

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0041.13
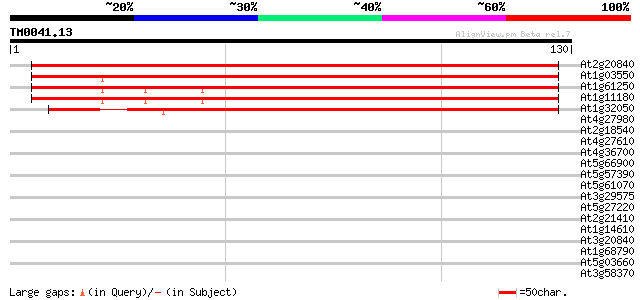
(130 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g20840 putative secretory carrier-associated membrane protein 191 7e-50
At1g03550 unknown protein 173 2e-44
At1g61250 unknown protein 167 2e-42
At1g11180 secretory carrier membrane protein, putative 154 1e-38
At1g32050 unknown protein 122 4e-29
At4g27980 putative protein 35 0.011
At2g18540 putative vicilin storage protein (globulin-like) 34 0.018
At4g27610 unknown protein 33 0.031
At4g36700 globulin-like protein 33 0.041
At5g66900 disease resistance protein-like 33 0.053
At5g57390 putative protein 32 0.090
At5g61070 HDA18 31 0.15
At3g29575 unknown protein 31 0.20
At5g27220 putative protein 30 0.26
At2g21410 putative vacuolar proton-ATPase subunit 30 0.26
At1g14610 valyl tRNA synthetase (valRS) 30 0.26
At3g20840 putative transcription factor 30 0.34
At1g68790 putative nuclear matrix constituent protein 1 (NMCP1) 30 0.34
At5g03660 unknown protein 30 0.45
At3g58370 putative protein 30 0.45
>At2g20840 putative secretory carrier-associated membrane protein
Length = 282
Score = 191 bits (486), Expect = 7e-50
Identities = 91/122 (74%), Positives = 107/122 (87%)
Query: 6 EMEPARNSGSVPLATSKLKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAELKRRE 65
E+ P N SVP ATSKL PLPPEPYDRGAT+DIPLD+ KD+KAKEKEL+ +EAELKRRE
Sbjct: 13 EINPFANPTSVPAATSKLSPLPPEPYDRGATMDIPLDSGKDLKAKEKELREKEAELKRRE 72
Query: 66 QELKRKEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGMLKFLF 125
QE+KRKEDAIA+AGIVIEEKNWPPFFP+IHH+IS EIP+HLQRIQYVAF + LG++ L
Sbjct: 73 QEIKRKEDAIAQAGIVIEEKNWPPFFPLIHHDISNEIPIHLQRIQYVAFTSMLGLVVCLL 132
Query: 126 FS 127
++
Sbjct: 133 WN 134
>At1g03550 unknown protein
Length = 283
Score = 173 bits (439), Expect = 2e-44
Identities = 84/123 (68%), Positives = 102/123 (82%), Gaps = 1/123 (0%)
Query: 6 EMEPARNSGSVPLAT-SKLKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAELKRR 64
E+ P N SVP A+ S LKPLPPEPYDRGAT+DIPLD+ D++AKE ELQA+E ELKR+
Sbjct: 13 EINPFANHTSVPPASNSYLKPLPPEPYDRGATVDIPLDSGNDLRAKEMELQAKENELKRK 72
Query: 65 EQELKRKEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGMLKFL 124
EQELKR+EDAIAR G+VIEEKNWP FFP+IHH+I EIP+HLQ+IQYVAF T LG++ L
Sbjct: 73 EQELKRREDAIARTGVVIEEKNWPEFFPLIHHDIPNEIPIHLQKIQYVAFTTLLGLVGCL 132
Query: 125 FFS 127
++
Sbjct: 133 LWN 135
>At1g61250 unknown protein
Length = 289
Score = 167 bits (422), Expect = 2e-42
Identities = 84/127 (66%), Positives = 104/127 (81%), Gaps = 5/127 (3%)
Query: 6 EMEPARNSGSVPLAT-SKLKPLPPEP--YDRGATIDIPLDT--TKDIKAKEKELQAREAE 60
E+ P N+ VP A+ S+L PLPPEP +D G T+DIPLD T+D+K KEKELQA+EAE
Sbjct: 15 EVNPFANARGVPPASNSRLSPLPPEPVGFDYGRTVDIPLDRAGTQDLKKKEKELQAKEAE 74
Query: 61 LKRREQELKRKEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGM 120
LKRREQ+LKRKEDA ARAGIVIE KNWPPFFP+IHH+I+ EIP+HLQR+QYV F T+LG+
Sbjct: 75 LKRREQDLKRKEDAAARAGIVIEVKNWPPFFPLIHHDIANEIPVHLQRLQYVTFATYLGL 134
Query: 121 LKFLFFS 127
+ LF++
Sbjct: 135 VLCLFWN 141
>At1g11180 secretory carrier membrane protein, putative
Length = 291
Score = 154 bits (389), Expect = 1e-38
Identities = 78/129 (60%), Positives = 101/129 (77%), Gaps = 7/129 (5%)
Query: 6 EMEPARNSGSVPLAT-SKLKPLPPEP--YDRGATIDIPLDT----TKDIKAKEKELQARE 58
E+ P N GSVP A+ S+L PLPPEP + G T+DIPLD +D+K KEKELQA+E
Sbjct: 15 EVNPFANPGSVPAASNSRLSPLPPEPAGFGYGRTVDIPLDRPGSGAQDLKKKEKELQAKE 74
Query: 59 AELKRREQELKRKEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWL 118
A+L+RREQ+LKRK+DA ARAGIVIE KNWP FFP+IHH+I+ EI + LQR+QY+AF T+L
Sbjct: 75 ADLRRREQDLKRKQDAAARAGIVIEAKNWPTFFPLIHHDIANEILVRLQRLQYIAFATYL 134
Query: 119 GMLKFLFFS 127
G++ LF++
Sbjct: 135 GLVLALFWN 143
>At1g32050 unknown protein
Length = 264
Score = 122 bits (307), Expect = 4e-29
Identities = 57/120 (47%), Positives = 86/120 (71%), Gaps = 8/120 (6%)
Query: 10 ARNSGSVPLATSKLKPLPPEPYDRG--ATIDIPLDTTKDIKAKEKELQAREAELKRREQE 67
++ G VP A+ P Y + AT+DIPLD D K+++L EAEL+++E +
Sbjct: 21 SKGGGRVPAASR------PVEYGQSLDATVDIPLDNMNDSSQKQRKLADWEAELRKKEMD 74
Query: 68 LKRKEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGMLKFLFFS 127
+KR+E+AIA+ G+ I++KNWPPFFPIIHH+I+KEIP+H Q++QY+AF +WLG++ L F+
Sbjct: 75 IKRREEAIAKFGVQIDDKNWPPFFPIIHHDIAKEIPVHAQKLQYLAFASWLGIVLCLVFN 134
>At4g27980 putative protein
Length = 565
Score = 35.0 bits (79), Expect = 0.011
Identities = 21/51 (41%), Positives = 33/51 (64%), Gaps = 10/51 (19%)
Query: 45 KDIKAKEKELQAREAELKRREQ--ELKRKEDA--------IARAGIVIEEK 85
KD++A E+ ++ + AELKR+E+ ELK KE+A + R G+ I+EK
Sbjct: 181 KDLRALEEAVKEKTAELKRKEETLELKMKEEAEKLREETELMRKGLEIKEK 231
Score = 31.2 bits (69), Expect = 0.15
Identities = 13/31 (41%), Positives = 23/31 (73%)
Query: 45 KDIKAKEKELQAREAELKRREQELKRKEDAI 75
++ +A+EK+L+A E +K + ELKRKE+ +
Sbjct: 174 EESEAREKDLRALEEAVKEKTAELKRKEETL 204
>At2g18540 putative vicilin storage protein (globulin-like)
Length = 699
Score = 34.3 bits (77), Expect = 0.018
Identities = 16/42 (38%), Positives = 23/42 (54%)
Query: 48 KAKEKELQAREAELKRREQELKRKEDAIARAGIVIEEKNWPP 89
K +E+E RE E KR E+ KR E+ + E++ WPP
Sbjct: 650 KRREEEAMRREEERKREEEAAKRAEEERRKKEEEEEKRRWPP 691
Score = 31.2 bits (69), Expect = 0.15
Identities = 14/32 (43%), Positives = 22/32 (68%)
Query: 42 DTTKDIKAKEKELQAREAELKRREQELKRKED 73
+ T+ K +E+E + RE E KR E+E KR+E+
Sbjct: 465 EETERKKREEEEARKREEERKREEEEAKRREE 496
Score = 30.0 bits (66), Expect = 0.34
Identities = 14/41 (34%), Positives = 26/41 (63%)
Query: 45 KDIKAKEKELQAREAELKRREQELKRKEDAIARAGIVIEEK 85
++ + +E+E + E E KRRE+E K++E+ +A EE+
Sbjct: 475 EEARKREEERKREEEEAKRREEERKKREEEAEQARKREEER 515
Score = 28.9 bits (63), Expect = 0.76
Identities = 12/30 (40%), Positives = 21/30 (70%)
Query: 48 KAKEKELQAREAELKRREQELKRKEDAIAR 77
+ +E+E + RE E ++RE+E KR+E+ R
Sbjct: 541 RREEQERKRREEEARKREEERKREEEMAKR 570
Score = 27.3 bits (59), Expect = 2.2
Identities = 12/25 (48%), Positives = 17/25 (68%)
Query: 48 KAKEKELQAREAELKRREQELKRKE 72
K +E+E + RE E KR E+ KR+E
Sbjct: 548 KRREEEARKREEERKREEEMAKRRE 572
Score = 27.3 bits (59), Expect = 2.2
Identities = 12/28 (42%), Positives = 19/28 (67%)
Query: 45 KDIKAKEKELQAREAELKRREQELKRKE 72
++ + +E+E + E KRREQE +RKE
Sbjct: 552 EEARKREEERKREEEMAKRREQERQRKE 579
Score = 26.2 bits (56), Expect = 5.0
Identities = 12/33 (36%), Positives = 23/33 (69%), Gaps = 3/33 (9%)
Query: 48 KAKEKELQAREAE---LKRREQELKRKEDAIAR 77
K +E+E Q +E E K+RE+E +++E+ +A+
Sbjct: 601 KRREQERQKKEREEMERKKREEEARKREEEMAK 633
Score = 26.2 bits (56), Expect = 5.0
Identities = 13/26 (50%), Positives = 20/26 (76%), Gaps = 1/26 (3%)
Query: 48 KAKEKELQAREAEL-KRREQELKRKE 72
K +E++ + RE E+ KRREQE ++KE
Sbjct: 586 KIREEQERKREEEMAKRREQERQKKE 611
Score = 25.8 bits (55), Expect = 6.5
Identities = 10/26 (38%), Positives = 18/26 (68%)
Query: 48 KAKEKELQAREAELKRREQELKRKED 73
+ +E+E + RE +R E+E KR+E+
Sbjct: 439 RKEEEEARKREEAKRREEEEAKRREE 464
>At4g27610 unknown protein
Length = 334
Score = 33.5 bits (75), Expect = 0.031
Identities = 22/71 (30%), Positives = 37/71 (51%), Gaps = 4/71 (5%)
Query: 9 PARNSGSVPLATSKLK-PLPPEPYDRGATI-DIPL--DTTKDIKAKEKELQAREAELKRR 64
P+ V ATS + LPP PY+ + D P +T+ DI +K + +Q +LKRR
Sbjct: 127 PSSKENGVMWATSPDRLELPPRPYNHNSNCSDSPCVSETSSDIFSKREVIQKLRQQLKRR 186
Query: 65 EQELKRKEDAI 75
+ ++ ++ I
Sbjct: 187 DDMIQEMQEQI 197
>At4g36700 globulin-like protein
Length = 499
Score = 33.1 bits (74), Expect = 0.041
Identities = 18/58 (31%), Positives = 31/58 (53%), Gaps = 1/58 (1%)
Query: 48 KAKEKELQAREAELKRREQELKRKEDAIARAGIVIEEKNWPPFFPIIHHEI-SKEIPL 104
K + E + R E K+ E+E KR+E+ + E+K WPP P E+ +++P+
Sbjct: 430 KKIDDERKRRHDERKKEEEEAKREEEERRKREEEEEKKRWPPQQPPQEEELRERQLPM 487
>At5g66900 disease resistance protein-like
Length = 809
Score = 32.7 bits (73), Expect = 0.053
Identities = 29/110 (26%), Positives = 50/110 (45%), Gaps = 2/110 (1%)
Query: 22 KLKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAELKRREQELKRKEDAIARAGIV 81
K K P D +T++I T+ I + +KEL ELK ++R + A+ + V
Sbjct: 30 KFKAFKPLSKDLVSTMEILFPLTQKIDSMQKELDFGVKELKELRDTIERADVAVRKFPRV 89
Query: 82 --IEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGMLKFLFFSND 129
E+ + I+ ++ K + LQ +Q+ +T LG+ L S D
Sbjct: 90 KWYEKSKYTRKIERINKDMLKFCQIDLQLLQHRNQLTLLGLTGNLVNSVD 139
>At5g57390 putative protein
Length = 555
Score = 32.0 bits (71), Expect = 0.090
Identities = 33/120 (27%), Positives = 46/120 (37%), Gaps = 11/120 (9%)
Query: 5 SEMEPARNSGSVPLATSKLKPLPPEPYDR-GATIDIPLDTTKDIKAKEKELQAREAELKR 63
SE+ PLA S+ P P + + G I T+ E + +R
Sbjct: 168 SEVSSVHKQQPNPLAVSEASPTPKKNVESFGQRTSIYRGVTRHRWTGRYEAHLWDNSCRR 227
Query: 64 REQELKR------KEDAIARAGIVIEEKNWPPF----FPIIHHEISKEIPLHLQRIQYVA 113
Q K KED ARA + K W P FPI ++E E H+ R ++VA
Sbjct: 228 EGQSRKGRQGGYDKEDKAARAYDLAALKYWGPTTTTNFPISNYESELEEMKHMTRQEFVA 287
>At5g61070 HDA18
Length = 682
Score = 31.2 bits (69), Expect = 0.15
Identities = 16/39 (41%), Positives = 23/39 (58%)
Query: 45 KDIKAKEKELQAREAELKRREQELKRKEDAIARAGIVIE 83
K++KAK KEL+A E EL+ ++ +ED I IE
Sbjct: 462 KELKAKNKELEANEKELEAGLMLIRAREDVICGLHAKIE 500
Score = 29.3 bits (64), Expect = 0.59
Identities = 16/48 (33%), Positives = 30/48 (62%), Gaps = 8/48 (16%)
Query: 45 KDIKAKEKELQAREAELKRREQEL--------KRKEDAIARAGIVIEE 84
KD++AKEKEL+AR + RE ++ + +++A+A+A + +E
Sbjct: 554 KDLEAKEKELEARLMLVHAREDKIHAKIERLQQERDEAVAKAERIDKE 601
Score = 26.9 bits (58), Expect = 2.9
Identities = 13/29 (44%), Positives = 19/29 (64%)
Query: 47 IKAKEKELQAREAELKRREQELKRKEDAI 75
I AK K+L+A+E EL+ R + +ED I
Sbjct: 549 IMAKNKDLEAKEKELEARLMLVHAREDKI 577
Score = 26.9 bits (58), Expect = 2.9
Identities = 13/34 (38%), Positives = 23/34 (67%), Gaps = 1/34 (2%)
Query: 49 AKEKELQAREAELKRREQELKRKEDAIARAGIVI 82
A++ EL+AR ELK + +EL+ E + AG+++
Sbjct: 452 ARDGELEARRKELKAKNKELEANEKEL-EAGLML 484
Score = 26.2 bits (56), Expect = 5.0
Identities = 10/24 (41%), Positives = 19/24 (78%)
Query: 46 DIKAKEKELQAREAELKRREQELK 69
+++A+ KEL+A+ EL+ E+EL+
Sbjct: 456 ELEARRKELKAKNKELEANEKELE 479
>At3g29575 unknown protein
Length = 231
Score = 30.8 bits (68), Expect = 0.20
Identities = 19/50 (38%), Positives = 29/50 (58%), Gaps = 2/50 (4%)
Query: 36 TIDIPLDTTKDIKAKEKELQA-REAELKRREQELKRKEDAIARAGIVIEE 84
T +P++T ++ + K KELQ+ R E KR+ E +RK A V+EE
Sbjct: 58 TCSLPVETEEEWR-KRKELQSLRRLEAKRKRSEKQRKHKACGGEEKVVEE 106
>At5g27220 putative protein
Length = 1181
Score = 30.4 bits (67), Expect = 0.26
Identities = 12/34 (35%), Positives = 24/34 (70%)
Query: 44 TKDIKAKEKELQAREAELKRREQELKRKEDAIAR 77
++ I+ K+K+L ARE L +++++LK E +A+
Sbjct: 583 SEKIELKDKKLDAREERLDKKDEQLKSAEQKLAK 616
Score = 26.2 bits (56), Expect = 5.0
Identities = 15/46 (32%), Positives = 26/46 (55%), Gaps = 2/46 (4%)
Query: 42 DTTKDIKAKEKELQAREAELKRREQE--LKRKEDAIARAGIVIEEK 85
D+ KD ++KE EL + L E+E LK+K+ + I +++K
Sbjct: 546 DSLKDFQSKEAELVKLKESLTEHEKELGLKKKQIHVRSEKIELKDK 591
>At2g21410 putative vacuolar proton-ATPase subunit
Length = 821
Score = 30.4 bits (67), Expect = 0.26
Identities = 21/81 (25%), Positives = 36/81 (43%), Gaps = 7/81 (8%)
Query: 21 SKLKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAELKRREQELKRKEDAIARAGI 80
SK P E DR ID+ D++ K +EL+A E+ +L+R + + +
Sbjct: 86 SKAGVTPKETLDRENDIDLD-----DVEVKLEELEAELVEINANNDKLQRSYNELVEYKL 140
Query: 81 VIEEKNWPPFFPIIHHEISKE 101
V+E+ FF H + +
Sbjct: 141 VLEKAG--EFFASAHRSATAQ 159
>At1g14610 valyl tRNA synthetase (valRS)
Length = 1108
Score = 30.4 bits (67), Expect = 0.26
Identities = 25/73 (34%), Positives = 38/73 (51%), Gaps = 11/73 (15%)
Query: 14 GSVPLATSKLKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAELKRREQELKRKED 73
GS ++ S+ K L E +R K+ KAKEKEL+ ++A K R ELK K+
Sbjct: 41 GSRTMSESEKKILTEEELER--------KKKKEEKAKEKELKKQKALEKERLAELKAKQ- 91
Query: 74 AIARAGIVIEEKN 86
A+ G + +K+
Sbjct: 92 --AKDGTNVPKKS 102
>At3g20840 putative transcription factor
Length = 529
Score = 30.0 bits (66), Expect = 0.34
Identities = 21/63 (33%), Positives = 31/63 (48%), Gaps = 4/63 (6%)
Query: 57 REAELKRREQELKRKEDAIARAGIVIEEKNWPPF----FPIIHHEISKEIPLHLQRIQYV 112
RE + ++ Q KED AR+ + K W P FPI ++E E H+ R ++V
Sbjct: 163 REGQSRKGRQGGYDKEDKAARSYDLAALKYWGPSTTTNFPITNYEKEVEEMKHMTRQEFV 222
Query: 113 AFI 115
A I
Sbjct: 223 AAI 225
>At1g68790 putative nuclear matrix constituent protein 1 (NMCP1)
Length = 1085
Score = 30.0 bits (66), Expect = 0.34
Identities = 17/41 (41%), Positives = 28/41 (67%), Gaps = 1/41 (2%)
Query: 46 DIKAKEKELQAREAELKRREQELKRKE-DAIARAGIVIEEK 85
+I KE++L REA L+++E+ +K+KE D AR V E++
Sbjct: 395 EISHKEEKLAKREAALEKKEEGVKKKEKDLDARLKTVKEKE 435
Score = 29.3 bits (64), Expect = 0.59
Identities = 10/38 (26%), Positives = 27/38 (70%)
Query: 48 KAKEKELQAREAELKRREQELKRKEDAIARAGIVIEEK 85
++ ++EL+ ++AE+++ + E+ KE+ +A+ +E+K
Sbjct: 376 RSLDEELEGKKAEIEQLQVEISHKEEKLAKREAALEKK 413
Score = 26.2 bits (56), Expect = 5.0
Identities = 13/40 (32%), Positives = 21/40 (52%)
Query: 45 KDIKAKEKELQAREAELKRREQELKRKEDAIARAGIVIEE 84
K +K KEK L+A E +L + L ++ + + IEE
Sbjct: 429 KTVKEKEKALKAEEKKLHMENERLLEDKECLRKLKDEIEE 468
Score = 25.8 bits (55), Expect = 6.5
Identities = 16/52 (30%), Positives = 29/52 (55%), Gaps = 3/52 (5%)
Query: 22 KLKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAELKRREQELKRKED 73
K+ E ++ +I I L+ DI KEK+ +A +A++ +E+EL E+
Sbjct: 291 KISVAKSELTEKEESIKIKLN---DISLKEKDFEAMKAKVDIKEKELHEFEE 339
>At5g03660 unknown protein
Length = 173
Score = 29.6 bits (65), Expect = 0.45
Identities = 15/48 (31%), Positives = 28/48 (58%), Gaps = 1/48 (2%)
Query: 38 DIPLDTTKDIKAKEKELQAREAELKRREQ-ELKRKEDAIARAGIVIEE 84
++ + KAKE+E++ ++ E++ R Q +L R ED R ++ EE
Sbjct: 38 ELSMKALSAFKAKEEEIEKKKMEIRERVQAQLGRVEDESKRLAMIREE 85
>At3g58370 putative protein
Length = 219
Score = 29.6 bits (65), Expect = 0.45
Identities = 12/41 (29%), Positives = 26/41 (63%)
Query: 46 DIKAKEKELQAREAELKRREQELKRKEDAIARAGIVIEEKN 86
++KAK+K+++ +A L+R E+EL++ +E++N
Sbjct: 163 EVKAKKKKVETGKARLQRAEEELQKLNQKCLELKAFLEKEN 203
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.138 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,070,088
Number of Sequences: 26719
Number of extensions: 131679
Number of successful extensions: 1105
Number of sequences better than 10.0: 119
Number of HSP's better than 10.0 without gapping: 67
Number of HSP's successfully gapped in prelim test: 52
Number of HSP's that attempted gapping in prelim test: 896
Number of HSP's gapped (non-prelim): 221
length of query: 130
length of database: 11,318,596
effective HSP length: 88
effective length of query: 42
effective length of database: 8,967,324
effective search space: 376627608
effective search space used: 376627608
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0041.13


