

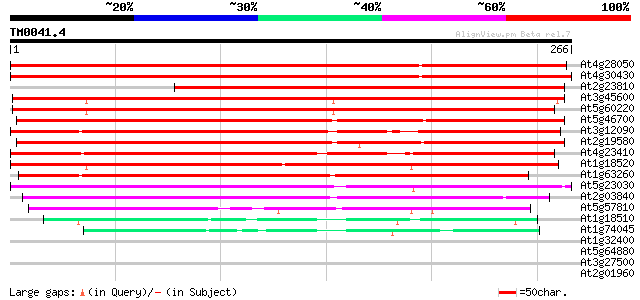
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0041.4
(266 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g28050 senescence-associated protein -like 386 e-108
At4g30430 senescence-associated protein homolog 353 4e-98
At2g23810 similar to senescence-associated protein 274 4e-74
At3g45600 unknown protein 269 1e-72
At5g60220 senescence-associated protein - like 262 2e-70
At5g46700 senescence-associated protein 5-like protein 253 1e-67
At3g12090 putative protein 246 1e-65
At2g19580 putative senescence-associated protein 5 235 2e-62
At4g23410 unknown protein 235 2e-62
At1g18520 unknown protein 232 1e-61
At1g63260 unknown protein 207 4e-54
At5g23030 senescence-associated protein 5-like protein 184 4e-47
At2g03840 putative senescence-associated protein 130 9e-31
At5g57810 unknown protein 84 1e-16
At1g18510 hypothetical protein 47 8e-06
At1g74045 putative protein 41 6e-04
At1g32400 unknown protein 39 0.003
At5g64880 unknown protein 32 0.27
At3g27500 hypothetical protein 32 0.46
At2g01960 unknown protein 31 0.78
>At4g28050 senescence-associated protein -like
Length = 263
Score = 386 bits (992), Expect = e-108
Identities = 172/264 (65%), Positives = 208/264 (78%), Gaps = 1/264 (0%)
Query: 1 MVRLSNNLIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAG 60
MV+ SNNL+G+LNF T LLSIPIL G+WL K T+CER+L++P++ LG+FL+ VS+AG
Sbjct: 1 MVQCSNNLLGILNFFTFLLSIPILSAGIWLGKNAATECERFLDKPMVVLGIFLMFVSIAG 60
Query: 61 LVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSN 120
LVGACCRVS LLW+YL MFLLI+L F FTIFAFAVTN+GAGE +S RGY+EY + DYSN
Sbjct: 61 LVGACCRVSCLLWLYLFAMFLLILLGFCFTIFAFAVTNRGAGEVISDRGYKEYHVADYSN 120
Query: 121 WLQKRVNSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCCKPSNDCG 180
WLQKRVN+ NW RIRSCL +CS +R R+ + E FY NL+ALQSGCCKPSNDC
Sbjct: 121 WLQKRVNNAKNWERIRSCLMYSDVCSTYRTRYASINVEDFYKSNLNALQSGCCKPSNDCN 180
Query: 181 FTYQGPSVWNKTEGVNYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVNVI 240
FTY P+ W KT G Y N DC+ WDN P LC++C++CKAGLL NIK WKKVA VN++
Sbjct: 181 FTYVNPTTWTKTPG-PYKNEDCNVWDNKPGTLCYDCEACKAGLLDNIKNSWKKVAKVNIV 239
Query: 241 FLVFLIIVYSIGCCAFRNNRNDNW 264
FL+FLIIVYS+GCCAFRNNR +W
Sbjct: 240 FLIFLIIVYSVGCCAFRNNRKRSW 263
>At4g30430 senescence-associated protein homolog
Length = 272
Score = 353 bits (907), Expect = 4e-98
Identities = 154/266 (57%), Positives = 203/266 (75%), Gaps = 1/266 (0%)
Query: 1 MVRLSNNLIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAG 60
MVR SN+L+G+LNF LLS+PIL TG+WLS + T CER+L++P+IALGVFL+++++AG
Sbjct: 1 MVRFSNSLVGILNFFVFLLSVPILSTGIWLSLKATTQCERFLDKPMIALGVFLMIIAIAG 60
Query: 61 LVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSN 120
+VG+CCRV+WLLW YL VMF LI++V FTIFAF VT+KG+GE + G+ Y+EYRL YS+
Sbjct: 61 VVGSCCRVTWLLWSYLFVMFFLILIVLCFTIFAFVVTSKGSGETIQGKAYKEYRLEAYSD 120
Query: 121 WLQKRVNSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCCKPSNDCG 180
WLQ+RVN+ +WN IRSCL K C N T FY E+L+A +SGCCKPSNDC
Sbjct: 121 WLQRRVNNAKHWNSIRSCLYESKFCYNLELVTANHTVSDFYKEDLTAFESGCCKPSNDCD 180
Query: 181 FTYQGPSVWNKTEGVNYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVNVI 240
FTY + WNKT G + N DC WDN+ + LC+NC++CKAG L N+K WK+VA+VN+I
Sbjct: 181 FTYITSTTWNKTSG-THKNSDCQLWDNEKHKLCYNCKACKAGFLDNLKAAWKRVAIVNII 239
Query: 241 FLVFLIIVYSIGCCAFRNNRNDNWKR 266
FLV L++VY++GCCAFRNN+ D + R
Sbjct: 240 FLVLLVVVYAMGCCAFRNNKEDRYGR 265
>At2g23810 similar to senescence-associated protein
Length = 195
Score = 274 bits (700), Expect = 4e-74
Identities = 119/185 (64%), Positives = 146/185 (78%)
Query: 79 MFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSNWLQKRVNSTGNWNRIRSC 138
MFLLI+LVF T+FAF VTNKGAGEA+ G+GY+EY+LGDYS WLQKRV + NWN+IRSC
Sbjct: 1 MFLLILLVFCITVFAFVVTNKGAGEAIEGKGYKEYKLGDYSTWLQKRVENGKNWNKIRSC 60
Query: 139 LGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCCKPSNDCGFTYQGPSVWNKTEGVNYS 198
L K+CS+ +FVN FY E+L+ALQSGCCKPS++CGF Y P+ W K ++
Sbjct: 61 LVESKVCSKLEAKFVNVPVNSFYKEHLTALQSGCCKPSDECGFEYVNPTTWTKNTTGTHT 120
Query: 199 NPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVNVIFLVFLIIVYSIGCCAFRN 258
NPDC WDN LCF+CQSCKAGLL N+K+ WKKVA+VN++FLVFLIIVYS+GCCAFRN
Sbjct: 121 NPDCQTWDNAKEKLCFDCQSCKAGLLDNVKSAWKKVAIVNIVFLVFLIIVYSVGCCAFRN 180
Query: 259 NRNDN 263
N+ D+
Sbjct: 181 NKRDD 185
>At3g45600 unknown protein
Length = 285
Score = 269 bits (687), Expect = 1e-72
Identities = 127/266 (47%), Positives = 178/266 (66%), Gaps = 4/266 (1%)
Query: 2 VRLSNNLIGLLNFLTLLLSIPILVTGVWLSKQPG-TDCERWLERPIIALGVFLLLVSLAG 60
+R SN+LIGL+NFLT LLSIPIL G+WLS + TDC R+L+ P+I +G+ +++VSLAG
Sbjct: 1 MRTSNHLIGLVNFLTFLLSIPILGGGIWLSSRANSTDCLRFLQWPLIVIGISIMVVSLAG 60
Query: 61 LVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSN 120
GAC R +L+W+YL+VM L+I + F IFA+AVT+KG+G + RGY +Y L DYS
Sbjct: 61 FAGACYRNKFLMWLYLVVMLLIIAALIGFIIFAYAVTDKGSGRTVLNRGYLDYYLEDYSG 120
Query: 121 WLQKRVNSTGNWNRIRSCLGSGKLCSEFRNRF--VNDTAEQFYAENLSALQSGCCKPSND 178
WL+ RV+ W +I SCL C + F V +TA+ F+ LS ++SGCCKP D
Sbjct: 121 WLKDRVSDDSYWGKISSCLRDSGACRKIGRNFNGVPETADMFFLRRLSPVESGCCKPPTD 180
Query: 179 CGFTYQGPSVWNKTEGVNYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVN 238
CGF+Y + W+ G+ N DC W ND ++LC+ C SCKAG+L ++K W+KV+V+N
Sbjct: 181 CGFSYVNETGWDTRGGMIGPNQDCMVWSNDQSMLCYQCSSCKAGVLGSLKKSWRKVSVIN 240
Query: 239 VIFLVFLIIVYSIGCCAFRN-NRNDN 263
++ L+ L+I Y I A+RN R DN
Sbjct: 241 IVVLIILVIFYVIAYAAYRNVKRIDN 266
>At5g60220 senescence-associated protein - like
Length = 327
Score = 262 bits (669), Expect = 2e-70
Identities = 119/260 (45%), Positives = 171/260 (65%), Gaps = 3/260 (1%)
Query: 2 VRLSNNLIGLLNFLTLLLSIPILVTGVWLSKQPG-TDCERWLERPIIALGVFLLLVSLAG 60
+R +NLIGL+NF T LLSIPIL G+WLS + TDC R+L+ P+I +G+ ++++SLAG
Sbjct: 1 MRSRSNLIGLINFFTFLLSIPILGGGIWLSSRANSTDCLRFLQWPLIIIGISIMVISLAG 60
Query: 61 LVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSN 120
+ GAC + +L+W+YL MF +I + FTIFA+ VT+KG+G + R Y +Y L DYS
Sbjct: 61 IAGACYQNKFLMWLYLFTMFFVIAALIGFTIFAYVVTDKGSGRFVMNRRYLDYYLNDYSG 120
Query: 121 WLQKRVNSTGNWNRIRSCLGSGKLCSEFRNRF--VNDTAEQFYAENLSALQSGCCKPSND 178
WL+ RV G W I SC+ +C + V +TA FY NLS ++SGCCKP D
Sbjct: 121 WLKDRVTDNGYWRDIGSCVRDSGVCKKIGRDLNGVPETAHMFYFRNLSPVESGCCKPPTD 180
Query: 179 CGFTYQGPSVWNKTEGVNYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVN 238
CG+TY +VW + NPDC W+ND +LC+ C SCKAG+L ++K W+KV+V+N
Sbjct: 181 CGYTYVNETVWIPGGEMVGPNPDCMLWNNDQRLLCYQCSSCKAGVLGSLKKSWRKVSVIN 240
Query: 239 VIFLVFLIIVYSIGCCAFRN 258
++ ++ L+I Y I C A++N
Sbjct: 241 IVVVIILVIFYVIACAAYQN 260
>At5g46700 senescence-associated protein 5-like protein
Length = 269
Score = 253 bits (645), Expect = 1e-67
Identities = 120/260 (46%), Positives = 166/260 (63%), Gaps = 3/260 (1%)
Query: 4 LSNNLIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAGLVG 63
LSNN+IG +NF+T+LLSIP++ G+WL+ C + L+ P+I LGV +LLV LAG +G
Sbjct: 3 LSNNVIGCINFITVLLSIPVIGAGIWLAIGTVNSCVKLLQWPVIILGVLILLVGLAGFIG 62
Query: 64 ACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSNWLQ 123
R++WLL VYL+ M +LIVL+ F + VT +G+G R Y EY L D+S WL+
Sbjct: 63 GFWRITWLLVVYLIAMLILIVLLGCLVGFIYMVTIRGSGHPEPSRAYLEYSLQDFSGWLR 122
Query: 124 KRVNSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCCKPSNDCGFTY 183
+RV + W RIR+CL + +C E R+ A+ F+ +L +QSGCCKP CGFT+
Sbjct: 123 RRVQRSYKWERIRTCLSTTTICPELNQRYT--LAQDFFNAHLDPIQSGCCKPPTKCGFTF 180
Query: 184 QGPSVWNKTEGVNYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVNVIFLV 243
P+ W ++ ++ DC W ND N LC+ C SCKAGLL NIK DW K + ++ L+
Sbjct: 181 VNPTYWISPIDMS-ADMDCLNWSNDQNTLCYTCDSCKAGLLANIKVDWLKADIFLLLALI 239
Query: 244 FLIIVYSIGCCAFRNNRNDN 263
LIIVY IGCCAFRN ++
Sbjct: 240 GLIIVYIIGCCAFRNAETED 259
>At3g12090 putative protein
Length = 282
Score = 246 bits (627), Expect = 1e-65
Identities = 117/261 (44%), Positives = 168/261 (63%), Gaps = 15/261 (5%)
Query: 1 MVRLSNNLIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAG 60
M R SN +IG+LN LTLL SIPI+ T ++ ++ T CE +L+ P++ +G +L+VSLAG
Sbjct: 1 MYRFSNTVIGVLNLLTLLASIPIIGTALYKARS-STTCENFLQTPLLVIGFIILIVSLAG 59
Query: 61 LVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSN 120
+GAC V+W LWVYL+VM LI + T+F VT++G G + GR Y+EYRLGDY
Sbjct: 60 FIGACFNVAWALWVYLVVMIFLIATLMGLTLFGLVVTSQGGGVEVPGRIYKEYRLGDYHP 119
Query: 121 WLQKRVNSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCCKPSNDCG 180
WL++RV WN IRSC+ S K C++ + T ++ +++++QSGCCKP C
Sbjct: 120 WLRERVRDPEYWNSIRSCILSSKTCTKIESW----TTLDYFQRDMTSVQSGCCKPPTAC- 174
Query: 181 FTYQGPSVWNKTEGVNYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVNVI 240
TY+ GV DC W+N +LC+ C +CKAG+L+ I+ DW+K++VVN++
Sbjct: 175 -TYEA--------GVVDGGGDCFRWNNGVEMLCYECDACKAGVLEEIRLDWRKLSVVNIL 225
Query: 241 FLVFLIIVYSIGCCAFRNNRN 261
LV LI VY+ GCCAF N R+
Sbjct: 226 VLVLLIAVYAAGCCAFHNTRH 246
>At2g19580 putative senescence-associated protein 5
Length = 270
Score = 235 bits (600), Expect = 2e-62
Identities = 112/261 (42%), Positives = 164/261 (61%), Gaps = 4/261 (1%)
Query: 4 LSNNLIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAGLVG 63
L+NNL +LN L LL SIPI +G+WL+ +P +C L P++ LGV +L+VS G +G
Sbjct: 3 LANNLTAILNLLALLCSIPITASGIWLASKPDNECVNLLRWPVVVLGVLILVVSATGFIG 62
Query: 64 ACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSNWLQ 123
A LL VYL M +LI L+ IFAF VT + GRGY+EYRL +SNWL+
Sbjct: 63 AYKYKETLLAVYLCCMAILIGLLLVVLIFAFVVTRPDGSYRVPGRGYKEYRLEGFSNWLK 122
Query: 124 KRVNSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAEN-LSALQSGCCKPSNDCGFT 182
+ V + NW R+R+CL +C + F+ TA+QF++ + ++ LQSGCCKP CG+
Sbjct: 123 ENVVDSKNWGRLRACLADTNVCPKLNQEFI--TADQFFSSSKITPLQSGCCKPPTACGYN 180
Query: 183 YQGPSVWNKTEGVNYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVNVIFL 242
+ P++W + ++ DC+ W ND + LC+NC SCKAGLL N++ +W+K ++ +I +
Sbjct: 181 FVNPTLWLNPTNM-AADADCYLWSNDQSQLCYNCNSCKAGLLGNLRKEWRKANLILIITV 239
Query: 243 VFLIIVYSIGCCAFRNNRNDN 263
V LI VY I C AFRN + ++
Sbjct: 240 VVLIWVYVIACSAFRNAQTED 260
>At4g23410 unknown protein
Length = 281
Score = 235 bits (599), Expect = 2e-62
Identities = 112/258 (43%), Positives = 167/258 (64%), Gaps = 14/258 (5%)
Query: 1 MVRLSNNLIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAG 60
M R+SN +IG LN LTL+ SI +L + +W+ + T CE +L++P++ LG+ +L++S+AG
Sbjct: 1 MNRMSNTVIGFLNILTLISSIVLLGSALWMGRSK-TTCEHFLQKPLLILGLAILILSVAG 59
Query: 61 LVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSN 120
LVGACC V+W+LWVYL M +IV + T+F F VT+ G + GR Y+E++L Y
Sbjct: 60 LVGACCDVAWVLWVYLFFMVFIIVALMGLTLFGFIVTSHSGGVVVDGRVYKEFKLEAYHP 119
Query: 121 WLQKRVNSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCCKPSNDCG 180
WL+ RV T W I++CL CS + T + ++LS LQSGCCKP C
Sbjct: 120 WLKTRVVDTNYWVTIKTCLLGSVTCS----KLALWTPLDYLQKDLSPLQSGCCKPPTSC- 174
Query: 181 FTYQGPSVWNKTEGVNYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVNVI 240
V+N T+ V +PDC+ W+N VLC++C +C+AG+L+ ++ DW K+++VNVI
Sbjct: 175 -------VYN-TDTVIQQDPDCYRWNNAATVLCYDCDTCRAGVLETVRRDWHKLSLVNVI 226
Query: 241 FLVFLIIVYSIGCCAFRN 258
++FLI VY +GCCAF+N
Sbjct: 227 VVIFLIAVYCVGCCAFKN 244
>At1g18520 unknown protein
Length = 271
Score = 232 bits (592), Expect = 1e-61
Identities = 114/264 (43%), Positives = 160/264 (60%), Gaps = 5/264 (1%)
Query: 1 MVRLSNNLIGLLNFLTLLLSIPILVTGVWLSKQPG-TDCERWLERPIIALGVFLLLVSLA 59
M R+SN ++GL N L +L+ + +++ G TDCE + P++ G+ L LVSL
Sbjct: 1 MFRVSNFMVGLANTLVMLVGASAIGYSIYMFVHQGVTDCESAIRIPLLTTGLILFLVSLL 60
Query: 60 GLVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYS 119
G++G+C + + + YL+++F IV + F+IF F VTNKGAG +SGRGY+EYR D+S
Sbjct: 61 GVIGSCFKENLAMVSYLIILFGGIVALMIFSIFLFFVTNKGAGRVVSGRGYKEYRTVDFS 120
Query: 120 NWLQKRVNSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCCKPSNDC 179
WL V W IRSCL +C + + V+ A+ FY +NLS +QSGCCKP +DC
Sbjct: 121 TWLNGFVGGK-RWVGIRSCLAEANVCDDLSDGRVSQIADAFYHKNLSPIQSGCCKPPSDC 179
Query: 180 GFTYQGPSVW---NKTEGVNYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAV 236
F ++ + W +K E N DC W N LCFNC +CKAG+L NI+ W+ + V
Sbjct: 180 NFEFRNATFWIPPSKNETAVAENGDCGTWSNVQTELCFNCNACKAGVLANIREKWRNLLV 239
Query: 237 VNVIFLVFLIIVYSIGCCAFRNNR 260
N+ L+ LI VYS GCCA RNNR
Sbjct: 240 FNICLLILLITVYSCGCCARRNNR 263
>At1g63260 unknown protein
Length = 284
Score = 207 bits (528), Expect = 4e-54
Identities = 95/242 (39%), Positives = 152/242 (62%), Gaps = 3/242 (1%)
Query: 5 SNNLIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAGLVGA 64
S +I +N LT+LL++ +++ GVW+S C R L P+IALG F+ L+S+ G +GA
Sbjct: 6 STFVIRWVNLLTMLLAVAVIIFGVWMSTH-NDGCRRSLTFPVIALGGFIFLISIIGFLGA 64
Query: 65 CCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSNWLQK 124
C R LLW+YL V+ ++++ + FT+ AF VTN G+G G Y+EY+L DYS+W K
Sbjct: 65 CKRSVALLWIYLAVLLIVLIAILVFTVLAFIVTNNGSGHTNPGLRYKEYKLNDYSSWFLK 124
Query: 125 RVNSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCCKPSNDCGFTYQ 184
++N+T NW R++SCL + C + ++ T +Q + L+ +++GCC+P ++CG+
Sbjct: 125 QLNNTSNWIRLKSCLVKSEQCRKLSKKY--KTIKQLKSAELTPIEAGCCRPPSECGYPAV 182
Query: 185 GPSVWNKTEGVNYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVNVIFLVF 244
S ++ + SN DC + N + C+NC SCKAG+ Q +KT+W+ VA+ NV+ V
Sbjct: 183 NASYYDLSFHSISSNKDCKLYKNLRTIKCYNCDSCKAGVAQYMKTEWRLVAIFNVVLFVV 242
Query: 245 LI 246
LI
Sbjct: 243 LI 244
>At5g23030 senescence-associated protein 5-like protein
Length = 264
Score = 184 bits (467), Expect = 4e-47
Identities = 92/267 (34%), Positives = 145/267 (53%), Gaps = 7/267 (2%)
Query: 1 MVRLSNNLIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAG 60
M+RLSN + N + L+ + L V++ Q + C+R+++ P+I L +S G
Sbjct: 1 MLRLSNAAVITTNAILALIGLAALSFSVYVYVQGPSQCQRFVQNPLIVTAALLFFISSLG 60
Query: 61 LVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSN 120
L+ A ++ +YL +FL I+L+ ++F F VTN AG+ALSGRG + GDY N
Sbjct: 61 LIAALYGSHIIITLYLFFLFLSILLLLVLSVFIFLVTNPTAGKALSGRGIGNVKTGDYQN 120
Query: 121 WLQKRVNSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCCKPSNDCG 180
W+ NW I CL ++C F R ++ F +++LS +Q GCC+P +CG
Sbjct: 121 WIGNHFLRGKNWEGITKCLSDSRVCKRFGPRDID-----FDSKHLSNVQFGCCRPPVECG 175
Query: 181 FTYQGPSVWN-KTEGVNYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVNV 239
F + + W DC AW N LC+ C+SCK G+L+ I+ W+ + VVN+
Sbjct: 176 FESKNATWWTVPATATTAIIGDCKAWSNTQRQLCYACESCKIGVLKGIRKRWRILIVVNL 235
Query: 240 IFLVFLIIVYSIGCCAFRNNRNDNWKR 266
+ ++ ++ +YS GCC +NNR WKR
Sbjct: 236 LLILLVVFLYSCGCCVRKNNRVP-WKR 261
>At2g03840 putative senescence-associated protein
Length = 278
Score = 130 bits (326), Expect = 9e-31
Identities = 71/250 (28%), Positives = 116/250 (46%), Gaps = 4/250 (1%)
Query: 7 NLIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAGLVGACC 66
N I L++ L++ V +C R++ P I + LL +SL G A
Sbjct: 24 NTIFLISSAIFLVTAAFWFVAVMTLHYRTDECNRFVTTPGIFISFSLLAMSLTGFYAAYF 83
Query: 67 RVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSNWLQKRV 126
+ L ++ + FL + +V + IF + + G E+R DYS W+ + V
Sbjct: 84 KSDCLFRIHFFIFFLWMFVVVSKAIFVIFLHKETNPRLFPGTKIYEFRYEDYSGWVSRLV 143
Query: 127 NSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCCKPSNDCGFTYQGP 186
W R R CL +C+ ++ A +FY NL+ +QSGCCKP CG Y+ P
Sbjct: 144 IKDDEWYRTRRCLVKDNVCNRLNHKM---PASEFYQMNLTPIQSGCCKPPLSCGLNYEKP 200
Query: 187 SVWNKTEGVNYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVNVIFLVFLI 246
+ W + N DC W+N + LCF+C SCKA ++ ++ + VN+I ++F +
Sbjct: 201 NNWTVSRYYNNLEVDCKRWNNSADTLCFDCDSCKAVIIADVHNTSFSI-TVNIIHIIFSL 259
Query: 247 IVYSIGCCAF 256
+ G A+
Sbjct: 260 CIGMTGWFAW 269
>At5g57810 unknown protein
Length = 317
Score = 83.6 bits (205), Expect = 1e-16
Identities = 57/254 (22%), Positives = 114/254 (44%), Gaps = 28/254 (11%)
Query: 10 GLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAGLVGACCRVS 69
G+L T +LS+ +L VWL DCE L P + + L+++ + A +
Sbjct: 60 GVLPIFTFVLSLTLLGYAVWLLYMRSYDCEDILGLPRVQTLASVGLLAVFVVSNAALFLR 119
Query: 70 WLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSNWLQKRV-NS 128
+ LV+ ++++L+ F A+A N+ + R + R+ W + ++ +
Sbjct: 120 RKFPMPALVVMVVVLLLMLFIGLAYAGVNE-----MQSRRFPATRM-----WFKLKIMDD 169
Query: 129 TGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCCKPSNDCGFTYQGPSV 188
WN I+SC+ C++ N+ + + + +++GCC P C +
Sbjct: 170 HVTWNNIKSCVYDKGACNDLIYGSPNE--KPYNRRKMPPIKNGCCMPPETCNMDAINATF 227
Query: 189 W--NKTEGVNYSN-------------PDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKK 233
W K EG S DC W ND ++LC++C+SCK G +++++ W +
Sbjct: 228 WYRRKDEGPPSSMNLMYGDEMMVGRISDCQLWRNDWSILCYDCRSCKFGFIRSVRRKWWQ 287
Query: 234 VAVVNVIFLVFLII 247
+ + ++ + L++
Sbjct: 288 LGIFLIVISILLLM 301
>At1g18510 hypothetical protein
Length = 238
Score = 47.4 bits (111), Expect = 8e-06
Identities = 46/239 (19%), Positives = 96/239 (39%), Gaps = 28/239 (11%)
Query: 17 LLLSIPILVTGVWLS-KQPGTDCERWLERPIIALGVFLLLVSLAGLVGACCRVSWLLWVY 75
+L+ I + +TG L ++ + C R + + +G+ LL++ L CC + +Y
Sbjct: 5 ILICIGLTMTGTGLYYRKTVSKCIRETDGSFVVIGLLLLVIPQFALYAICCHSKRMFTIY 64
Query: 76 LLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSNWLQKRVNSTGNWNRI 135
+ M + +++ +++ F + N G A + + + L R+ ++
Sbjct: 65 IYAMIFVSIVLGGYSLKCF-IYNTTFGIAKNPAEEKR-----TAKQLVGRLVPESKLAKV 118
Query: 136 RSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCCKPSNDCGFT--YQGPSVWNKTE 193
C+ C+ F A S + CC CG T + P W+
Sbjct: 119 TECIIHNHDCN-------------FNASQNSNVWRYCCAQPRGCGVTTMFGQPGEWS--- 162
Query: 194 GVNYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVN--VIFLVFLIIVYS 250
+ + + H + C +C+ C+ +L+ I WK +++ + +FLV L + S
Sbjct: 163 -WKHQHVENHVPEECSYEYCLSCRGCQMSILKAIVHQWKYLSMFSYPALFLVCLSLAIS 220
>At1g74045 putative protein
Length = 215
Score = 41.2 bits (95), Expect = 6e-04
Identities = 50/220 (22%), Positives = 82/220 (36%), Gaps = 29/220 (13%)
Query: 36 TDCERWLERPIIALGVFLLLVSLAGLVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFA 95
+ C R LG+ LLL+ GL G CCR L + M +LI++V ++I +
Sbjct: 2 SSCIRETSSQFTLLGLLLLLIPQIGLYGICCRSKRLFNFFFYGMVVLIIIVSYYSIKC-S 60
Query: 96 VTNKGAGEALSGRGYREYRLGDYSNWLQKRVNSTGNWNRIRSCLGSGKLCSEFRNRFVND 155
+ N G A + ++ R L R+ S + ++ C+ C+
Sbjct: 61 IYNTTFGIAKNPA--KDNRTVPQ---LLGRLVSKEKFEKVTYCIIHKHDCN--------- 106
Query: 156 TAEQFYAENLSALQSGCCKPSNDCG----FTYQGPSVWNKTEGVNYSNPDCHAWDNDPNV 211
+ A S + CC CG F G W N +C
Sbjct: 107 ----YNASKNSNVWKYCCAQPVGCGTITMFDKPGEWSWKHQYERNQVPEECSY------E 156
Query: 212 LCFNCQSCKAGLLQNIKTDWKKVAVVNVIFLVFLIIVYSI 251
C +C+ C+ +L+ I WK +++ LV I +I
Sbjct: 157 YCLDCRGCQLSILKAIVHQWKYLSMFAYPALVLSCISLAI 196
>At1g32400 unknown protein
Length = 280
Score = 38.9 bits (89), Expect = 0.003
Identities = 20/54 (37%), Positives = 27/54 (49%)
Query: 41 WLERPIIALGVFLLLVSLAGLVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAF 94
W I +GV L ++S G VG C R L Y L++ LLI++ F F F
Sbjct: 80 WFIYLFIGIGVALFVISCCGCVGTCSRSVCCLSCYSLLLILLILVELGFAAFIF 133
>At5g64880 unknown protein
Length = 171
Score = 32.3 bits (72), Expect = 0.27
Identities = 11/31 (35%), Positives = 21/31 (67%)
Query: 68 VSWLLWVYLLVMFLLIVLVFAFTIFAFAVTN 98
+ WL+W+ +LV LL++L+ IF+F + +
Sbjct: 17 IPWLIWIQMLVFVLLLLLLCVIGIFSFDIVD 47
>At3g27500 hypothetical protein
Length = 609
Score = 31.6 bits (70), Expect = 0.46
Identities = 12/28 (42%), Positives = 15/28 (52%)
Query: 196 NYSNPDCHAWDNDPNVLCFNCQSCKAGL 223
NY+NP CH D L ++C CK L
Sbjct: 193 NYTNPRCHICGEDTGNLLYHCDICKFNL 220
Score = 27.3 bits (59), Expect = 8.6
Identities = 13/43 (30%), Positives = 23/43 (53%), Gaps = 1/43 (2%)
Query: 182 TYQGPSVWNKTEGV-NYSNPDCHAWDNDPNVLCFNCQSCKAGL 223
++ G S+ T G ++++P CH N+ L F+C C+ L
Sbjct: 64 SHDGHSLKLLTTGAPDHTDPKCHLCGNNTKRLLFHCSVCQLNL 106
>At2g01960 unknown protein
Length = 260
Score = 30.8 bits (68), Expect = 0.78
Identities = 44/195 (22%), Positives = 79/195 (39%), Gaps = 44/195 (22%)
Query: 67 RVSWLLWVYLLVMFLL---------------IVLVFAFTIFAFAVTNKGAGEALSGRGYR 111
RV L+ V LL +FLL +V+VF F FA++ K + R +
Sbjct: 54 RVQTLVSVSLLALFLLSNIGMFLRPRRLSYFLVIVF-FIGFAYSGVYK-----MESRRFS 107
Query: 112 EYRL---GDYSNWLQKRVNSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENL-SA 167
+ G+Y+N +R T + ++ G+L RFVN A Y L +
Sbjct: 108 PTPMCFKGEYNNGQGER--KTEQYQVVKIEQSQGRL-QRVHLRFVNSYALPPYDRRLLPS 164
Query: 168 LQSGCCKPSNDCGFTYQGPSVWNKTE---------------GVNYSNPDCH-AWDNDPNV 211
+++GCC +C ++W G N D + W ++ +V
Sbjct: 165 VKTGCCNRPGNCKLETVNATLWVTRNREGPPLETAMIYDRYGGNADIKDYYDMWRHELSV 224
Query: 212 LCFNCQSCKAGLLQN 226
L ++C +C+ ++++
Sbjct: 225 LYYDCMTCQVRIIKS 239
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.141 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,630,855
Number of Sequences: 26719
Number of extensions: 288414
Number of successful extensions: 969
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 903
Number of HSP's gapped (non-prelim): 41
length of query: 266
length of database: 11,318,596
effective HSP length: 98
effective length of query: 168
effective length of database: 8,700,134
effective search space: 1461622512
effective search space used: 1461622512
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0041.4


