: 2 DSIWVPPPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDGS 61
+++ PPPPPPP + H S SS+P PP + + K D
Sbjct: 828 ETLLPPPPPPPPWKSLYASTFETHEACST----SSSPPPPPPPPP--FSPLNTTKAND-- 879
Query: 62 SKTAIAKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNAAQ 121
I YT + P+ S +K L G+ K P + A
Sbjct: 880 ---YILPPPPLPYTSIAPSPS------VKILPLHGISSAPSPPVKTAPPPPPPPPFSNAH 930
Query: 122 PVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSLAQPPVP-VGS 180
V PP G P PP P S G PP + P PP P GS
Sbjct: 931 SVLSPPPPSYGSPP----PPPPPPPSYGSPPPPPPPPPS-----YGSPPPPPPPPPGYGS 981
Query: 181 PRPRGRPRKNAAALPPPHAVGGVDSSLMVPGRPQKLAVRGRPKNPAGRPV 230
P P P + + PPP S + P P G P P P+
Sbjct: 982 PPPPPPPPPSYGSPPPPPPPPFSHVSSIPPPPPPPPMHGGAPPPPPPPPM 1031
Score = 36.2 bits (82), Expect = 0.028
Identities = 51/205 (24%), Positives = 65/205 (30%), Gaps = 40/205 (19%)
Query: 7 PPPPPPPTAVPFTQEAINH--APDSNIPINSSAPANHPPYAEMIYRAIEALKEKDGSSKT 64
PPPPPPP P N P +P S AP+ +++ L SS
Sbjct: 861 PPPPPPPPFSPLNTTKANDYILPPPPLPYTSIAPSP----------SVKILPLHGISS-- 908
Query: 65 AIAKYIEEAYTDLPP-----AHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNA 119
A + ++ A PP AHS L SY P P
Sbjct: 909 APSPPVKTAPPPPPPPPFSNAHSVLSPP--------------PPSYGSPPPPPPPPPSYG 954
Query: 120 AQPVRRGRPPRSGIPKRR-------GRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSLA 172
+ P PP G P G PP P PP + + P
Sbjct: 955 SPPPPPPPPPSYGSPPPPPPPPPGYGSPPPPPPPPPSYGSPPPPPPPPFSHVSSIPPPPP 1014
Query: 173 QPPVPVGSPRPRGRPRKNAAALPPP 197
PP+ G+P P P + A PPP
Sbjct: 1015 PPPMHGGAPPPPPPPPMHGGAPPPP 1039
Score = 32.3 bits (72), Expect = 0.40
Identities = 23/81 (28%), Positives = 29/81 (35%), Gaps = 14/81 (17%)
Query: 122 PVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSLAQPPVPVGSP 181
P+ G PP PP P + G PP + P P PP+ G+P
Sbjct: 1017 PMHGGAPP----------PPPPPPMHGGAPPPPPPPPMHGGAPPPPPP----PPMHGGAP 1062
Query: 182 RPRGRPRKNAAALPPPHAVGG 202
P P A PPP + G
Sbjct: 1063 PPPPPPMFGGAQPPPPPPMRG 1083
Score = 32.3 bits (72), Expect = 0.40
Identities = 60/252 (23%), Positives = 82/252 (31%), Gaps = 38/252 (15%)
Query: 7 PPPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDGSSKTAI 66
PPPPPPP +P +H S +P P PP++ + L
Sbjct: 647 PPPPPPPPPLP----TYSHYQTSQLP---PPPPPPPPFSSERPNSGTVLPPPP-PPPPPF 698
Query: 67 AKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLI------MLKKSYKLPSSLPSDITQNAA 120
+ + T LPP L +R ++G ++ K Y ++P+ I +
Sbjct: 699 SSERPNSGTVLPPPPPPPLPFSSER-PNSGTVLPPPPSPPWKSVYASALAIPA-ICSTSQ 756
Query: 121 QPVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRRP-PKHQLQATVIPFADPSLAQ------ 173
P PP PP P S G K QL + P P A
Sbjct: 757 APTSSPTPP----------PPPPAYYSVGQKSSDLQTSQLPSPPPPPPPPPFASVRRNSE 806
Query: 174 ---PPVPVGSPRPRGRPRKNAAALPPPHAVGGVDSSLMVP--GRPQKLAVRGRPKNPAGR 228
PP P P P R+N+ L PP SL + + P P
Sbjct: 807 TLLPPPPPPPPPPFASVRRNSETLLPPPPPPPPWKSLYASTFETHEACSTSSSPPPPPPP 866
Query: 229 PVGRPKGTTSAN 240
P P TT AN
Sbjct: 867 PPFSPLNTTKAN 878
Score = 31.2 bits (69), Expect = 0.89
Identities = 18/58 (31%), Positives = 23/58 (39%), Gaps = 4/58 (6%)
Query: 140 PPKPKSLSNGLKRRPPKHQLQATVIPFADPSLAQPPVPVGSPRPRGRPRKNAAALPPP 197
PP P + G PP + P P PP+ G+P P P + A PPP
Sbjct: 1012 PPPPPPMHGGAPPPPPPPPMHGGAPPPPPP----PPMHGGAPPPPPPPPMHGGAPPPP 1065
Score = 28.5 bits (62), Expect = 5.8
Identities = 15/39 (38%), Positives = 19/39 (48%), Gaps = 8/39 (20%)
Query: 6 VPPPPPPPTAVPFTQEAINHA------PDSNIPINSSAP 38
+PPPPPPP PF+ E N P +P +S P
Sbjct: 688 LPPPPPPPP--PFSSERPNSGTVLPPPPPPPLPFSSERP 724
>At1g61080 hypothetical protein
Length = 907
Score = 43.9 bits (102), Expect = 1e-04
Identities = 52/204 (25%), Positives = 71/204 (34%), Gaps = 26/204 (12%)
Query: 7 PPPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEM-IYRAIEALKEKDGSSKTA 65
PPPPPPP + F + A ++P+ S P PP A++ I +
Sbjct: 418 PPPPPPPPPLSFIKTA-------SLPLPSPPPT--PPIADIAISMPPPPPPPPPPPAVMP 468
Query: 66 IAKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNAAQPVRR 125
+ + LPPA L H T K P P + A P
Sbjct: 469 LKHFAPPPPPPLPPAVMPL--KHFAPPPPTPPAFKPLKGSAPPPPPPPPLPTTIAAPPPP 526
Query: 126 GRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSL-----AQPPVPVGS 180
PPR+ + PP P + PP QA P P + + PP+P+G+
Sbjct: 527 PPPPRAAVAP--PPPPPPPGTAAAPPPPPPPPGTQAAPPPPPPPPMQNRAPSPPPMPMGN 584
Query: 181 -------PRPRGRPRKNAAALPPP 197
P P P N A PPP
Sbjct: 585 SGSGGPPPPPPPMPLANGATPPPP 608
Score = 36.6 bits (83), Expect = 0.021
Identities = 61/240 (25%), Positives = 78/240 (32%), Gaps = 39/240 (16%)
Query: 5 WVPPPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDGSSKT 64
+ PPPP PP P A P +P +AP PP RA A T
Sbjct: 490 FAPPPPTPPAFKPLKGSAPPPPPPPPLPTTIAAPPPPPPPP----RAAVAPPPPPPPPGT 545
Query: 65 AIAKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNAAQPVR 124
A A PP T + ++ P +P + + P
Sbjct: 546 AAAP------PPPPPPPGT------QAAPPPPPPPPMQNRAPSPPPMPMGNSGSGGPP-- 591
Query: 125 RGRPPRSGIPKRRGR----PPKPKSLSNGLKRRPPKHQLQATVIPFADPSLAQPPVPVGS 180
PP +P G PP P +++NG PP A PP P G+
Sbjct: 592 ---PPPPPMPLANGATPPPPPPPMAMANGAAGPPPPPPRMGMA-----NGAAGPPPPPGA 643
Query: 181 PRPRGRPRKNAAALPPPHAVGGVDSSLMVPGRPQKLAVRGR-PKNPAGRPVGRPKGTTSA 239
R RP+K A L +G + L K V GR P G GR G SA
Sbjct: 644 ARSL-RPKKAATKLKRSTQLGNLYRIL-------KGKVEGRDPNAKTGSGSGRKAGAGSA 695
Score = 33.1 bits (74), Expect = 0.23
Identities = 42/156 (26%), Positives = 53/156 (33%), Gaps = 22/156 (14%)
Query: 104 SYKLPSSLPS----DITQNAAQPVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQL 159
S LPS P+ DI + P PP + +P + PP P L + P KH
Sbjct: 434 SLPLPSPPPTPPIADIAISMPPPPPPPPPPPAVMPLKHFAPPPPPPLPPAVM--PLKHFA 491
Query: 160 QATVIP--------FADPSLAQPPVPV---GSPRPRGRPRKNAAALPPPHAVGGVDSSLM 208
P A P PP+P P P PR A PPP G +
Sbjct: 492 PPPPTPPAFKPLKGSAPPPPPPPPLPTTIAAPPPPPPPPRAAVAPPPPPPPPGTAAAPPP 551
Query: 209 VPGRPQKLAVRGRP-----KNPAGRPVGRPKGTTSA 239
P P A P +N A P P G + +
Sbjct: 552 PPPPPGTQAAPPPPPPPPMQNRAPSPPPMPMGNSGS 587
Score = 28.1 bits (61), Expect = 7.5
Identities = 15/41 (36%), Positives = 20/41 (48%), Gaps = 5/41 (12%)
Query: 3 SIWVPPPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPP 43
+I PPPPPPP + A+ P P ++AP PP
Sbjct: 519 TIAAPPPPPPP-----PRAAVAPPPPPPPPGTAAAPPPPPP 554
>At5g58160 strong similarity to unknown protein (gb|AAD23008.1)
Length = 1307
Score = 42.7 bits (99), Expect = 3e-04
Identities = 60/265 (22%), Positives = 87/265 (32%), Gaps = 59/265 (22%)
Query: 8 PPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDGSSKTAIA 67
PPPPPP +P +S P+ ++ + +A E L + + A +
Sbjct: 548 PPPPPP-----------------LPAAASKPSEQLQHS--VVQATEPLSQGNSWMSLAGS 588
Query: 68 KY----IEEAYTDLPPAHSTLLTHHLK---RLKDTGLLIMLKKSYKLPSSLPS------- 113
+ E+ LPP T H K T L++ ++ + PS
Sbjct: 589 TFQTVPNEKNLITLPPTPPLASTSHASPEPSSKTTNSLLLSPQASPATPTNPSKTVSVDF 648
Query: 114 -------------DITQNAAQPVRRGRPPRSGIPKRRG--RPPKPKSLSNGLKRRPPKHQ 158
++ N QP R PP S K+ RPP P P +H
Sbjct: 649 FGAATSPHLGASDNVASNLGQPA-RSPPPISNSDKKPALPRPPPPP------PPPPMQHS 701
Query: 159 LQATVIPFADPSLAQPPVPV----GSPRPRGRPRKNAAALPPPHAVGGVDSSLMVPGRPQ 214
V P P+ PP P+ P P P A P + + + SS P P
Sbjct: 702 TVTKVPPPPPPAPPAPPTPIVHTSSPPPPPPPPPPPAPPTPQSNGISAMKSSPPAPPAPP 761
Query: 215 KLAVRGRPKNPAGRPVGRPKGTTSA 239
+L P P P G T A
Sbjct: 762 RLPTHSASPPPPTAPPPPPLGQTRA 786
Score = 33.9 bits (76), Expect = 0.14
Identities = 34/116 (29%), Positives = 42/116 (35%), Gaps = 18/116 (15%)
Query: 123 VRRGRPPRSGIPKRRGRPPKPKSLS-NGLKRRPPKHQLQATVIPFADPSL----AQPPVP 177
V PP P PP P+S + +K PP P A P L A PP P
Sbjct: 722 VHTSSPPPPPPPPPPPAPPTPQSNGISAMKSSPPA--------PPAPPRLPTHSASPPPP 773
Query: 178 VGSPRPR-GRPRKNAAALPPPHAVGGVDSSLMVPGRPQKLAVRGRPKNPAGRPVGR 232
P P G+ R +A PPP +G + + P P P P GR
Sbjct: 774 TAPPPPPLGQTRAPSAPPPPPPKLG----TKLSPSGPNVPPTPALPTGPLSSGKGR 825
Score = 28.9 bits (63), Expect = 4.4
Identities = 13/39 (33%), Positives = 19/39 (48%)
Query: 7 PPPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYA 45
PPPPPPP P T ++ + + P AP P ++
Sbjct: 729 PPPPPPPPPAPPTPQSNGISAMKSSPPAPPAPPRLPTHS 767
>At1g31810 hypothetical protein
Length = 1201
Score = 41.2 bits (95), Expect = 9e-04
Identities = 57/234 (24%), Positives = 76/234 (32%), Gaps = 26/234 (11%)
Query: 7 PPPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDGSSKTAI 66
PPPPPPP++ +IP S+ P PP + +
Sbjct: 601 PPPPPPPSS-------------RSIPSPSAPPPPPPPPPSFGSTGNKRQAQPPPPPPPPP 647
Query: 67 AKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNAAQPVRRG 126
I A PP +H +G + + S P P + P
Sbjct: 648 PTRIPAAKCAPPPPPPPPTSH-------SGSIRVGPPSTPPPPPPPPPKANISNAPKPPA 700
Query: 127 RPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSLAQPPVPVGSPRPRGR 186
PP R G PP P PP L T +P P L + P P G
Sbjct: 701 PPPLPPSSTRLGAPPPPPPPPLSKTPAPPPPPLSKTPVPPPPPGLGRG--TSSGPPPLGA 758
Query: 187 PRKNAAALPPPHAVGGVDSSLMVPGRPQKLAV-RGRPKNPAGRPVGRPKGTTSA 239
NA PPP G L GR + ++V PK A +P+ K T +A
Sbjct: 759 KGSNAPPPPPPAGRGRASLGL---GRGRGVSVPTAAPKKTALKPLHWSKVTRAA 809
Score = 40.4 bits (93), Expect = 0.001
Identities = 59/244 (24%), Positives = 78/244 (31%), Gaps = 39/244 (15%)
Query: 6 VPPPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDGSSKTA 65
+PPPPPPP FT S + S P PP + +
Sbjct: 481 LPPPPPPPPPPLFT---------STTSFSPSQPPPPPPPPPLFMSTTSFSPSQPPPPPPP 531
Query: 66 IAKYIEE-AYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDI-TQNAAQPV 123
+ +++ P L R T L + K+ P P + +++ P+
Sbjct: 532 PPLFTSTTSFSPSQPPPPPPLPSFSNRDPLTTLHQPINKTPPPPPPPPPPLPSRSIPPPL 591
Query: 124 RRGRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSL--------AQPP 175
+ PPR P PP P S S PP P PS AQPP
Sbjct: 592 AQPPPPRPPPPP----PPPPSSRSIPSPSAPPPP-------PPPPPSFGSTGNKRQAQPP 640
Query: 176 VPVGSPRPRGRPRKNAAALPPPHAVGGVDSSLMV---------PGRPQKLAVRGRPKNPA 226
P P P P A PPP S+ V P P K + PK PA
Sbjct: 641 PPPPPPPPTRIPAAKCAPPPPPPPPTSHSGSIRVGPPSTPPPPPPPPPKANISNAPKPPA 700
Query: 227 GRPV 230
P+
Sbjct: 701 PPPL 704
>At1g17520 myb-related DNA-binding like protein
Length = 296
Score = 39.7 bits (91), Expect = 0.003
Identities = 22/61 (36%), Positives = 34/61 (55%), Gaps = 1/61 (1%)
Query: 38 PANHPPYAEMIYRAIEALKEKDGSSKTAIAKYIEEAYTDLPPAHSTLLTHHLKRLKDTGL 97
P N P Y MI+ A+ L + +GS +AI +IE+ ++PP +L+ L+RL G
Sbjct: 124 PKNAPRYDGMIFEALSNLTDANGSDVSAIFNFIEQR-QEVPPNFRRMLSSRLRRLAAQGK 182
Query: 98 L 98
L
Sbjct: 183 L 183
>At3g05680 unknown protein
Length = 2070
Score = 39.3 bits (90), Expect = 0.003
Identities = 53/217 (24%), Positives = 82/217 (37%), Gaps = 44/217 (20%)
Query: 7 PPPP--PPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDGSSKT 64
PP P PPP+ P I H+ DS +S ++ + R + L +
Sbjct: 1760 PPLPLVPPPSVSP----VIPHSSDSLSNQSSPFISHGTQSSGGPTRLMPPLP-------S 1808
Query: 65 AIAKYIEEAYTDLPPAHSTL---------------------LTHHLKRLKDTGLLIMLKK 103
AI +Y Y LPP ST+ + H L TG+
Sbjct: 1809 AIPQYSSNPYASLPPRTSTVQSFGYNHAGVGTTEQQQSGPTIDHQSGNLSVTGMT----- 1863
Query: 104 SYKLPSSLPS-DITQNAAQPVR-RGRPPRSGIPKRRGR---PPKPKSLSNGLKRRPPKHQ 158
SY P+ +PS + ++ ++ PV G P G K + P P+SL+ + P Q
Sbjct: 1864 SYPPPNLMPSHNFSRPSSLPVPFYGNPSHQGGDKPQTMLLVPSIPQSLNTQSIPQLPSMQ 1923
Query: 159 LQATVIPFADPSLAQPPVPVGSPRPRGRPRKNAAALP 195
L P P +PP+ + P +G +N +P
Sbjct: 1924 LSQLQRPMQPPQHVRPPIQISQPSEQGVSMQNPFQIP 1960
>At1g68690 protein kinase, putative
Length = 708
Score = 39.3 bits (90), Expect = 0.003
Identities = 39/135 (28%), Positives = 46/135 (33%), Gaps = 14/135 (10%)
Query: 118 NAAQPVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSLA----- 172
N A P +PP S P PP+P S PP + + P A P A
Sbjct: 65 NGAPPPPLPKPPESSSP-----PPQPVIPSPPPSTSPPPQPVIPSPPPSASPPPALVPPL 119
Query: 173 --QPPVPVGSPRPRGRPRKNAAALPPPHAVGGVDSSLMVPG-RP-QKLAVRGRPKNPAGR 228
PP P P PR P PP +V + S P RP Q P P+ R
Sbjct: 120 PSSPPPPASVPPPRPSPSPPILVRSPPPSVRPIQSPPPPPSDRPTQSPPPPSPPSPPSER 179
Query: 229 PVGRPKGTTSANIAQ 243
P P S Q
Sbjct: 180 PTQSPPSPPSERPTQ 194
Score = 34.3 bits (77), Expect = 0.11
Identities = 59/255 (23%), Positives = 88/255 (34%), Gaps = 32/255 (12%)
Query: 9 PPPPPTAVPFTQEAINHAPDSNIPINSSAP--ANHPPYAEMIYRAIEALKEKDGSSKTAI 66
PPP + +P + N AP P+ +S P AN P + K + SS
Sbjct: 32 PPPVTSPLPPSAPPPNRAPPPPPPVTTSPPPVANGAPPPPL-------PKPPESSSPPPQ 84
Query: 67 AKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSD-----------I 115
+ PP + + L+ L S P+S+P
Sbjct: 85 PVIPSPPPSTSPPPQPVIPSPPPSASPPPALVPPLPSSPPPPASVPPPRPSPSPPILVRS 144
Query: 116 TQNAAQPVRRGRPPRSGIPKRRGRPPKPKS-LSNGLKRRPPKHQLQATVIPFADPSLAQP 174
+ +P++ PP S P + PP P S S + PP + PS P
Sbjct: 145 PPPSVRPIQSPPPPPSDRPTQSPPPPSPPSPPSERPTQSPPSPPSERPTQSPPPPSPPSP 204
Query: 175 PV--PVGSPRPRGRPRKNAAALPPPHAVGGVDSSLMVPGR-PQKLAVRG----RPKNPAG 227
P P SP P P ++ PP + + P R P ++ V G NP
Sbjct: 205 PSDRPSQSPPP---PPEDTKPQPPRRSPNSPPPTFSSPPRSPPEILVPGSNNPSQNNPTL 261
Query: 228 R-PVGRPKGTTSANI 241
R P+ P T ++ I
Sbjct: 262 RPPLDAPNSTNNSGI 276
Score = 27.7 bits (60), Expect = 9.9
Identities = 29/126 (23%), Positives = 41/126 (32%), Gaps = 34/126 (26%)
Query: 108 PSSLPSDITQNAAQPVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFA 167
P L + + PV PP + P R PP P + S
Sbjct: 20 PPPLNNATSPATPPPVTSPLPPSAPPPNRAPPPPPPVTTS-------------------- 59
Query: 168 DPSLAQPPVPVGSPRPRGRPRKNAAALPPPHAVGGVDSSLMVPGRPQKLAVRGRPKNPAG 227
PPV G+P P P+ ++ PPP V +P P + +P P+
Sbjct: 60 -----PPPVANGAPPPP-LPKPPESSSPPPQPV--------IPSPPPSTSPPPQPVIPSP 105
Query: 228 RPVGRP 233
P P
Sbjct: 106 PPSASP 111
>At1g59910 hypothetical protein
Length = 929
Score = 38.5 bits (88), Expect = 0.006
Identities = 35/132 (26%), Positives = 44/132 (32%), Gaps = 26/132 (19%)
Query: 108 PSSLPSDITQNAAQPVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFA 167
P SLP + P G PP P +N PP
Sbjct: 340 PVSLPPGQYMPGNAALSASTPLTPGQFTTANAPPAPPGPANQTSPPPP------------ 387
Query: 168 DPSLAQPPVPVGSPRPRGRPRKNAAALPPPHAVGGVDSSLMVPGRPQKLAVRGRPKNPAG 227
PP +P P P+K AA PPP G + P P ++ +G PK
Sbjct: 388 ------PPPSAAAPPPPPPPKKGPAAPPPPPPPGKKGAG---PPPPPPMSKKGPPK---- 434
Query: 228 RPVGRPKGTTSA 239
P G PKG T +
Sbjct: 435 -PPGNPKGPTKS 445
>At3g44340 putative protein
Length = 1122
Score = 38.1 bits (87), Expect = 0.007
Identities = 38/130 (29%), Positives = 43/130 (32%), Gaps = 17/130 (13%)
Query: 108 PSSLPSDITQNAAQPVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIP-- 165
PS +P N P S P G P P LSNG PP + P
Sbjct: 184 PSGMPGGPLSNGPPPSGMHGGHLSNGPPPSGMPGGP--LSNG----PPPPMMGPGAFPRG 237
Query: 166 --FADPSLAQPPVPVGSPRPRGRPRKNAAALPPPHAVGGVDSSLMVPGRPQKLAVRGRPK 223
F + PP P G P G N+ PP + PG P GRP
Sbjct: 238 SQFTSGPMMAPPPPYGQPPNAGPFTGNSPLSSPP--AHSIPPPTNFPGVPY-----GRPP 290
Query: 224 NPAGRPVGRP 233
P G P G P
Sbjct: 291 MPGGFPYGAP 300
Score = 31.2 bits (69), Expect = 0.89
Identities = 36/130 (27%), Positives = 46/130 (34%), Gaps = 14/130 (10%)
Query: 112 PSDITQNAAQPVRRGRPPRSGIPKRRGRPPKPKS-LSNGLKRRPPKHQLQATVIPFADPS 170
P+ + N Q + RPP P G P+P + P + Q Q P A P
Sbjct: 33 PNSLAANM-QNLNINRPP----PPMPGSGPRPSPPFGQSPQSFPQQQQQQPRPSPMARPG 87
Query: 171 LAQPPVPVGSPRPRGRPRKNAAALPPPHAVGGVDSSLMVP--GRPQKLAVRGRPKN---P 225
PP P RP G P+ + PP S P GRP + G + P
Sbjct: 88 ---PPPPAAMARPGGPPQVSQPGGFPPVGRPVAPPSNQPPFGGRPSTGPLVGGGSSFPQP 144
Query: 226 AGRPVGRPKG 235
G P P G
Sbjct: 145 GGFPASGPPG 154
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,280,064
Number of Sequences: 26719
Number of extensions: 474863
Number of successful extensions: 4034
Number of sequences better than 10.0: 179
Number of HSP's better than 10.0 without gapping: 68
Number of HSP's successfully gapped in prelim test: 112
Number of HSP's that attempted gapping in prelim test: 2526
Number of HSP's gapped (non-prelim): 825
length of query: 352
length of database: 11,318,596
effective HSP length: 100
effective length of query: 252
effective length of database: 8,646,696
effective search space: 2178967392
effective search space used: 2178967392
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0042.15


