

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0042.1
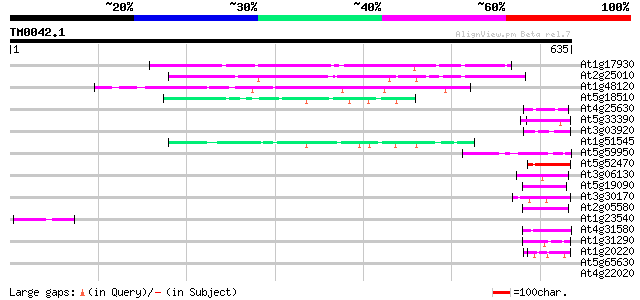
(635 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g17930 unknown protein 130 2e-30
At2g25010 unknown protein 118 1e-26
At1g48120 serine/threonine phosphatase PP7, putative 105 1e-22
At5g18510 putative protein 58 1e-08
At4g25630 fibrillarin - like protein 50 3e-06
At5g33390 unknown protein (At5g33390) 49 7e-06
At3g03920 putative GAR1 protein 48 1e-05
At1g51545 unknown protein 45 9e-05
At5g59950 transcriptional coactivator - like protein 45 2e-04
At5g52470 fibrillarin homolog (Fbr1) 45 2e-04
At3g06130 unknown protein 45 2e-04
At5g19090 unknown protein 44 3e-04
At3g30170 unknown protein 44 3e-04
At2g05580 unknown protein 44 3e-04
At1g23540 putative serine/threonine protein kinase 44 4e-04
At4g31580 splicing factor 9G8-like SR protein / SRZ-22 43 5e-04
At1g31290 hypothetical protein 43 5e-04
At1g20220 unknown protein 43 6e-04
At5g65630 unknown protein 42 0.001
At4g22020 glycine-rich protein 42 0.001
>At1g17930 unknown protein
Length = 478
Score = 130 bits (328), Expect = 2e-30
Identities = 123/425 (28%), Positives = 197/425 (45%), Gaps = 34/425 (8%)
Query: 159 LIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMSFGEMTITLDDVSALLHLPMG 218
L+E+ G + LI ALVERW ET++FH GEMTITLD+VS +L L +
Sbjct: 45 LVEKAGFGWFRLVGSISLNNSLISALVERWRRETNTFHFPCGEMTITLDEVSLILGLAVD 104
Query: 219 SRFYTPGRGERDECAALCAQLMGGSVGIYEAEFDTNR--SQTIRFGVLQTRYEAALAEHR 276
+ + + ++ + +C +L+G + + E NR ++ ++ + A + E
Sbjct: 105 GKPVVGVKEKDEDPSQVCLRLLG---KLPKGELSGNRVTAKWLKESFAECPKGATMKEIE 161
Query: 277 YEDAARIWLVNQLGATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQLSR 336
Y R +L+ +G+T+FA+ + Y I + ED + EYAWGA ALA LY Q+
Sbjct: 162 YH--TRAYLIYIVGSTIFATTDPSKISVDYLI-LFEDFEKAGEYAWGAAALAFLYRQIGN 218
Query: 337 ASRRGTAQMGGFSSLLLEWVYEYLS-DRVIIRRADPEYSQDQPRARRWAMSRVGHAGLD- 394
AS+R + +GG +LL W Y +L+ DR +R ++ P A W + + D
Sbjct: 219 ASQRSQSIIGGCLTLLQCWSYFHLNIDRP--KRTTRQF----PLALLWKGRQQSRSKNDL 272
Query: 395 -ERRVMLDELTVDDIIWTPFEDHRAHRPRD--PRAMYSRYIRSPFG-RVVRRHLPERVLR 450
+ R LD+L ++ W PFE P+ + R G +VV H P+R ++
Sbjct: 273 FKYRKALDDLDPSNVSWCPFEGDLDIVPQSFKDNLLLGRSRTKLIGPKVVEWHFPDRCMK 332
Query: 451 QFGFIQ----DVPRHPSEIQTSGSLAETADAAFAEFAPHLRPQGIPATYPGEAVE-DYMR 505
QFG Q +VP +E L E + A E+ R + I G E +YM+
Sbjct: 333 QFGLCQVIPGEVPPRKNEKNHDEDLLEDMNTADEEWM--RRRENIVENGGGNGDESEYMQ 390
Query: 506 WYNAVSHRFIIPDDRREEFSAVTVMRRAVDLLEQSLEVSDAPAEGTH--SRSLTERALDL 563
W+N+++ +P R+ +M +L+ S E H R E A+ +
Sbjct: 391 WFNSIT----VPKLHRDTSLEADIMNVQAAILQFDEVASTLSLEDLHPEEREAIEEAV-M 445
Query: 564 IRSNA 568
SNA
Sbjct: 446 SMSNA 450
>At2g25010 unknown protein
Length = 509
Score = 118 bits (295), Expect = 1e-26
Identities = 127/429 (29%), Positives = 184/429 (42%), Gaps = 46/429 (10%)
Query: 180 LILALVERWHEETSSFHMSFGEMTITLDDVSALLHLPMGSRFYTPGRGERDECAALCAQL 239
LI ALVERW ET++FH+ GEMTITLD+V+ +L L + + + +C +L
Sbjct: 75 LISALVERWRRETNTFHLPLGEMTITLDEVALVLGLEIDGDPIVGSKVGDEVAMDMCGRL 134
Query: 240 MGGSVGIYEAEFDTNRSQTIRFGVLQTRYEAALAEHRYEDA---ARIWLVNQLGATLFAS 296
+G E + +R ++ L+ + + ++ R +L+ +G+T+FA+
Sbjct: 135 LGKLPSAANKEVNCSR---VKLNWLKRTFSECPEDASFDVVKCHTRAYLLYLIGSTIFAT 191
Query: 297 KSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQLSRASRRGTAQMGGFSSLLLEWV 356
G +V ++ + ED + YAWGA ALA LY L AS + + + G +LL W
Sbjct: 192 TDGD-KVSVKYLPLFEDFDQAGRYAWGAAALACLYRALGNASLKSQSNICGCLTLLQCWS 250
Query: 357 YEYLSDRVIIRRADPEYSQD-QPRARRW-AMSRVGHAGLDERRVMLDELTVDDIIWTPFE 414
Y +L I R PE S+ P A W L E R LD+L I W P+E
Sbjct: 251 YFHLD----IGR--PEKSEACFPLALLWKGKGSRSKTDLSEYRRELDDLDPSKITWCPYE 304
Query: 415 DHRAHRPRDPRAMY----SRYIRSPFGRVVRRHLPERVLRQFGFIQDVP----------R 460
P +A S+ F + + H P+R LRQFG Q +P R
Sbjct: 305 RFENLIPPHIKAKLILGRSKTTLVCFEK-IELHFPDRCLRQFGKRQPIPLKVKRRDRKNR 363
Query: 461 HPSEIQTSGSLAETADAAFAEFAPHLRPQGIPATYPGEAVED--YMRWYNAVSHRFIIPD 518
++ TS SL A +AE H+ + + G V+D YM WY +S I
Sbjct: 364 RLDDLDTSMSL---ACEEWAERGDHI----VDSPGGGNVVDDGAYMEWYARIS----ITK 412
Query: 519 DRREEFSAVTVMRRAVDLLEQSLEVSDAPAE--GTHSRSLTERALD-LIRSNAFIGTQGV 575
RE F VM + E S E R + E D S F G Q V
Sbjct: 413 LYREAFLESQVMNMIACMREFEEAASGIALERLSPAEREVMESVKDTFANSLTFGGWQEV 472
Query: 576 AFAAVRGAR 584
A + G R
Sbjct: 473 AVNSGYGKR 481
>At1g48120 serine/threonine phosphatase PP7, putative
Length = 1338
Score = 105 bits (261), Expect = 1e-22
Identities = 118/456 (25%), Positives = 187/456 (40%), Gaps = 54/456 (11%)
Query: 97 GPVELSLLTHYADHKAPWAWHALLRTDERYVDRRQL--KVATAGGKVWNLACDGDSDSHR 154
GPV+ S+L +H++ W E V R+L + G + W L
Sbjct: 14 GPVDQSILVWQHEHRSAAIW-------EDEVPPRELTCRHKLLGMRDWPL--------DP 58
Query: 155 RVRELIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMSFGEMTITLDDVSALLH 214
V + + + GL+ + ++ D LI ALVERW ET +FH+ GE+T+TL DV+ LL
Sbjct: 59 LVCQKLIEFGLYGVYKVAFIQLDYALITALVERWRPETHTFHLPAGEITVTLQDVNILLG 118
Query: 215 LPMGSRFYTPGRGERDECAALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQTRYEAALA- 273
L + T + A LC L+G G + + L+ + A
Sbjct: 119 LRVDGPAVT--GSTKYNWADLCEDLLGHRPGPKDL-----HGSHVSLAWLRENFRNLPAD 171
Query: 274 --EHRYEDAARIWLVNQLGATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLY 331
E + R +++ + L+ KS + + ++ +L D V + +WG+ LA LY
Sbjct: 172 PDEVTLKCHTRAFVLALMSGFLYGDKS-KHDVALTFLPLLRDFDEVAKLSWGSATLALLY 230
Query: 332 DQLSRASRRGTAQMGGFSSLLLEWVYEYLSDRVIIRRADPEYSQ--------DQPRARRW 383
+L RAS+R + + G LL W +E L R D S P R
Sbjct: 231 RELCRASKRTVSTICGPLVLLQLWAWERLHVGRPGRLKDVGASYMDGIDGPLPDPLGCRA 290
Query: 384 AMSRVGH--AGLDERRVMLDELTVDDIIWTPF-EDHRAHRPR---DPRAMYSRYIRSPFG 437
++S + GLD R D+ + +IW P+ D A P ++
Sbjct: 291 SLSHKENPRGGLDFYRDQFDQQKDEQVIWQPYTPDLLAKIPLICVSGENIWRTVAPLICF 350
Query: 438 RVVRRHLPERVLRQFGFIQDVPRHPSEIQTSGSLAETADAAFAEFAPHLRPQGI------ 491
VV H P+RVLRQFG Q +P + ++ + + + A H R G+
Sbjct: 351 DVVEWHRPDRVLRQFGLHQTIPAPCDNEKALHAIDKRGKSEYDWSARHSRHIGLWEARVS 410
Query: 492 ------PATYPGEAVEDYMRWYNAVSHRFIIPDDRR 521
P P + + YM WY ++ R I P + R
Sbjct: 411 SVVSGEPECSPMDYNDPYMEWYRRITRRIISPMNER 446
>At5g18510 putative protein
Length = 702
Score = 58.2 bits (139), Expect = 1e-08
Identities = 85/311 (27%), Positives = 123/311 (39%), Gaps = 48/311 (15%)
Query: 175 VTDAGLILALVERWHEETSSFHMSFGEMTITLDDVSALLHLP-MGSRFYTPGR-GERDEC 232
+ D IL++ E+W ET SF +GE TITL+DV LL +GS ++P E +
Sbjct: 92 IKDTSSILSIAEKWCSETKSFIFPWGEATITLEDVMVLLGFSVLGSPVFSPLECSEMRDS 151
Query: 233 AALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQTRYEAALAEHRYEDAARIWLVNQLGAT 292
A ++ S+G +A+ + RS T F EH +LV L
Sbjct: 152 AEKLEKVRRDSLG--KAKRVSQRSWTSSF-----MGRGGQMEH------EAFLVLWLSLF 198
Query: 293 LFASK-SGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQL------SRASRRGTAQM 345
+F K T V I + L R A LA LY L SR G +
Sbjct: 199 VFPGKFCRSISTNV--IPIAVRLARGERIALAPAVLAFLYKDLDRICDFSRGKCAGKVNL 256
Query: 346 GGFSSLLLEWVYEYLSDRVIIRRADPEYSQDQPRARRW----AMSRVGHAGLD--ERRVM 399
L+ W +E S+ IR E + +PR +W +S+ D E R
Sbjct: 257 KSLFKLVQVWTWERFSN---IRPKAKEIPKGEPRIAQWDGLQQISKNVKLSFDVFEWRPY 313
Query: 400 LDELT--------VDDIIWTPFEDHRAHRPRDPRAMYSRYIRSPF---GRVVRRHLPERV 448
L VD+ +W +D D A ++R ++ + V + P RV
Sbjct: 314 TKPLKNWNPLRFYVDEAMWLTVDD----SVDDAFASFARCVKVSYLAGNGFVEDYFPYRV 369
Query: 449 LRQFGFIQDVP 459
RQFG QD+P
Sbjct: 370 ARQFGLSQDLP 380
>At4g25630 fibrillarin - like protein
Length = 320
Score = 50.4 bits (119), Expect = 3e-06
Identities = 29/51 (56%), Positives = 29/51 (56%), Gaps = 5/51 (9%)
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRG 632
G R GGRG G RG RGGR RG G P GARGG RGP GR G G
Sbjct: 32 GGRGGGGRGGG---RGFSDRGGRGRGRGPPRGGARGG--RGPAGRGGMKGG 77
Score = 44.7 bits (104), Expect = 2e-04
Identities = 26/51 (50%), Positives = 29/51 (55%), Gaps = 3/51 (5%)
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRG 632
G +GGRGRG + GGRG GG + G G G GGR RG RG GRG
Sbjct: 9 GGGFSGGRGRGGYS-GGRGDGGFSGGRGGGGRG--GGRGFSDRGGRGRGRG 56
Score = 39.7 bits (91), Expect = 0.005
Identities = 25/50 (50%), Positives = 27/50 (54%), Gaps = 5/50 (10%)
Query: 590 GRGDRARGGRGRGG--RARGEGAPAEGARGGRARGPRG---RRGAGRGRG 634
G G GGRGRGG RG+G + G GG G RG R G GRGRG
Sbjct: 7 GSGGGFSGGRGRGGYSGGRGDGGFSGGRGGGGRGGGRGFSDRGGRGRGRG 56
Score = 28.9 bits (63), Expect = 9.2
Identities = 17/34 (50%), Positives = 18/34 (52%), Gaps = 5/34 (14%)
Query: 581 RGARAAGGRGRG-----DRARGGRGRGGRARGEG 609
RG GGRGRG ARGGRG GR +G
Sbjct: 43 RGFSDRGGRGRGRGPPRGGARGGRGPAGRGGMKG 76
>At5g33390 unknown protein (At5g33390)
Length = 118
Score = 49.3 bits (116), Expect = 7e-06
Identities = 29/61 (47%), Positives = 31/61 (50%), Gaps = 5/61 (8%)
Query: 579 AVRGARAAGGRGRGDRARGGRGRGGRA--RGEGAPAEGARGGRARG---PRGRRGAGRGR 633
+V G +GG G G GG G GGRA RG G G GGR G RG G GRGR
Sbjct: 18 SVSGGSGSGGSGSGGSGSGGSGSGGRANGRGNGGRGSGRGGGRGDGRGDGRGIGGGGRGR 77
Query: 634 G 634
G
Sbjct: 78 G 78
Score = 43.1 bits (100), Expect = 5e-04
Identities = 23/49 (46%), Positives = 24/49 (48%)
Query: 586 AGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
A GRG G R G G G RG+G G GR R P G GRGRG
Sbjct: 44 ANGRGNGGRGSGRGGGRGDGRGDGRGIGGGGRGRGRIPAAFMGRGRGRG 92
Score = 41.6 bits (96), Expect = 0.001
Identities = 22/53 (41%), Positives = 23/53 (42%), Gaps = 1/53 (1%)
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
G + G G G GG G GG G GGRA G RG G G GRG
Sbjct: 6 GGSGSSGSGSGGSVSGGSGSGGSGSGGSGSGGSGSGGRANG-RGNGGRGSGRG 57
Score = 38.9 bits (89), Expect = 0.009
Identities = 28/60 (46%), Positives = 30/60 (49%), Gaps = 3/60 (5%)
Query: 571 GTQGVAFAAVRGARAAG-GRGRGDRARGGRGRGGRARGEG-APAEGARGGRARGPRGRRG 628
G+ G A G R +G G GRGD GRG GG RG G PA GR RG RG G
Sbjct: 39 GSGGRANGRGNGGRGSGRGGGRGDGRGDGRGIGGGGRGRGRIPAAFMGRGRGRG-RGNHG 97
Score = 37.0 bits (84), Expect = 0.034
Identities = 22/50 (44%), Positives = 25/50 (50%), Gaps = 2/50 (4%)
Query: 586 AGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGR-RGAGRGRG 634
+GG G GG G GG G G+ + G GR G RG RG GRG G
Sbjct: 15 SGGSVSGGSGSGGSGSGGSGSG-GSGSGGRANGRGNGGRGSGRGGGRGDG 63
>At3g03920 putative GAR1 protein
Length = 202
Score = 48.1 bits (113), Expect = 1e-05
Identities = 30/54 (55%), Positives = 32/54 (58%), Gaps = 7/54 (12%)
Query: 582 GARAAGGRGRGDRARGGRGRGGR-ARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
G R GRGRGD GRGRGG +RG GAP RGGR G RG+ RGRG
Sbjct: 153 GGRGGAGRGRGDSR--GRGRGGSFSRGRGAP----RGGRFPPRGGSRGSFRGRG 200
Score = 33.1 bits (74), Expect = 0.49
Identities = 20/37 (54%), Positives = 20/37 (54%), Gaps = 1/37 (2%)
Query: 597 GGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGR 633
G RGRGGR G G G GGR G GR G G GR
Sbjct: 10 GFRGRGGRDGGGGGRFGGG-GGRFGGGGGRFGGGGGR 45
Score = 30.8 bits (68), Expect = 2.4
Identities = 22/55 (40%), Positives = 23/55 (41%), Gaps = 6/55 (10%)
Query: 580 VRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
+RG GRG D GGR GG R G GGR G GR G R G
Sbjct: 5 MRGGGGFRGRGGRDGGGGGRFGGGGGR------FGGGGGRFGGGGGRFGGFRDEG 53
Score = 30.0 bits (66), Expect = 4.1
Identities = 17/41 (41%), Positives = 20/41 (48%), Gaps = 7/41 (17%)
Query: 594 RARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
++ GGRG GR RG+ GR RG RG G RG
Sbjct: 150 QSTGGRGGAGRGRGDSR-------GRGRGGSFSRGRGAPRG 183
>At1g51545 unknown protein
Length = 629
Score = 45.4 bits (106), Expect = 9e-05
Identities = 89/377 (23%), Positives = 137/377 (35%), Gaps = 59/377 (15%)
Query: 180 LILALVERWHEETSSFHMSFGEMTITLDDVSALLHLPM-GSRFYTPGRGERDECAALCAQ 238
L+LALVE+W ET SF +GE TITL+DV LL + GS + P L +
Sbjct: 31 LLLALVEKWCPETKSFLFPWGEATITLEDVLVLLGFSVQGSPVFAP----------LESS 80
Query: 239 LMGGSVGIYEAEFDTNRSQ--TIRFGVLQTRYEAALAEHRYEDAARIWLVNQLGATLFAS 296
M SV E NR Q +R + + + + +E WL +Q
Sbjct: 81 EMRDSVEKLEKARLENRGQDGLVRQNLWVSSFLGRGDQMEHEAFLAFWL-SQFVFPDMCR 139
Query: 297 KSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQL------SRASRRGTAQMGGFSS 350
+S + + M L R A+ LA LY L +R +
Sbjct: 140 RS----ISTKVLPMAVRLARGERIAFAPAVLARLYRDLGQIQASAREKSTPNVTLKSLFK 195
Query: 351 LLLEWVYEYLSDRVIIRRADPEYSQDQPRARRWAMSRVGHAGLD----ERRVMLDELTV- 405
L+ W +E R P + +PR RW + L+ + R L +
Sbjct: 196 LVQLWAWERFKSTSPKARVIP---KGEPRISRWHSQTSKNVRLNLVDFDWRPYTKPLQIW 252
Query: 406 -------DDIIWTPFEDHRAHRPRDPRAMYSRYIRSP---FGRVVRRHLPERVLRQFGFI 455
++ +W +D+ D ++R +R +V + P RV QFG
Sbjct: 253 NPPRFYPEEAMWMTVDDNL----DDGFVSFARCMRVSQLVGVGIVEDYYPNRVAMQFGLA 308
Query: 456 QDVP----RHPS--EIQTSGSLAETADAAFAEFAPHLRPQGIPATYPGEAVEDYMRWYNA 509
QD+P H S E + ++ D P + T E D+ W +
Sbjct: 309 QDLPGLVTDHSSFTEKEAWDGYNKSLDGLMLYI-----PSRVATTSVTERYRDW--WLKS 361
Query: 510 VSHRFIIPDDRREEFSA 526
+S F+ + E F A
Sbjct: 362 ISKFFLDSSESTETFDA 378
>At5g59950 transcriptional coactivator - like protein
Length = 244
Score = 44.7 bits (104), Expect = 2e-04
Identities = 39/125 (31%), Positives = 56/125 (44%), Gaps = 20/125 (16%)
Query: 513 RFIIPDDR--REEFSAVTVMRRAVDLLEQSLEVSDAPAEGTHSRSLTERALDLIRSNAFI 570
R+ + DR R + +A V R D L + +D +G + ++++ +N
Sbjct: 116 RYTVHFDRSGRSKGTAEVVYSRRGDALAAVKKYNDVQLDGKPMK------IEIVGTN--- 166
Query: 571 GTQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAG 630
+ AA R A G G RGG+GRGG+ RG G GGR G RGRR G
Sbjct: 167 ----LQTAAAPSGRPANGNSNGAPWRGGQGRGGQQRGGGR----GGGGRGGGGRGRR-PG 217
Query: 631 RGRGE 635
+G E
Sbjct: 218 KGPAE 222
>At5g52470 fibrillarin homolog (Fbr1)
Length = 308
Score = 44.7 bits (104), Expect = 2e-04
Identities = 31/51 (60%), Positives = 31/51 (60%), Gaps = 4/51 (7%)
Query: 587 GGRGRGDRARGGRGRGGRARGEGAPAEGARGG-RAR-GPRGR-RGAGRGRG 634
GGRG G RGGR GGR G G G R G R R GPRGR RGA RGRG
Sbjct: 7 GGRGGGG-FRGGRDGGGRGFGGGRSFGGGRSGDRGRSGPRGRGRGAPRGRG 56
Score = 35.8 bits (81), Expect = 0.075
Identities = 29/66 (43%), Positives = 30/66 (44%), Gaps = 12/66 (18%)
Query: 572 TQGVAFAAVRGARAAGGRG-RGDRARGG-----RGRGG-RARGEGAPAEGARGGRARGPR 624
T G RG R GGRG G R+ GG RGR G R RG GAP GR PR
Sbjct: 6 TGGRGGGGFRGGRDGGGRGFGGGRSFGGGRSGDRGRSGPRGRGRGAPR-----GRGGPPR 60
Query: 625 GRRGAG 630
G G
Sbjct: 61 GGMKGG 66
>At3g06130 unknown protein
Length = 473
Score = 44.7 bits (104), Expect = 2e-04
Identities = 26/64 (40%), Positives = 32/64 (49%), Gaps = 5/64 (7%)
Query: 574 GVAFAAVRGARAAGGRGRGDRARGGRGR----GGRARGEGAPAEGARGGRARGPR-GRRG 628
G A G A GG+G G A+GG G GG+ G G P +G GG GP G++G
Sbjct: 257 GKGAPAAGGGGAGGGKGAGGGAKGGPGNQNQGGGKNGGGGHPQDGKNGGGGGGPNAGKKG 316
Query: 629 AGRG 632
G G
Sbjct: 317 NGGG 320
Score = 35.0 bits (79), Expect = 0.13
Identities = 22/54 (40%), Positives = 27/54 (49%), Gaps = 6/54 (11%)
Query: 581 RGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
+GA AAGG G G GG+G GG A+G P +GG G G G+ G
Sbjct: 258 KGAPAAGGGGAG----GGKGAGGGAKG--GPGNQNQGGGKNGGGGHPQDGKNGG 305
>At5g19090 unknown protein
Length = 587
Score = 43.9 bits (102), Expect = 3e-04
Identities = 19/50 (38%), Positives = 26/50 (52%)
Query: 581 RGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAG 630
+G GG G++ +GG G+ G G G P +G GG GP G +G G
Sbjct: 323 KGGPGGGGGNMGNQNQGGGGKNGGKGGGGHPLDGKMGGGGGGPNGNKGGG 372
Score = 30.8 bits (68), Expect = 2.4
Identities = 16/39 (41%), Positives = 20/39 (51%), Gaps = 1/39 (2%)
Query: 597 GGRG-RGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
GG G GG+ G+G P GG GP G++G G G
Sbjct: 292 GGPGPAGGKIEGKGMPFPVQMGGGGGGPGGKKGGPGGGG 330
Score = 29.6 bits (65), Expect = 5.4
Identities = 19/64 (29%), Positives = 22/64 (33%), Gaps = 1/64 (1%)
Query: 570 IGTQGVAFAAVRGARAAGGRG-RGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRG 628
+G Q G + GG G GG G G G G G G +G G G
Sbjct: 333 MGNQNQGGGGKNGGKGGGGHPLDGKMGGGGGGPNGNKGGGGVQMNGGPNGGKKGGGGGGG 392
Query: 629 AGRG 632
G G
Sbjct: 393 GGGG 396
Score = 29.3 bits (64), Expect = 7.0
Identities = 16/38 (42%), Positives = 19/38 (49%), Gaps = 3/38 (7%)
Query: 597 GGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
GG+G GG G G PA +G + G G G G G G
Sbjct: 102 GGKGGGG---GGGGPANNNKGQKIGGGGGGGGGGGGGG 136
>At3g30170 unknown protein
Length = 995
Score = 43.9 bits (102), Expect = 3e-04
Identities = 32/75 (42%), Positives = 41/75 (54%), Gaps = 11/75 (14%)
Query: 570 IGTQGVAFAAVRGARAAG----GRGRGDRARGGRGRGGRAR-----GEGAPAEGAR-GGR 619
+G+QG + AA RG + G GRG G + GRG A+ G G A+G R GR
Sbjct: 841 LGSQGPS-AAGRGLGSQGPSAAGRGLGSQEPSAAGRGRGAQGPSAAGRGRVAQGPRAAGR 899
Query: 620 ARGPRGRRGAGRGRG 634
RG +G+R AGRG G
Sbjct: 900 GRGAQGQRAAGRGCG 914
Score = 39.3 bits (90), Expect = 0.007
Identities = 33/91 (36%), Positives = 41/91 (44%), Gaps = 23/91 (25%)
Query: 566 SNAFIGTQGVAFAAVRGARAAG----GRGRGDRARGGRGRG---------GRARGEGAPA 612
S A + +QG + AA RG + G GRG G + GRG GR G P+
Sbjct: 813 SQADLESQGPS-AAGRGLGSQGPSAAGRGLGSQGPSAAGRGLGSQGPSAAGRGLGSQEPS 871
Query: 613 EGARGGRARGP---------RGRRGAGRGRG 634
RG A+GP +G R AGRGRG
Sbjct: 872 AAGRGRGAQGPSAAGRGRVAQGPRAAGRGRG 902
Score = 37.4 bits (85), Expect = 0.026
Identities = 23/54 (42%), Positives = 29/54 (53%), Gaps = 4/54 (7%)
Query: 581 RGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
RGA+ GRG A+G R G RG GA + A GR G +G+ +G GRG
Sbjct: 877 RGAQGPSAAGRGRVAQGPRAAG---RGRGAQGQRA-AGRGCGAQGQTASGGGRG 926
Score = 36.6 bits (83), Expect = 0.044
Identities = 27/74 (36%), Positives = 38/74 (50%), Gaps = 10/74 (13%)
Query: 571 GTQGVAFAA----VRGARAAGGRGRGDRARGGRGRGGRARGE-----GAPAEGARGGRAR 621
G QG + A +G RAAG RGRG + + GRG A+G+ G A+ GG ++
Sbjct: 878 GAQGPSAAGRGRVAQGPRAAG-RGRGAQGQRAAGRGCGAQGQTASGGGRGAQRQAGGPSQ 936
Query: 622 GPRGRRGAGRGRGE 635
GP+ R G + E
Sbjct: 937 GPQPYRRHGSAQAE 950
>At2g05580 unknown protein
Length = 302
Score = 43.9 bits (102), Expect = 3e-04
Identities = 20/52 (38%), Positives = 29/52 (55%)
Query: 581 RGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRG 632
+G + GG G+G + GG G+GG+ G G G +GG G G++G G G
Sbjct: 106 QGGQKGGGGGQGGQKGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGG 157
Score = 42.0 bits (97), Expect = 0.001
Identities = 22/62 (35%), Positives = 32/62 (51%), Gaps = 1/62 (1%)
Query: 571 GTQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAG 630
G +G G + GG G+G + GG G+GG+ +G G G +GG G G++G G
Sbjct: 86 GQKGGGGGGQGGQKGGGGGGQGGQKGGGGGQGGQ-KGGGGGQGGQKGGGGGGQGGQKGGG 144
Query: 631 RG 632
G
Sbjct: 145 GG 146
Score = 40.8 bits (94), Expect = 0.002
Identities = 22/55 (40%), Positives = 30/55 (54%), Gaps = 2/55 (3%)
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPR-GRRGAGRGRGE 635
G + GG+G G + GGRG+GG G G G +GG G + G +G G G G+
Sbjct: 179 GQKGGGGQG-GQKGGGGRGQGGMKGGGGGGQGGHKGGGGGGGQGGHKGGGGGGGQ 232
Score = 40.0 bits (92), Expect = 0.004
Identities = 19/54 (35%), Positives = 27/54 (49%)
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRGE 635
G + GG G + GG G+GG+ G G G +GG G G++G G G+
Sbjct: 118 GQKGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGQGGQ 171
Score = 40.0 bits (92), Expect = 0.004
Identities = 20/49 (40%), Positives = 28/49 (56%), Gaps = 1/49 (2%)
Query: 587 GGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRGE 635
GG G+G + +GG G+GG+ G G G +GG G G +G G G G+
Sbjct: 173 GGGGQGGQ-KGGGGQGGQKGGGGRGQGGMKGGGGGGQGGHKGGGGGGGQ 220
Score = 39.3 bits (90), Expect = 0.007
Identities = 22/48 (45%), Positives = 24/48 (49%), Gaps = 1/48 (2%)
Query: 587 GGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
GG+G G GG G+GG G G G GGR G GR G G G G
Sbjct: 241 GGQGGGGHKGGGGGQGGHKGGGGGGHVGG-GGRGGGGGGRGGGGSGGG 287
Score = 39.3 bits (90), Expect = 0.007
Identities = 18/47 (38%), Positives = 26/47 (55%)
Query: 588 GRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
GRG+G + GG G G +G G +G + G G G++G G G+G
Sbjct: 81 GRGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGGQGGQKGGGGGQG 127
Score = 38.5 bits (88), Expect = 0.012
Identities = 23/69 (33%), Positives = 33/69 (47%), Gaps = 8/69 (11%)
Query: 571 GTQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAE-------GARGGRARGP 623
G +G G + GG G+G + +GG G+GG+ G G + G +GG RG
Sbjct: 139 GQKGGGGGGQGGQKGGGGGGQGGQ-KGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGRGQ 197
Query: 624 RGRRGAGRG 632
G +G G G
Sbjct: 198 GGMKGGGGG 206
Score = 38.5 bits (88), Expect = 0.012
Identities = 24/55 (43%), Positives = 26/55 (46%), Gaps = 5/55 (9%)
Query: 582 GARAAGGRGRGDRARGGRGR--GGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
G GG G+G GG G GG RG G G RGG G G G+GRG G
Sbjct: 246 GGHKGGGGGQGGHKGGGGGGHVGGGGRGGGG---GGRGGGGSGGGGGGGSGRGGG 297
Score = 38.5 bits (88), Expect = 0.012
Identities = 22/59 (37%), Positives = 27/59 (45%), Gaps = 6/59 (10%)
Query: 582 GARAAGGRGRGDRARGGRGR------GGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
G + GGRG+G GG G GG G+G G GG G +G G G+G G
Sbjct: 188 GQKGGGGRGQGGMKGGGGGGQGGHKGGGGGGGQGGHKGGGGGGGQGGHKGGGGGGQGGG 246
Score = 38.1 bits (87), Expect = 0.015
Identities = 23/69 (33%), Positives = 31/69 (44%), Gaps = 5/69 (7%)
Query: 571 GTQGVAFAAVRGARAAGGRGRGDR--ARGGRGRGGRARGEGAPAEGAR---GGRARGPRG 625
G +G G + GG G+G GG G+GG G G +G GG +G G
Sbjct: 188 GQKGGGGRGQGGMKGGGGGGQGGHKGGGGGGGQGGHKGGGGGGGQGGHKGGGGGGQGGGG 247
Query: 626 RRGAGRGRG 634
+G G G+G
Sbjct: 248 HKGGGGGQG 256
Score = 38.1 bits (87), Expect = 0.015
Identities = 20/42 (47%), Positives = 20/42 (47%)
Query: 587 GGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRG 628
GG G G GGRG GG RG G G GG RG G G
Sbjct: 260 GGGGGGHVGGGGRGGGGGGRGGGGSGGGGGGGSGRGGGGGGG 301
Score = 37.7 bits (86), Expect = 0.020
Identities = 24/64 (37%), Positives = 27/64 (41%), Gaps = 1/64 (1%)
Query: 571 GTQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAG 630
G QG G G +G G +GG G G G+G G GG G GR G G
Sbjct: 218 GGQGGHKGGGGGGGQGGHKGGGGGGQGGGGHKGGGGGQGGHKGGGGGGHV-GGGGRGGGG 276
Query: 631 RGRG 634
GRG
Sbjct: 277 GGRG 280
Score = 37.4 bits (85), Expect = 0.026
Identities = 24/76 (31%), Positives = 32/76 (41%), Gaps = 1/76 (1%)
Query: 557 TERALDLIRSNAFIGTQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGAR 616
T+ LD QG G + GG G+G + GG G G +G G G +
Sbjct: 61 TDEVLDGEEEEESKNNQGGIGRGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGGQGGQK 120
Query: 617 GGRARGPRGRRGAGRG 632
GG G G++G G G
Sbjct: 121 GG-GGGQGGQKGGGGG 135
Score = 35.4 bits (80), Expect = 0.098
Identities = 21/64 (32%), Positives = 25/64 (38%)
Query: 571 GTQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAG 630
G +G +G GG G G G G GG+ G G +GG G G G
Sbjct: 210 GHKGGGGGGGQGGHKGGGGGGGQGGHKGGGGGGQGGGGHKGGGGGQGGHKGGGGGGHVGG 269
Query: 631 RGRG 634
GRG
Sbjct: 270 GGRG 273
Score = 35.4 bits (80), Expect = 0.098
Identities = 22/71 (30%), Positives = 30/71 (41%), Gaps = 7/71 (9%)
Query: 571 GTQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRG----- 625
G +G G + GG G+G + GG G G +G G GG G +G
Sbjct: 128 GQKGGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGQG 187
Query: 626 --RRGAGRGRG 634
+ G GRG+G
Sbjct: 188 GQKGGGGRGQG 198
Score = 35.0 bits (79), Expect = 0.13
Identities = 20/64 (31%), Positives = 28/64 (43%)
Query: 571 GTQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAG 630
G +G G + GG G +GG G GG+ +G G GG +G G +G
Sbjct: 199 GMKGGGGGGQGGHKGGGGGGGQGGHKGGGGGGGQGGHKGGGGGGQGGGGHKGGGGGQGGH 258
Query: 631 RGRG 634
+G G
Sbjct: 259 KGGG 262
Score = 33.1 bits (74), Expect = 0.49
Identities = 19/59 (32%), Positives = 27/59 (45%), Gaps = 5/59 (8%)
Query: 582 GARAAGGRGRGDRARGGRG-----RGGRARGEGAPAEGARGGRARGPRGRRGAGRGRGE 635
G + GG G+G + GG G +GG G+G G G +G G+ G G G+
Sbjct: 128 GQKGGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGQ 186
Score = 33.1 bits (74), Expect = 0.49
Identities = 17/42 (40%), Positives = 18/42 (42%)
Query: 581 RGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARG 622
+G G G G R GG GRGG G G RGG G
Sbjct: 259 KGGGGGGHVGGGGRGGGGGGRGGGGSGGGGGGGSGRGGGGGG 300
Score = 31.2 bits (69), Expect = 1.8
Identities = 16/37 (43%), Positives = 16/37 (43%)
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGG 618
G GGRG G RGG G GG G G GG
Sbjct: 265 GHVGGGGRGGGGGGRGGGGSGGGGGGGSGRGGGGGGG 301
>At1g23540 putative serine/threonine protein kinase
Length = 731
Score = 43.5 bits (101), Expect = 4e-04
Identities = 25/69 (36%), Positives = 30/69 (43%), Gaps = 6/69 (8%)
Query: 5 PLVDYPPSPTVEIPTIDSPPSPMVESSGEESSGEESSGEESSGEASSGMGGSDEDSIPPP 64
PL D PP P+ P +DS PSP +S E S E S E D+ PPP
Sbjct: 73 PLTDSPPPPSDSSPPVDSTPSPPPPTSNESPSPPEDS------ETPPAPPNESNDNNPPP 126
Query: 65 TVDADVLPP 73
+ D PP
Sbjct: 127 SQDLQSPPP 135
Score = 30.0 bits (66), Expect = 4.1
Identities = 23/84 (27%), Positives = 28/84 (32%), Gaps = 22/84 (26%)
Query: 10 PPSPTVEIPTIDSPPSPMVESSGEESSGEESSGEESSGEASSGMGGSDEDSIPPPTVDAD 69
PP P++ P DSPP P S +S+ S PPPT +
Sbjct: 65 PPLPSILPPLTDSPPPPSDSSPPVDST----------------------PSPPPPTSNES 102
Query: 70 VLPPEQGEQGAQGGEEDLIQRLPP 93
PPE E E PP
Sbjct: 103 PSPPEDSETPPAPPNESNDNNPPP 126
Score = 29.6 bits (65), Expect = 5.4
Identities = 23/90 (25%), Positives = 35/90 (38%), Gaps = 9/90 (10%)
Query: 3 EVPLVDYPPSPTVEIPTIDSPPSPMVESSGEESSGEESSGEESSGEASSGMGGSDEDSIP 62
E P + PP+P PT P SP+ ++ + SG +S A+ S ++P
Sbjct: 148 ESPPLQSPPAPPASDPTNSPPASPLDPTNPPPI---QPSGPATSPPANPNAPPSPFPTVP 204
Query: 63 P------PTVDADVLPPEQGEQGAQGGEED 86
P P V + P +G G D
Sbjct: 205 PKTPSSGPVVSPSLTSPSKGTPTPNQGNGD 234
>At4g31580 splicing factor 9G8-like SR protein / SRZ-22
Length = 200
Score = 43.1 bits (100), Expect = 5e-04
Identities = 28/56 (50%), Positives = 30/56 (53%), Gaps = 2/56 (3%)
Query: 581 RGARAAGGRGRGDRARGGRGRGGRARGEGAPAE-GARGGRARGPRGRRGAGRGRGE 635
RG R GGRG GDR GG GRGGR + E G G AR R R G GR R +
Sbjct: 72 RGERGGGGRG-GDRGGGGGGRGGRGGSDLKCYECGETGHFARECRNRGGTGRRRSK 126
>At1g31290 hypothetical protein
Length = 1194
Score = 43.1 bits (100), Expect = 5e-04
Identities = 27/56 (48%), Positives = 28/56 (49%), Gaps = 3/56 (5%)
Query: 581 RGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAG--RGRG 634
RG R GRGRG GRG GR G G +G RG RG RG G RGRG
Sbjct: 7 RGGRG-DGRGRGGGGDRGRGYSGRGDGRGRGGDGDRGYSGRGDGHGRGGGGDRGRG 61
Score = 42.4 bits (98), Expect = 8e-04
Identities = 31/63 (49%), Positives = 31/63 (49%), Gaps = 13/63 (20%)
Query: 581 RGARAAGGRGRGDRARG-GRGRGG--------RARGEGAPAEGARGGRARGPRGRRGAGR 631
RG G RGRG RG GRGRGG R G G G G R RG G RG GR
Sbjct: 14 RGRGGGGDRGRGYSGRGDGRGRGGDGDRGYSGRGDGHG---RGGGGDRGRGYSG-RGDGR 69
Query: 632 GRG 634
GRG
Sbjct: 70 GRG 72
Score = 40.0 bits (92), Expect = 0.004
Identities = 27/55 (49%), Positives = 28/55 (50%), Gaps = 4/55 (7%)
Query: 582 GARAAGGRGRGDRARGG--RGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
G R GRG G GG RGRG RG+G G G R RG G RG G GRG
Sbjct: 39 GDRGYSGRGDGHGRGGGGDRGRGYSGRGDGR-GRGGGGDRGRGYSG-RGDGHGRG 91
Score = 38.1 bits (87), Expect = 0.015
Identities = 28/66 (42%), Positives = 29/66 (43%), Gaps = 12/66 (18%)
Query: 581 RGARAAG-GRGRGDRARGGRGRGGRARGEGAPAEGARG--------GRARGPRGRRG--- 628
RG G G GRG GRG GR G G G RG G RG G RG
Sbjct: 41 RGYSGRGDGHGRGGGGDRGRGYSGRGDGRGRGGGGDRGRGYSGRGDGHGRGGGGDRGRGY 100
Query: 629 AGRGRG 634
+GRGRG
Sbjct: 101 SGRGRG 106
Score = 35.8 bits (81), Expect = 0.075
Identities = 24/54 (44%), Positives = 26/54 (47%), Gaps = 10/54 (18%)
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRG-AGRGRG 634
G R G GRG G RGRG RG+G R RG G RG +GRG G
Sbjct: 5 GYRGGRGDGRGRGGGGDRGRGYSGRGDG---------RGRGGDGDRGYSGRGDG 49
Score = 32.7 bits (73), Expect = 0.63
Identities = 22/39 (56%), Positives = 22/39 (56%), Gaps = 5/39 (12%)
Query: 596 RGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
RGG RGGR G G G G R RG G RG GRGRG
Sbjct: 3 RGGY-RGGRGDGRG---RGGGGDRGRGYSG-RGDGRGRG 36
>At1g20220 unknown protein
Length = 315
Score = 42.7 bits (99), Expect = 6e-04
Identities = 29/55 (52%), Positives = 29/55 (52%), Gaps = 9/55 (16%)
Query: 587 GGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGR-------RGAGRGRG 634
G RGRG R RG RGRGGR G A E GGR RG RG GRGRG
Sbjct: 153 GRRGRGGRGRG-RGRGGRGNGP-ANVEYDDGGRGRGGRGNGYVNNEYDDGGRGRG 205
Score = 42.4 bits (98), Expect = 8e-04
Identities = 30/66 (45%), Positives = 33/66 (49%), Gaps = 14/66 (21%)
Query: 582 GARAAGGRGRG----DRARGGRGRGGRARG-------EGAPAEGARGGRARGPRGRRG-- 628
G R GGRG G + GGRGRGGR G +G + GR RG RGR G
Sbjct: 181 GGRGRGGRGNGYVNNEYDDGGRGRGGRGSGYVNNEYNDGGMEQDRSYGRGRG-RGRGGGR 239
Query: 629 AGRGRG 634
GRGRG
Sbjct: 240 GGRGRG 245
Score = 40.0 bits (92), Expect = 0.004
Identities = 26/55 (47%), Positives = 27/55 (48%), Gaps = 16/55 (29%)
Query: 588 GRGRGDRARGGR-GRGGRARGEGAPAEGARGGRARGP---------RGRRGAGRG 632
G+ R RGGR GRGGR RG G RGGR GP RGR G G G
Sbjct: 143 GQDGSPRGRGGRRGRGGRGRGRG------RGGRGNGPANVEYDDGGRGRGGRGNG 191
Score = 34.7 bits (78), Expect = 0.17
Identities = 29/77 (37%), Positives = 31/77 (39%), Gaps = 35/77 (45%)
Query: 588 GRGRGDRARGGRGRGGRARG--------------------------------EGAPAEGA 615
GRGRG RGG GRGGR RG +G P +G
Sbjct: 228 GRGRGR-GRGG-GRGGRGRGGYNGPPPPYYEAQQDGGDYGYNNVAPPADHGYDGPPPQGR 285
Query: 616 RGGRARGPRGRRGAGRG 632
GR RG RG RG GRG
Sbjct: 286 GRGRGRGGRG-RGGGRG 301
Score = 33.5 bits (75), Expect = 0.37
Identities = 19/32 (59%), Positives = 20/32 (62%), Gaps = 5/32 (15%)
Query: 588 GRGRGDRARGGRGRGGRA----RGEGAPAEGA 615
GRGRG R RGGRGRGG R GAP + A
Sbjct: 284 GRGRG-RGRGGRGRGGGRGGFNRSNGAPIQAA 314
>At5g65630 unknown protein
Length = 590
Score = 42.0 bits (97), Expect = 0.001
Identities = 29/72 (40%), Positives = 36/72 (49%), Gaps = 10/72 (13%)
Query: 3 EVPLVDYPPSPTVEIPTIDSPPSPMVESSGEESSGEESSGEESSGEASSGMGG------S 56
++P+ DYP +VEI D SSG SSG SS SS + SG GG S
Sbjct: 518 DIPIEDYP---SVEIER-DGTAVAAAASSGSSSSGSSSSSGGSSSSSDSGSGGSSSGSDS 573
Query: 57 DEDSIPPPTVDA 68
D DS+ P V+A
Sbjct: 574 DADSVQSPFVEA 585
>At4g22020 glycine-rich protein
Length = 396
Score = 42.0 bits (97), Expect = 0.001
Identities = 27/65 (41%), Positives = 29/65 (44%), Gaps = 1/65 (1%)
Query: 571 GTQGVAFAAVRGARA-AGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGA 629
G+ A A V G+ AGG G G GG GG G GA A GG A G G G
Sbjct: 161 GSGAGAGAGVGGSSGGAGGGGGGGGGEGGGANGGSGHGSGAGAGAGVGGAAGGVGGGGGG 220
Query: 630 GRGRG 634
G G G
Sbjct: 221 GGGEG 225
Score = 38.9 bits (89), Expect = 0.009
Identities = 25/68 (36%), Positives = 27/68 (38%), Gaps = 1/68 (1%)
Query: 566 SNAFIGTQGVAFAAVRGARAAGGRGRGDRARGGRGRG-GRARGEGAPAEGARGGRARGPR 624
S A G G G GG G G + GG G G G G GA G+ GG G
Sbjct: 123 SGAGAGVGGTTGGVGGGGGGGGGGGEGGGSSGGSGHGSGSGAGAGAGVGGSSGGAGGGGG 182
Query: 625 GRRGAGRG 632
G G G G
Sbjct: 183 GGGGEGGG 190
Score = 38.1 bits (87), Expect = 0.015
Identities = 22/62 (35%), Positives = 25/62 (39%), Gaps = 1/62 (1%)
Query: 574 GVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGR 633
G + G GGRG G+ G GG G G+ A GG G G G G G
Sbjct: 88 GAGMSGYGGVGGGGGRGGGEGG-GSSANGGSGHGSGSGAGAGVGGTTGGVGGGGGGGGGG 146
Query: 634 GE 635
GE
Sbjct: 147 GE 148
Score = 38.1 bits (87), Expect = 0.015
Identities = 25/69 (36%), Positives = 29/69 (41%), Gaps = 8/69 (11%)
Query: 574 GVAFAAVRGARAAGGRGRGDRARGGRGRG-------GRARGEGAPAEGARGGRARGPRGR 626
GV F G GG G G GG G G G G+G+ + G GG G G
Sbjct: 278 GVGFGN-SGGGGGGGGGGGGGGGGGNGSGYGSGSGYGSGMGKGSGSGGGGGGGGGGSGGG 336
Query: 627 RGAGRGRGE 635
G+G G GE
Sbjct: 337 NGSGSGSGE 345
Score = 38.1 bits (87), Expect = 0.015
Identities = 22/53 (41%), Positives = 23/53 (42%), Gaps = 1/53 (1%)
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
G GG G G A GG G G + G GA G GG G G G G G G
Sbjct: 100 GGGRGGGEGGGSSANGGSGHGSGS-GAGAGVGGTTGGVGGGGGGGGGGGEGGG 151
Score = 37.7 bits (86), Expect = 0.020
Identities = 23/51 (45%), Positives = 23/51 (45%), Gaps = 1/51 (1%)
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRG 632
G GG G G A GG G G A G GA GA GG G G G G G
Sbjct: 178 GGGGGGGGGEGGGANGGSGHGSGA-GAGAGVGGAAGGVGGGGGGGGGEGGG 227
Score = 37.4 bits (85), Expect = 0.026
Identities = 25/65 (38%), Positives = 27/65 (41%), Gaps = 1/65 (1%)
Query: 571 GTQGVAFAAVRGARAA-GGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGA 629
G+ A A V GA GG G G GG GG G G+ A G G A G G G
Sbjct: 198 GSGAGAGAGVGGAAGGVGGGGGGGGGEGGGANGGSGHGSGSGAGGGVSGAAGGGGGGGGG 257
Query: 630 GRGRG 634
G G
Sbjct: 258 GGSGG 262
Score = 37.0 bits (84), Expect = 0.034
Identities = 22/53 (41%), Positives = 23/53 (42%), Gaps = 3/53 (5%)
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
G A GG G G A G G GG A G G G GG G G+G G G
Sbjct: 188 GGGANGGSGHGSGAGAGAGVGGAAGGVGG---GGGGGGGEGGGANGGSGHGSG 237
Score = 35.8 bits (81), Expect = 0.075
Identities = 22/63 (34%), Positives = 23/63 (35%), Gaps = 2/63 (3%)
Query: 574 GVAFAAVRGARAAGGRGRGDRARGGRGRG--GRARGEGAPAEGARGGRARGPRGRRGAGR 631
G G GG G G GG G G G GA A GG + G G G G
Sbjct: 126 GAGVGGTTGGVGGGGGGGGGGGEGGGSSGGSGHGSGSGAGAGAGVGGSSGGAGGGGGGGG 185
Query: 632 GRG 634
G G
Sbjct: 186 GEG 188
Score = 35.8 bits (81), Expect = 0.075
Identities = 21/64 (32%), Positives = 27/64 (41%), Gaps = 3/64 (4%)
Query: 571 GTQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAG 630
G G + + G + G+G G GG G GG G G+ G+ G G G G G
Sbjct: 300 GGNGSGYGSGSGYGSGMGKGSGSGGGGGGGGGGSGGGNGS---GSGSGEGYGMGGGAGTG 356
Query: 631 RGRG 634
G G
Sbjct: 357 NGGG 360
Score = 34.7 bits (78), Expect = 0.17
Identities = 25/63 (39%), Positives = 27/63 (42%), Gaps = 2/63 (3%)
Query: 574 GVAFAAVRGARAAGGRGR-GDRARGGRGRG-GRARGEGAPAEGARGGRARGPRGRRGAGR 631
GV+ AA G GG G G + GG G G G G G G GG G G G G
Sbjct: 243 GVSGAAGGGGGGGGGGGSGGSKVGGGYGHGSGFGGGVGFGNSGGGGGGGGGGGGGGGGGN 302
Query: 632 GRG 634
G G
Sbjct: 303 GSG 305
Score = 34.7 bits (78), Expect = 0.17
Identities = 25/72 (34%), Positives = 27/72 (36%), Gaps = 3/72 (4%)
Query: 566 SNAFIGTQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARG--EGAPAEGARGGRARGP 623
+ A G G A G GG G G G G G A G GA G GG G
Sbjct: 201 AGAGAGVGGAAGGVGGGGGGGGGEGGGANGGSGHGSGSGAGGGVSGAAGGGGGGGGGGGS 260
Query: 624 RGRR-GAGRGRG 634
G + G G G G
Sbjct: 261 GGSKVGGGYGHG 272
Score = 34.7 bits (78), Expect = 0.17
Identities = 20/53 (37%), Positives = 23/53 (42%), Gaps = 1/53 (1%)
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
G+ A GG G G + G G GG G G G GG G G+G G G
Sbjct: 110 GSSANGGSGHGSGSGAGAGVGGTTGGVGGGGGGG-GGGGEGGGSSGGSGHGSG 161
Score = 33.5 bits (75), Expect = 0.37
Identities = 22/53 (41%), Positives = 22/53 (41%), Gaps = 3/53 (5%)
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
GA G G G A G G GG A G G G GG G G G G G G
Sbjct: 190 GANGGSGHGSG--AGAGAGVGGAAGGVGG-GGGGGGGEGGGANGGSGHGSGSG 239
Score = 32.3 bits (72), Expect = 0.83
Identities = 21/58 (36%), Positives = 24/58 (41%), Gaps = 5/58 (8%)
Query: 582 GARAAGGRGRGDRARGGRGRG-----GRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
G ++GG G G + G G G G A G G G GG G GAG G G
Sbjct: 149 GGGSSGGSGHGSGSGAGAGAGVGGSSGGAGGGGGGGGGEGGGANGGSGHGSGAGAGAG 206
Score = 31.6 bits (70), Expect = 1.4
Identities = 19/58 (32%), Positives = 23/58 (38%), Gaps = 5/58 (8%)
Query: 582 GARAAGGRGRGDRARGG-----RGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
G++ GG G G GG G GG G G G G G G+G G+G
Sbjct: 262 GSKVGGGYGHGSGFGGGVGFGNSGGGGGGGGGGGGGGGGGNGSGYGSGSGYGSGMGKG 319
Score = 31.2 bits (69), Expect = 1.8
Identities = 22/70 (31%), Positives = 25/70 (35%), Gaps = 3/70 (4%)
Query: 568 AFIGTQGVAFAAVRGARAAGGRGRGDRARG---GRGRGGRARGEGAPAEGARGGRARGPR 624
+F G + G GG G G+ G G G G A G G GGR G
Sbjct: 49 SFYGKGAKRYGGGGGGGGGGGGGGGEGGDGYGHGEGYGAGAGMSGYGGVGGGGGRGGGEG 108
Query: 625 GRRGAGRGRG 634
G A G G
Sbjct: 109 GGSSANGGSG 118
Score = 30.8 bits (68), Expect = 2.4
Identities = 22/65 (33%), Positives = 24/65 (36%), Gaps = 1/65 (1%)
Query: 571 GTQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEG-APAEGARGGRARGPRGRRGA 629
G G + G +G G G GG G GG G G G GG G G G
Sbjct: 230 GGSGHGSGSGAGGGVSGAAGGGGGGGGGGGSGGSKVGGGYGHGSGFGGGVGFGNSGGGGG 289
Query: 630 GRGRG 634
G G G
Sbjct: 290 GGGGG 294
Score = 29.6 bits (65), Expect = 5.4
Identities = 20/46 (43%), Positives = 20/46 (43%), Gaps = 2/46 (4%)
Query: 588 GRGRGDRARGGRGRG-GRARGEGAPAEGARGGRARGPRGRRGAGRG 632
G G G GG G G GR GEG A GG G GAG G
Sbjct: 86 GAGAGMSGYGGVGGGGGRGGGEGG-GSSANGGSGHGSGSGAGAGVG 130
Score = 29.3 bits (64), Expect = 7.0
Identities = 20/48 (41%), Positives = 21/48 (43%), Gaps = 4/48 (8%)
Query: 587 GGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
GG G G + GG G G GEG G GG G G G G G G
Sbjct: 325 GGGGGGGGSGGGNG-SGSGSGEG---YGMGGGAGTGNGGGGGVGFGMG 368
Score = 28.9 bits (63), Expect = 9.2
Identities = 20/54 (37%), Positives = 22/54 (40%), Gaps = 1/54 (1%)
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEG-ARGGRARGPRGRRGAGRGRG 634
G A GG G G + G G G A G G G GG G G+G G G
Sbjct: 225 GGGANGGSGHGSGSGAGGGVSGAAGGGGGGGGGGGSGGSKVGGGYGHGSGFGGG 278
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,023,670
Number of Sequences: 26719
Number of extensions: 703725
Number of successful extensions: 6696
Number of sequences better than 10.0: 217
Number of HSP's better than 10.0 without gapping: 127
Number of HSP's successfully gapped in prelim test: 99
Number of HSP's that attempted gapping in prelim test: 3263
Number of HSP's gapped (non-prelim): 1833
length of query: 635
length of database: 11,318,596
effective HSP length: 105
effective length of query: 530
effective length of database: 8,513,101
effective search space: 4511943530
effective search space used: 4511943530
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0042.1


