

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
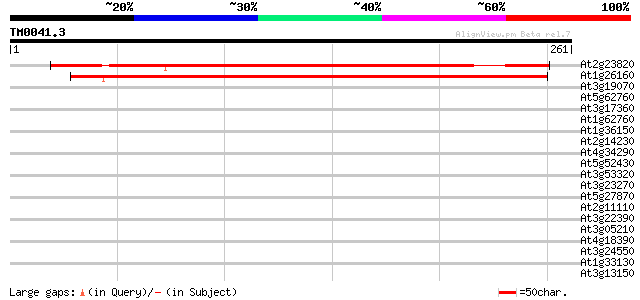
Query= TM0041.3
(261 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g23820 unknown protein 289 9e-79
At1g26160 unknown protein 282 1e-76
At3g19070 hypothetical protein 36 0.024
At5g62760 ZAP - like protein 35 0.031
At3g17360 kinesin-like protein 35 0.031
At1g62760 hypothetical protein 35 0.041
At1g36150 hypothetical protein 34 0.069
At2g14230 hypothetical protein 34 0.090
At4g34290 unknown protein 33 0.12
At5g52430 putative protein 33 0.15
At3g53320 unknown protein 33 0.15
At3g23270 hypothetical protein 33 0.20
At5g27870 pectin methyl-esterase - like protein 32 0.26
At2g11110 En/Spm-like transposon protein 32 0.26
At3g22390 unknown protein 32 0.34
At3g05210 putative nucleotide repair protein 32 0.34
At4g18390 teosinte branched1 like protein 32 0.45
At3g24550 protein kinase, putative 32 0.45
At1g33130 hypothetical protein 32 0.45
At3g13150 hypothetical protein 31 0.59
>At2g23820 unknown protein
Length = 243
Score = 289 bits (740), Expect = 9e-79
Identities = 153/238 (64%), Positives = 174/238 (72%), Gaps = 23/238 (9%)
Query: 20 SLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPS------AS 73
S + + +RF SP+R H S SP S + S P +A PS AS
Sbjct: 21 SFTRSHIRFTYSAAGASSPNRAIH---CMASDSPQSGDGSVSSPPNVAAVPSSSSSSSAS 77
Query: 74 SAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMA 133
SAIDFLS+C RLKTT RAGWI++DV+DPESIADHMYRM LMALI++DIPGV+RDKC+KMA
Sbjct: 78 SAIDFLSLCTRLKTTPRAGWIKRDVKDPESIADHMYRMGLMALISSDIPGVNRDKCMKMA 137
Query: 134 IVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEANSS 193
IVHDIAEAIVGDITP+ G+ KEEKNR E EAL+HMCK+LGGG RAKE+AELW EYE NSS
Sbjct: 138 IVHDIAEAIVGDITPSCGISKEEKNRRESEALEHMCKLLGGGERAKEIAELWREYEENSS 197
Query: 194 PEAKFVKDLDKVEMILQALEYEHEQGKDLDEFFQSTAGKFQTEIGKAWASEIVSRRKK 251
PEAK VKD DKVE+ILQALEYE GKFQT IGKAWASEIVSRR+K
Sbjct: 198 PEAKVVKDFDKVELILQALEYEQ--------------GKFQTNIGKAWASEIVSRRRK 241
>At1g26160 unknown protein
Length = 258
Score = 282 bits (721), Expect = 1e-76
Identities = 147/230 (63%), Positives = 170/230 (73%), Gaps = 8/230 (3%)
Query: 29 KSVTLSPHSPSRVF--------HMTTAAGSSSPPSSSVTTSPPSAASATPSASSAIDFLS 80
+S+ S HS SR F T + P S +S S S SS+IDFL+
Sbjct: 21 RSILASFHSSSRNFLFLGKPTPSSTIVSVRCQKPVSDGVSSMESMNHVASSVSSSIDFLT 80
Query: 81 ICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMAIVHDIAE 140
+CHRLKTTKR GWI + + PESIADHMYRM+LMALIA D+ GVDR++C+KMAIVHDIAE
Sbjct: 81 LCHRLKTTKRKGWINQGINGPESIADHMYRMALMALIAGDLTGVDRERCIKMAIVHDIAE 140
Query: 141 AIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEANSSPEAKFVK 200
AIVGDITP+DGVPKEEK+R E AL MC+VLGGG RA+E+ ELW EYE N+S EA VK
Sbjct: 141 AIVGDITPSDGVPKEEKSRRETAALKEMCEVLGGGLRAEEITELWLEYENNASLEANIVK 200
Query: 201 DLDKVEMILQALEYEHEQGKDLDEFFQSTAGKFQTEIGKAWASEIVSRRK 250
D DKVEMILQALEYE E GK LDEFF STAGKFQTEIGK+WA+EI +RRK
Sbjct: 201 DFDKVEMILQALEYEAEHGKVLDEFFISTAGKFQTEIGKSWAAEINARRK 250
>At3g19070 hypothetical protein
Length = 346
Score = 35.8 bits (81), Expect = 0.024
Identities = 23/65 (35%), Positives = 33/65 (50%)
Query: 11 SLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATP 70
S+ SSS+ SST S +L P +P F ++ + S PPSSS + S + P
Sbjct: 73 SMSSSSLQPPSSSTLAPSSSSSLQPPAPLIDFFRSSVSYSHQPPSSSTLATSSSPSLQPP 132
Query: 71 SASSA 75
S SS+
Sbjct: 133 SMSSS 137
>At5g62760 ZAP - like protein
Length = 383
Score = 35.4 bits (80), Expect = 0.031
Identities = 22/57 (38%), Positives = 30/57 (52%), Gaps = 6/57 (10%)
Query: 38 PSRVFHMTTAAGSSSPPSSSVTTSP---PSAASATPSASSAIDFLSICHRLKTTKRA 91
PS +F +TT + + PPSSS P PS+A P+ S ID + H LK R+
Sbjct: 218 PSSLFPVTTNSSPTIPPSSSYPQMPNASPSSAQLAPTRSKVID---VSHLLKPPHRS 271
>At3g17360 kinesin-like protein
Length = 2008
Score = 35.4 bits (80), Expect = 0.031
Identities = 20/64 (31%), Positives = 35/64 (54%), Gaps = 1/64 (1%)
Query: 155 EEKNRLEQEALDHMCKVLGGGSRAKEVAELWAE-YEANSSPEAKFVKDLDKVEMILQALE 213
EE N +EA +C+++ ++ V E+W + +E S E + DL++V+ IL A E
Sbjct: 1224 EEVNATMKEADLTICELVKANEKSNSVTEMWLQTHEELISKEKNLMDDLEQVKSILSACE 1283
Query: 214 YEHE 217
E +
Sbjct: 1284 EEKQ 1287
>At1g62760 hypothetical protein
Length = 312
Score = 35.0 bits (79), Expect = 0.041
Identities = 23/71 (32%), Positives = 38/71 (53%)
Query: 9 LSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASA 68
LSSL S PS SS+ +LSP SP + ++ P SS +++ PS++S+
Sbjct: 78 LSSLSPSLSPSPPSSSPSSAPPSSLSPSSPPPLSLSPSSPPPPPPSSSPLSSLSPSSSSS 137
Query: 69 TPSASSAIDFL 79
T S + +D++
Sbjct: 138 TYSNQTNLDYI 148
Score = 32.7 bits (73), Expect = 0.20
Identities = 27/73 (36%), Positives = 36/73 (48%), Gaps = 13/73 (17%)
Query: 14 SSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP------PSAAS 67
SS P SS+ L S +LSP PS ++ SS+PPSS +SP PS+
Sbjct: 66 SSPPPPPPSSSPLSSLSPSLSPSPPS-------SSPSSAPPSSLSPSSPPPLSLSPSSPP 118
Query: 68 ATPSASSAIDFLS 80
P +SS + LS
Sbjct: 119 PPPPSSSPLSSLS 131
Score = 31.2 bits (69), Expect = 0.59
Identities = 27/75 (36%), Positives = 38/75 (50%), Gaps = 16/75 (21%)
Query: 12 LFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP------PSA 65
LF ++ SSLS + S +LSP PS ++ SS+PPSS +SP PS+
Sbjct: 18 LFITTSSSSLSPSS---SSPSLSPSPPS-------SSPSSAPPSSLSPSSPPPLSLSPSS 67
Query: 66 ASATPSASSAIDFLS 80
P +SS + LS
Sbjct: 68 PPPPPPSSSPLSSLS 82
Score = 29.3 bits (64), Expect = 2.2
Identities = 27/67 (40%), Positives = 38/67 (56%), Gaps = 7/67 (10%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSA--AS 67
SS SS+ PSSLS + ++LSP SP ++ S SP S++ SPPS+ +S
Sbjct: 42 SSSPSSAPPSSLSPSSP--PPLSLSPSSPPPPPPSSSPLSSLSP---SLSPSPPSSSPSS 96
Query: 68 ATPSASS 74
A PS+ S
Sbjct: 97 APPSSLS 103
Score = 28.9 bits (63), Expect = 2.9
Identities = 22/58 (37%), Positives = 34/58 (57%), Gaps = 11/58 (18%)
Query: 21 LSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSS--VTTSPPSA--ASATPSASS 74
LS TQ+ S+ + + +TT++ S SP SSS ++ SPPS+ +SA PS+ S
Sbjct: 4 LSQTQILHLSIAI-------LLFITTSSSSLSPSSSSPSLSPSPPSSSPSSAPPSSLS 54
Score = 28.5 bits (62), Expect = 3.8
Identities = 30/96 (31%), Positives = 45/96 (46%), Gaps = 3/96 (3%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS SS+ PSSLS + ++LSP SP ++ S SP SSS T S +
Sbjct: 91 SSSPSSAPPSSLSPSSP--PPLSLSPSSPPPPPPSSSPLSSLSPSSSSSTYSNQTNLDYI 148
Query: 70 PSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIA 105
++ + + +IC+ + A IR + Q IA
Sbjct: 149 KTSCNITLYKTICYN-SLSPYASTIRSNPQKLAVIA 183
Score = 28.1 bits (61), Expect = 5.0
Identities = 20/64 (31%), Positives = 31/64 (48%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS S PS SS+ +LSP SP + ++ P SS +++ PS + +
Sbjct: 30 SSSSPSLSPSPPSSSPSSAPPSSLSPSSPPPLSLSPSSPPPPPPSSSPLSSLSPSLSPSP 89
Query: 70 PSAS 73
PS+S
Sbjct: 90 PSSS 93
>At1g36150 hypothetical protein
Length = 256
Score = 34.3 bits (77), Expect = 0.069
Identities = 27/64 (42%), Positives = 33/64 (51%), Gaps = 3/64 (4%)
Query: 15 SSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPS--SSVTTSPPSAASATPSA 72
SS P S SS + S S SP+ V H + +SSP SS T S P AAS +PS
Sbjct: 168 SSPPVSHSSPPVSHSSPPTSRSSPA-VSHSSPVVAASSPVKAVSSSTASSPRAASPSPSP 226
Query: 73 SSAI 76
S +I
Sbjct: 227 SPSI 230
Score = 30.0 bits (66), Expect = 1.3
Identities = 21/58 (36%), Positives = 30/58 (51%), Gaps = 3/58 (5%)
Query: 18 PSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSASSA 75
PSS S + S +S HSP V H + SSPP S +SPP++ S+ + S+
Sbjct: 143 PSSAKSPAITPSSPAVS-HSPPPVRHSSPPVSHSSPPVSH--SSPPTSRSSPAVSHSS 197
>At2g14230 hypothetical protein
Length = 365
Score = 33.9 bits (76), Expect = 0.090
Identities = 27/88 (30%), Positives = 45/88 (50%), Gaps = 5/88 (5%)
Query: 17 IPSSLSSTQLRFKSVTLSP---HSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSAS 73
+PSS +T R++S P +SP +++ GS PPSS T+ PS + + S +
Sbjct: 1 MPSS-ENTYARYRSQQRDPLHVNSPVSAGNISGVQGSRHPPSSPTLTTQPS-VNHSDSQA 58
Query: 74 SAIDFLSICHRLKTTKRAGWIRKDVQDP 101
++ L++ L + RAG R D + P
Sbjct: 59 ERLNNLTLEKLLDSPGRAGLTRLDPKRP 86
>At4g34290 unknown protein
Length = 144
Score = 33.5 bits (75), Expect = 0.12
Identities = 33/111 (29%), Positives = 57/111 (50%), Gaps = 12/111 (10%)
Query: 8 LLSSLFSSSI----PSSLSSTQLRFKSVTLSPHSPSR---VFHMTTAAGSSSPPSSSVTT 60
L S +FS++ + L S+ L S+ LSP+ P+ V +T+AA +SS P+++ T
Sbjct: 3 LSSGIFSTTFLCVDTAPLRSSMLSPSSLRLSPNHPTNLRMVRAVTSAAAASSDPTTTTKT 62
Query: 61 SPPSAASATPSASSAI-DFLSI--CHRLKTTKRA-GWIRK-DVQDPESIAD 106
P S A+ D + + R + KR +I++ D+QDP++ D
Sbjct: 63 REPRGIMKPRPVSQAMQDVVGVPEIPRTQALKRIWAYIKEHDLQDPQNKRD 113
>At5g52430 putative protein
Length = 438
Score = 33.1 bits (74), Expect = 0.15
Identities = 29/89 (32%), Positives = 40/89 (44%), Gaps = 14/89 (15%)
Query: 2 ATATRVLLSSL--------FSSSIPSSLSSTQ---LRFKSVTLSPHSPSRVFHMTTAAGS 50
AT+T V+L + F S PSS+S + L S T SP P VF + A
Sbjct: 78 ATSTTVVLPFIAPPSSPASFLQSDPSSVSHSPVGPLSLTSNTFSPKEPQSVFTVGPYANE 137
Query: 51 S---SPPSSSVTTSPPSAASATPSASSAI 76
+ +PP S + PS A TP S++
Sbjct: 138 TQPVTPPVFSAFITEPSTAPYTPPPESSV 166
>At3g53320 unknown protein
Length = 553
Score = 33.1 bits (74), Expect = 0.15
Identities = 31/99 (31%), Positives = 45/99 (45%), Gaps = 10/99 (10%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSS---------PPSSSVTT 60
SSL S + SS +S + S+ S SR+ A S+S PP + T
Sbjct: 325 SSLESCASASSSASHKPSIDSIKKKNDSSSRLSSQPLANRSTSRGIMGQPRIPPQQTNKT 384
Query: 61 SPPSAASATPSASSAIDFLSICHRL-KTTKRAGWIRKDV 98
S P +S+ P+A S D+ S R +T+K A +K V
Sbjct: 385 SKPKLSSSVPTAGSISDYSSESSRASETSKMANGNQKTV 423
>At3g23270 hypothetical protein
Length = 1045
Score = 32.7 bits (73), Expect = 0.20
Identities = 25/73 (34%), Positives = 33/73 (44%), Gaps = 2/73 (2%)
Query: 9 LSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPP--SSSVTTSPPSAA 66
L L + PSSLS+ Q FK V +P R + ++ S PP SSS PS
Sbjct: 730 LQQLKDIAFPSSLSAIQNAFKPVVAPTTTPPRTLVIGPSSPSPPPPPRSSSPYARRPSPP 789
Query: 67 SATPSASSAIDFL 79
+ + S ID L
Sbjct: 790 RTSGFSRSVIDSL 802
>At5g27870 pectin methyl-esterase - like protein
Length = 732
Score = 32.3 bits (72), Expect = 0.26
Identities = 22/59 (37%), Positives = 31/59 (52%)
Query: 15 SSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSAS 73
S PSSL S + L S + +T +A S+ PS+S + SP + SA+PSAS
Sbjct: 652 SDTPSSLVSPSTSPPAGHLGSPSDTPSSVVTPSASPSTSPSASPSVSPSAFPSASPSAS 710
Score = 29.3 bits (64), Expect = 2.2
Identities = 16/37 (43%), Positives = 24/37 (64%)
Query: 37 SPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSAS 73
SPS ++ +A S+ PS+S + SP + SA+PSAS
Sbjct: 690 SPSASPSVSPSAFPSASPSASPSASPSVSPSASPSAS 726
Score = 27.7 bits (60), Expect = 6.5
Identities = 16/44 (36%), Positives = 24/44 (54%)
Query: 30 SVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSAS 73
SV SPS + + S+ PS+S + SP ++ S +PSAS
Sbjct: 679 SVVTPSASPSTSPSASPSVSPSAFPSASPSASPSASPSVSPSAS 722
>At2g11110 En/Spm-like transposon protein
Length = 356
Score = 32.3 bits (72), Expect = 0.26
Identities = 26/88 (29%), Positives = 44/88 (49%), Gaps = 5/88 (5%)
Query: 17 IPSSLSSTQLRFKSVTLSP---HSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSAS 73
+PSS +T R++S P +SP +++ GS PPS T+ PS + + S +
Sbjct: 1 MPSS-ENTYARYRSQQRDPLHVNSPVSAGNISGVQGSCHPPSPPTPTTQPS-VNHSDSQA 58
Query: 74 SAIDFLSICHRLKTTKRAGWIRKDVQDP 101
++ L++ L + RAG R D + P
Sbjct: 59 ERLNNLTLEELLDSPGRAGLTRLDPKRP 86
>At3g22390 unknown protein
Length = 873
Score = 32.0 bits (71), Expect = 0.34
Identities = 20/59 (33%), Positives = 30/59 (49%), Gaps = 16/59 (27%)
Query: 36 HSPSRVFHM--------TTAAGSSSPPSSSVTTSPPSA--------ASATPSASSAIDF 78
H P+ +F + T AA S PP+S +T+S P+A ASATP+ + + F
Sbjct: 78 HGPTFIFPLGQQPHAAATIAAASVRPPNSGITSSGPTATSTSMNGSASATPAGAPTMSF 136
>At3g05210 putative nucleotide repair protein
Length = 410
Score = 32.0 bits (71), Expect = 0.34
Identities = 27/95 (28%), Positives = 43/95 (44%), Gaps = 15/95 (15%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
S ++S PSS +++ + + PHS S+ H T A SSS P + T S PS +
Sbjct: 66 SDVYSPPPPSSAAASSSQPSGASQVPHSSSQT-HQTDGASSSSTPVA--TGSVPSNTTQN 122
Query: 70 PSASSAIDFLSICHR------LKTTKRAGWIRKDV 98
+A + + HR LK + W+ D+
Sbjct: 123 RNA------ILVSHRQKGNPLLKHIRNVKWVFSDI 151
>At4g18390 teosinte branched1 like protein
Length = 365
Score = 31.6 bits (70), Expect = 0.45
Identities = 18/68 (26%), Positives = 32/68 (46%), Gaps = 3/68 (4%)
Query: 36 HSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSASSAIDFLSICHRL---KTTKRAG 92
H+ SR+ ++ A+G S +T+ P S S+A+ F + RL + +K
Sbjct: 28 HNSSRIIRVSRASGGKDRHSKVLTSKGPRDRRVRLSVSTALQFYDLQDRLGYDQPSKAVE 87
Query: 93 WIRKDVQD 100
W+ K +D
Sbjct: 88 WLIKAAED 95
>At3g24550 protein kinase, putative
Length = 652
Score = 31.6 bits (70), Expect = 0.45
Identities = 13/30 (43%), Positives = 20/30 (66%)
Query: 45 TTAAGSSSPPSSSVTTSPPSAASATPSASS 74
T + SPP++S TT+PP AAS+ P ++
Sbjct: 10 TPSPSPPSPPTNSTTTTPPPAASSPPPTTT 39
Score = 28.1 bits (61), Expect = 5.0
Identities = 19/51 (37%), Positives = 26/51 (50%), Gaps = 6/51 (11%)
Query: 32 TLSPHSPSRVFHMTTAA---GSSSPPSSSVTTSP---PSAASATPSASSAI 76
T SP PS + TT +SSPP ++ +SP PS S +P SS +
Sbjct: 10 TPSPSPPSPPTNSTTTTPPPAASSPPPTTTPSSPPPSPSTNSTSPPPSSPL 60
>At1g33130 hypothetical protein
Length = 356
Score = 31.6 bits (70), Expect = 0.45
Identities = 25/86 (29%), Positives = 42/86 (48%), Gaps = 4/86 (4%)
Query: 19 SSLSSTQLRFKSV---TLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSASSA 75
SS +T R++S +L +SP +++ GS PPS T+ PS + S +
Sbjct: 2 SSSENTYARYRSQQGDSLHVNSPVSAGNISGVQGSRHPPSPPTPTTQPS-VNDCDSQAER 60
Query: 76 IDFLSICHRLKTTKRAGWIRKDVQDP 101
++ L++ L + RAG R D + P
Sbjct: 61 LNNLTLEELLDSPGRAGLTRLDPKRP 86
>At3g13150 hypothetical protein
Length = 551
Score = 31.2 bits (69), Expect = 0.59
Identities = 26/75 (34%), Positives = 38/75 (50%), Gaps = 11/75 (14%)
Query: 14 SSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPS--------SSVTTSPPSA 65
SS + SS +T V+ SP + S + ++ AA SSS S SSV++SP S+
Sbjct: 415 SSPVSSSAKTTST---PVSSSPDTSSFLLSLSLAADSSSSDSDSSSPDSSSSVSSSPDSS 471
Query: 66 ASATPSASSAIDFLS 80
+S + S S F S
Sbjct: 472 SSVSSSPDSYSSFSS 486
Score = 28.5 bits (62), Expect = 3.8
Identities = 23/65 (35%), Positives = 33/65 (50%), Gaps = 4/65 (6%)
Query: 11 SLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATP 70
S FSSS SS S + F S + SP + + SS S SV++S ++S P
Sbjct: 482 SSFSSSPDSSSSVSSSLFSSSRENSSSPD----YSNSVSSSLDYSGSVSSSSDYSSSVFP 537
Query: 71 SASSA 75
SA+S+
Sbjct: 538 SANSS 542
Score = 28.1 bits (61), Expect = 5.0
Identities = 26/70 (37%), Positives = 37/70 (52%), Gaps = 15/70 (21%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPP---SAA 66
S L S S+ + SS+ S + SP S S V SSSP SSS +S P S+
Sbjct: 437 SFLLSLSLAADSSSSD----SDSSSPDSSSSV--------SSSPDSSSSVSSSPDSYSSF 484
Query: 67 SATPSASSAI 76
S++P +SS++
Sbjct: 485 SSSPDSSSSV 494
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.125 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,635,389
Number of Sequences: 26719
Number of extensions: 230450
Number of successful extensions: 1564
Number of sequences better than 10.0: 99
Number of HSP's better than 10.0 without gapping: 33
Number of HSP's successfully gapped in prelim test: 67
Number of HSP's that attempted gapping in prelim test: 1341
Number of HSP's gapped (non-prelim): 235
length of query: 261
length of database: 11,318,596
effective HSP length: 97
effective length of query: 164
effective length of database: 8,726,853
effective search space: 1431203892
effective search space used: 1431203892
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0041.3


