

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0039.8
(415 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
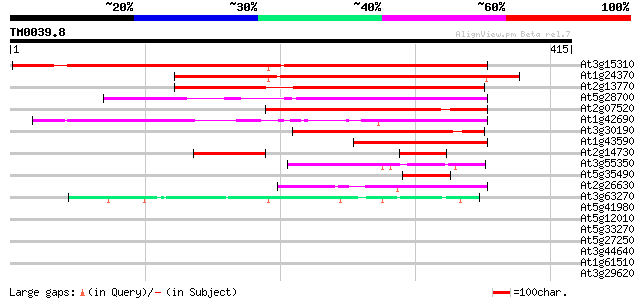
Sequences producing significant alignments: (bits) Value
At3g15310 unknown protein 347 9e-96
At1g24370 hypothetical protein 272 2e-73
At2g13770 hypothetical protein 248 4e-66
At5g28700 putative protein 207 7e-54
At2g07520 pseudogene 197 7e-51
At1g42690 unknown protein 162 3e-40
At3g30190 hypothetical protein 154 7e-38
At1g43590 hypothetical protein 124 1e-28
At2g14730 hypothetical protein 62 8e-10
At3g55350 unknown protein 52 5e-07
At5g35490 putative protein 44 1e-04
At2g26630 En/Spm-like transposon protein 44 2e-04
At3g63270 unknown protein 44 2e-04
At5g41980 unknown protein 40 0.003
At5g12010 putative protein 39 0.004
At5g33270 putative protein 35 0.076
At5g27250 putative protein 35 0.100
At3g44640 putative protein 34 0.13
At1g61510 hypothetical protein 34 0.17
At3g29620 hypothetical protein 32 0.65
>At3g15310 unknown protein
Length = 415
Score = 347 bits (889), Expect = 9e-96
Identities = 178/368 (48%), Positives = 235/368 (63%), Gaps = 27/368 (7%)
Query: 3 PNNMADLDIYDVIIDDIMNDTTIEDMMQEEMEFYQQHANTVRPKRTRKVIKRDREAGNER 62
P N A + +D ++D + +E EE+ + R R R RE G+
Sbjct: 8 PFNDAMEESFDQLLDQQFENVFVELSNSEEV---------AKKNRKRAYFDRKREEGHVL 58
Query: 63 LWKDYFSENPVYTEELFRRRFRMQKHVFLRIVGALRSHNPYFLMSVDEVGRQGLSPLQKC 122
LW DYFS+N ++ + FRRRFRM+K +FLRIV L S +F D GR G SP+QKC
Sbjct: 59 LWNDYFSDNAIFPLQTFRRRFRMKKPLFLRIVDRLSSELMFFQHRRDATGRFGHSPIQKC 118
Query: 123 TAVIRMLVYGSPADSVDEYVRIGESTAIECLNFFVEGVCAVFGETYLRRPNQEDITRLLQ 182
TA IR+L YG +D+VDEY+R+GE+TA+ CL F +G+ + FG+ YLR P ++ RLL
Sbjct: 119 TAAIRLLAYGYASDAVDEYLRMGETTAMSCLENFTKGIISFFGDEYLRAPTATNLRRLLN 178
Query: 183 WGESRGFP-----------------VAWKGQFTRGDHGKPTIMLEAVASQDLWIWHAFFG 225
G+ RGFP AWKGQ+TRG GKPTI+LEA+ASQDLWIWH FFG
Sbjct: 179 IGKIRGFPGMIGSLDCMHWEWKNCPTAWKGQYTRGS-GKPTIVLEAIASQDLWIWHVFFG 237
Query: 226 IAGSNNDINVLNQSSVFNEVLSGNAPMVNFSVNETMYNMRYYLADGIYPPWATFVKTIPM 285
G+ NDIN+L++S +F+++L G AP V + VN Y++ YYL DGIYP WATF+++I +
Sbjct: 238 PPGTLNDINILDRSPIFDDILQGRAPNVKYKVNGREYHLAYYLTDGIYPKWATFIQSIRL 297
Query: 286 PQGEKRQKFMKRQEAARKDVERAFNVLQSRFAIVRGPSRFWHPNEMKSIMYACIILHNMI 345
PQ K F QEA RKDVERAF VLQ+RF I++ P+ W ++ +IM ACIILHNMI
Sbjct: 298 PQNRKATLFATHQEADRKDVERAFGVLQARFHIIKNPALVWDKEKIGNIMKACIILHNMI 357
Query: 346 VEDERNTY 353
VEDER+ Y
Sbjct: 358 VEDERDGY 365
>At1g24370 hypothetical protein
Length = 413
Score = 272 bits (696), Expect = 2e-73
Identities = 143/275 (52%), Positives = 173/275 (62%), Gaps = 21/275 (7%)
Query: 123 TAVIRMLVYGSPADSVDEYVRIGESTAIECLNFFVEGVCAVFGETYLRRPNQEDITRLLQ 182
TA RML YG PADS DEY++IGESTA+E L F + VF YLR P+ D+ RLL
Sbjct: 2 TAAFRMLAYGVPADSTDEYIKIGESTALESLKRFCRAIVEVFACRYLRSPDANDVARLLH 61
Query: 183 WGESRGFP-----------------VAWKGQFTRGDHGKPTIMLEAVASQDLWIWHAFFG 225
GESRGFP AW GQ+ G PTI+LEAVA DLWIWHA+FG
Sbjct: 62 IGESRGFPRMLGSLDCMHWKWKNCPTAWGGQYA-GRSRSPTIILEAVADYDLWIWHAYFG 120
Query: 226 IAGSNNDINVLNQSSVFNEVLSGNAPMVNFSVNETMYNMRYYLADGIYPPWATFVKTIPM 285
+ GSNNDINVL S +F + G AP ++ +NE YNM YYLADGIYP W+T V+TI
Sbjct: 121 LPGSNNDINVLEASHLFANLAEGTAPPASYVINEKPYNMSYYLADGIYPKWSTLVQTIHD 180
Query: 286 PQGEKRQKFMKRQEAARKDVERAFNVLQSRFAIVRGPSRFWHPNEMKSIMYACIILHNMI 345
P+G K++ F +QEA RKDVERAF VLQSRFAIV GPSR W+ + IM +CII+HNMI
Sbjct: 181 PRGPKKKLFAMKQEACRKDVERAFGVLQSRFAIVAGPSRLWNKTVLHDIMTSCIIMHNMI 240
Query: 346 VEDERN---TYRGSARVRIKRAEKTRVVTVRPVVR 377
+EDER+ V I+ E T T+ + R
Sbjct: 241 IEDERDIDAPIEERVEVPIEEVEMTGETTIDVIKR 275
>At2g13770 hypothetical protein
Length = 244
Score = 248 bits (633), Expect = 4e-66
Identities = 125/229 (54%), Positives = 150/229 (64%), Gaps = 19/229 (8%)
Query: 123 TAVIRMLVYGSPADSVDEYVRIGESTAIECLNFFVEGVCAVFGETYLRRPNQEDITRLLQ 182
TA RML YG PADS DEY++IGESTA+E L F + VF YLR P+ D+ RLL
Sbjct: 2 TAAFRMLAYGVPADSTDEYIKIGESTALESLKRFCRAIVEVFACCYLRSPDANDVARLLH 61
Query: 183 WGESRGFPVAWKGQFTRGDHGKPTIMLEAVASQDLWIWHAFFGIAGSNNDINVLNQSSVF 242
GESRGFP EAVA DLWIWHA+FG+ GSNN INVL S +F
Sbjct: 62 IGESRGFP-------------------EAVADYDLWIWHAYFGLPGSNNGINVLEASHLF 102
Query: 243 NEVLSGNAPMVNFSVNETMYNMRYYLADGIYPPWATFVKTIPMPQGEKRQKFMKRQEAAR 302
+ G AP ++ +N YNM YYLADGIYP W+T V+TI P+G K++ F +QEA R
Sbjct: 103 ANLAEGTAPPASYVINGKPYNMGYYLADGIYPKWSTLVQTIHDPRGPKKKLFAMKQEACR 162
Query: 303 KDVERAFNVLQSRFAIVRGPSRFWHPNEMKSIMYACIILHNMIVEDERN 351
KDVERAF VLQ RFAIV GPSR W+ + IM +CII+HNMI+EDER+
Sbjct: 163 KDVERAFGVLQLRFAIVAGPSRLWNKTVLHDIMTSCIIMHNMIIEDERD 211
>At5g28700 putative protein
Length = 292
Score = 207 bits (528), Expect = 7e-54
Identities = 122/284 (42%), Positives = 154/284 (53%), Gaps = 60/284 (21%)
Query: 70 ENPVYTEELFRRRFRMQKHVFLRIVGALRSHNPYFLMSVDEVGRQGLSPLQKCTAVIRML 129
EN Y +FR RFRM K +F+RIV + PYF D GR S LQK TA IRML
Sbjct: 30 ENATYPPHIFRHRFRMNKPLFMRIVERFSNEVPYFKQRRDATGRLDFSALQKSTAAIRML 89
Query: 130 VYGSPADSVDEYVRIGESTAIECLNFFVEGVCAVFGETYLRRPNQEDITRLLQWGESRGF 189
Y G+ A + YLR
Sbjct: 90 AY---------------------------GIAADAVDEYLRI------------------ 104
Query: 190 PVAWKGQFTRGDHGKPTIMLEAVASQDLWIWHAFFGIAGSNNDINVLNQSSVFNEVLSGN 249
G+ T+++ VASQDLWIWHA FG G+ NDINVL++S VF+++L G
Sbjct: 105 -------------GESTLLV--VASQDLWIWHAIFGPPGTLNDINVLDRSPVFDDILQGR 149
Query: 250 APMVNFSVNETMYNMRYYLADGIYPPWATFVKTIPMPQGEKRQKFMKRQEAARKDVERAF 309
A V + VN YN+ YYL DGIYP WATF+++I PQG+K F+ Q+A RKDVERAF
Sbjct: 150 ASKVKYVVNGKDYNLAYYLTDGIYPKWATFIQSISNPQGDKASLFVTTQKACRKDVERAF 209
Query: 310 NVLQSRFAIVRGPSRFWHPNEMKSIMYACIILHNMIVEDERNTY 353
V Q+RF+IV+ P+ F ++ +IM ACIILHNMIVEDER+ Y
Sbjct: 210 GVFQARFSIVKHPALFHDKVKIGNIMRACIILHNMIVEDERDGY 253
>At2g07520 pseudogene
Length = 222
Score = 197 bits (502), Expect = 7e-51
Identities = 95/164 (57%), Positives = 119/164 (71%), Gaps = 6/164 (3%)
Query: 190 PVAWKGQFTRGDHGKPTIMLEAVASQDLWIWHAFFGIAGSNNDINVLNQSSVFNEVLSGN 249
P+AW+GQFTRGD G TI EAVAS DLWIWH FFG A + NDIN L++S VF+++ GN
Sbjct: 19 PIAWEGQFTRGDKGTSTISFEAVASHDLWIWHTFFGCASTLNDINFLDRSPVFDDIEQGN 78
Query: 250 APMVNFSVNETMYNMRYYLADGIYPPWATFVKTIPMPQGEKRQKFMKRQEAARKDVERAF 309
P VNF V + YNM YYLADGIYP + TFVK+I +PQ E + F++ QE RKD+ERAF
Sbjct: 79 TPRVNFFVYQRQYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFVQLQEGCRKDIERAF 138
Query: 310 NVLQSRFAIVRGPSRFWHPNEMKSIMYACIILHNMIVEDERNTY 353
VL +RF I+ W+ ++ IM +CIILHNMIVE+ER+TY
Sbjct: 139 GVLHARFKII------WNIADLAIIMRSCIILHNMIVENERDTY 176
>At1g42690 unknown protein
Length = 333
Score = 162 bits (410), Expect = 3e-40
Identities = 119/339 (35%), Positives = 157/339 (46%), Gaps = 86/339 (25%)
Query: 18 DIMNDTTIEDMMQEEMEFYQQHANTVRPKRTRKVIKRDREAGNERLWKDYFSENPVYTEE 77
D M D + + +E Y V+P++ + I+R+RE G+ +L DYF+ENP Y
Sbjct: 11 DDMFDDKFDQCFDQALESYGNRQR-VKPRKKKLYIERNREEGHIQLVNDYFTENPTYPPH 69
Query: 78 LFRRRFRMQKHVFLRIVGALRSHNPYFLMSVDEVGRQGLSPLQKCTAVIRMLVYGSPADS 137
+FRRRFRM K +F+RIV + PYF D R G S LQK TA IRML YG AD+
Sbjct: 70 IFRRRFRMNKSLFMRIVERFSNEVPYFKQRRDATRRLGFSALQKSTAAIRMLAYGIAADA 129
Query: 138 VDEYVRIGESTAIECLNFFVEGVCAVFGETYLRRPNQEDITRLLQWGESRGFPVAWKGQF 197
YLRRP ++D+ RLL GE
Sbjct: 130 ------------------------------YLRRPTRDDLIRLLHIGEQ----------- 148
Query: 198 TRGDHGKPTIMLEAVASQDLWIWHAFFGIAGSNNDINVLNQSSVFNEVLSGNAPMVNFSV 257
RG G M+ ++ W W N P
Sbjct: 149 -RGFPG----MIGSIDCMH-WEWK---------------------------NCP------ 169
Query: 258 NETMYNMRYYLADG---IYPPWATFVKTIPMPQGEKRQKFMKRQEAARKDVERAFNVLQS 314
T + +Y G I ATF+++I + QG K F QEA RKDVERAF VLQ+
Sbjct: 170 --TAWKGQYTRGSGKPTIVLEAATFIQSISILQGNKASLFATTQEACRKDVERAFGVLQA 227
Query: 315 RFAIVRGPSRFWHPNEMKSIMYACIILHNMIVEDERNTY 353
RFAI++ P+ F ++ +IM ACIILHNMIVED+R+ Y
Sbjct: 228 RFAIIKHPALFHDKVKIGNIMRACIILHNMIVEDKRDGY 266
>At3g30190 hypothetical protein
Length = 263
Score = 154 bits (390), Expect = 7e-38
Identities = 76/142 (53%), Positives = 95/142 (66%), Gaps = 6/142 (4%)
Query: 210 EAVASQDLWIWHAFFGIAGSNNDINVLNQSSVFNEVLSGNAPMVNFSVNETMYNMRYYLA 269
+AVA D+WIWHA+FG+ GSNNDINVL +F + AP ++ N YNM YYLA
Sbjct: 104 QAVADYDMWIWHAYFGLPGSNNDINVLEAYHLFANLAEDTAPPASYVSNGKPYNMGYYLA 163
Query: 270 DGIYPPWATFVKTIPMPQGEKRQKFMKRQEAARKDVERAFNVLQSRFAIVRGPSRFWHPN 329
DGIY W+T V+TI P+G K++ F + E RKDVE AF VL SRFAIV PSR W+
Sbjct: 164 DGIYSKWSTLVQTIHDPRGPKKKLFAMKLETCRKDVEPAFEVLHSRFAIVAEPSRLWNK- 222
Query: 330 EMKSIMYACIILHNMIVEDERN 351
M +CII+HNMI+EDER+
Sbjct: 223 -----MTSCIIMHNMIIEDERD 239
>At1g43590 hypothetical protein
Length = 168
Score = 124 bits (311), Expect = 1e-28
Identities = 60/99 (60%), Positives = 72/99 (72%)
Query: 255 FSVNETMYNMRYYLADGIYPPWATFVKTIPMPQGEKRQKFMKRQEAARKDVERAFNVLQS 314
F VN YN+ YYL D IYP WATF+++I +PQ EK F QEA RKDVERAF VLQ+
Sbjct: 3 FVVNGNEYNLAYYLTDEIYPKWATFIQSISLPQDEKASLFATNQEACRKDVERAFGVLQA 62
Query: 315 RFAIVRGPSRFWHPNEMKSIMYACIILHNMIVEDERNTY 353
RFAIV+ P+ W ++ +IM ACIILHNMIVEDER+ Y
Sbjct: 63 RFAIVKHPALIWDKIKIGNIMRACIILHNMIVEDERDGY 101
>At2g14730 hypothetical protein
Length = 117
Score = 61.6 bits (148), Expect = 8e-10
Identities = 27/53 (50%), Positives = 40/53 (74%)
Query: 137 SVDEYVRIGESTAIECLNFFVEGVCAVFGETYLRRPNQEDITRLLQWGESRGF 189
+VD+Y+RIGE+T + C+ VE + +FG+ YLRRP ++D+ RLL+ GE RGF
Sbjct: 19 AVDKYLRIGENTLMSCMIHSVEAIIYLFGKEYLRRPTRQDLKRLLRIGELRGF 71
Score = 43.5 bits (101), Expect = 2e-04
Identities = 21/35 (60%), Positives = 24/35 (68%)
Query: 289 EKRQKFMKRQEAARKDVERAFNVLQSRFAIVRGPS 323
+K F QE RKDVERAF VLQ+RFAIV P+
Sbjct: 81 DKDSLFATNQEVCRKDVERAFGVLQARFAIVTNPT 115
>At3g55350 unknown protein
Length = 406
Score = 52.4 bits (124), Expect = 5e-07
Identities = 45/155 (29%), Positives = 67/155 (43%), Gaps = 14/155 (9%)
Query: 206 TIMLEAVASQDLWIWHAFFGIAGSNNDINVLNQSSVFNEVLSGNAPM-VNFSVNETMYNM 264
++ L+AV D+ G GS ND VL S + V G ++E
Sbjct: 214 SMTLQAVVDPDMRFLDVIAGWPGSLNDDVVLKNSGFYKLVEKGKRLNGEKLPLSERTELR 273
Query: 265 RYYLADGIYP--PWATFV---KTIPMPQGEKRQKFMKRQEAARKDVERAFNVLQSRFAIV 319
Y + D +P PW K +PQ E F KR A K + A + L+ R+ I+
Sbjct: 274 EYIVGDSGFPLLPWLLTPYQGKPTSLPQTE----FNKRHSEATKAAQMALSKLKDRWRII 329
Query: 320 RGPSRFWHP--NEMKSIMYACIILHNMIVEDERNT 352
G W P N + I++ C +LHN+I++ E T
Sbjct: 330 NGV--MWMPDRNRLPRIIFVCCLLHNIIIDMEDQT 362
>At5g35490 putative protein
Length = 106
Score = 44.3 bits (103), Expect = 1e-04
Identities = 20/36 (55%), Positives = 25/36 (68%)
Query: 291 RQKFMKRQEAARKDVERAFNVLQSRFAIVRGPSRFW 326
++ F +QEA RKDVER F VLQS+FAI+ S W
Sbjct: 69 KKMFAAKQEACRKDVERVFRVLQSKFAIIARSSNCW 104
>At2g26630 En/Spm-like transposon protein
Length = 292
Score = 43.9 bits (102), Expect = 2e-04
Identities = 39/167 (23%), Positives = 69/167 (40%), Gaps = 24/167 (14%)
Query: 199 RGDHGKPTIMLEAVASQDLWIWHAFFGIAGSNNDINVLNQSSVFNEVLSGNAPMVNFSVN 258
+G +PT+ + A+ + D+ +A+ G+ G +D VLN + NE + P
Sbjct: 66 KGRKLEPTMNVLAICNFDMKFIYAYVGVPGRAHDTKVLNYCAT-NEPYFSHPP------- 117
Query: 259 ETMYNMRYYLADGIYPPWATFVKTIPM------------PQGEKRQKFMKRQEAARKDVE 306
N +YYL D YP ++ P R+ F ++ R +E
Sbjct: 118 ----NGKYYLVDSGYPTRTGYLGPHRRMRYHLGQFGRGGPPVTARELFNRKHSGLRSVIE 173
Query: 307 RAFNVLQSRFAIVRGPSRFWHPNEMKSIMYACIILHNMIVEDERNTY 353
R F V ++++ IV + + I+ A + LHN I + R +
Sbjct: 174 RTFGVWKAKWRIVDRKHPKYGLAKWIKIVTATMALHNFIRDSHREDH 220
>At3g63270 unknown protein
Length = 396
Score = 43.5 bits (101), Expect = 2e-04
Identities = 78/340 (22%), Positives = 131/340 (37%), Gaps = 48/340 (14%)
Query: 44 RPKRTRKVIKRDREAGNERLWKDYFSEN-----PVYTEELFRRRFRMQKHVFLRIVGALR 98
+ K+ + D EA + W ++ N P + F+ FR K F I +R
Sbjct: 25 KEKKRVNAVPLDPEAIDCDWWDTFWLRNSSPSVPSDEDYAFKHFFRASKTTFSYICSLVR 84
Query: 99 ----SHNPYFLMSVDEVGRQGLSPLQKCTAVIRMLVYGSPADSVDEYVRIGESTAIECLN 154
S P L++++ GR LS ++ +R L G SV +G+ST +
Sbjct: 85 EDLISRPPSGLINIE--GRL-LSVEKQVAIALRRLASGDSQVSVGAAFGVGQSTVSQVTW 141
Query: 155 FFVEGVCAVFGETYLRRPNQEDITRLL-QWGESRGFP---------------VAWKGQFT 198
F+E + + +LR P+ + I + ++ E G P A +
Sbjct: 142 RFIEAL-EERAKHHLRWPDSDRIEEIKSKFEEMYGLPNCCGAIDTTHIIMTLPAVQASDD 200
Query: 199 RGDHGKP-TIMLEAVASQDLWIWHAFFGIAGSNNDINVLNQSSVFN-----EVLSGNAPM 252
D K ++ L+ V ++ + G G +L S F ++L GN
Sbjct: 201 WCDQEKNYSMFLQGVFDHEMRFLNMVTGWPGGMTVSKLLKFSGFFKLCENAQILDGNPKT 260
Query: 253 VNFSVNETMYNMRYYLADGI-YP--PWATFVKTIPMPQGEKRQKFMKRQEAARKDVERAF 309
++ +R Y+ GI YP PW P + F +R E R AF
Sbjct: 261 LSQGAQ-----IREYVVGGISYPLLPWLITPHDSDHPS-DSMVAFNERHEKVRSVAATAF 314
Query: 310 NVLQSRFAIVRGPSRFWHPNEMK--SIMYACIILHNMIVE 347
L+ + I+ W P+ K SI+ C +LHN+I++
Sbjct: 315 QQLKGSWRILS--KVMWRPDRRKLPSIILVCCLLHNIIID 352
>At5g41980 unknown protein
Length = 374
Score = 39.7 bits (91), Expect = 0.003
Identities = 38/156 (24%), Positives = 64/156 (40%), Gaps = 19/156 (12%)
Query: 194 KGQFTRGDHGKPTIMLEAVASQDLWIWHAFFGIAGSNNDINVLNQSSVFNEVLSGNAPMV 253
+G F G+ G T + A +S DL + G GS +D VLN + L
Sbjct: 161 QGPFRNGN-GLLTQNVLAASSFDLRFNYVLAGWEGSASDQQVLNAALTRRNKLQ------ 213
Query: 254 NFSVNETMYNMRYYLADGIYPPWATFVKTI----PMPQGEKRQKFMKRQEAARKDVERAF 309
+ +YY+ D YP F+ + E ++ F +R + + + R F
Sbjct: 214 -------VPQGKYYIVDNKYPNLPGFIAPYHGVSTNSREEAKEMFNERHKLLHRAIHRTF 266
Query: 310 NVLQSRFAIVRGPSRFWHPNEMKSIMYACIILHNMI 345
L+ RF I+ + ++K ++ AC LHN +
Sbjct: 267 GALKERFPILLSAPPYPLQTQVKLVIAAC-ALHNYV 301
>At5g12010 putative protein
Length = 502
Score = 39.3 bits (90), Expect = 0.004
Identities = 62/294 (21%), Positives = 109/294 (36%), Gaps = 38/294 (12%)
Query: 74 YTEELFRRRFRMQKHVFLRIVGALRSHNPYFLMSVDEVGRQGLSPLQKCTAVIRMLVYGS 133
Y EE F++ FRM K F I L S + D R + Q+ I L G
Sbjct: 170 YPEEDFKKAFRMSKSTFELICDELNS----AVAKEDTALRNAIPVRQRVAVCIWRLATGE 225
Query: 134 PADSVDEYVRIGESTAIECLNFFVEGVCAVFGETYLRRPNQEDITRLLQWGES-RGFPVA 192
P V + +G ST + + + + V YL+ P+ E + + + ES G P
Sbjct: 226 PLRLVSKKFGLGISTCHKLVLEVCKAIKDVLMPKYLQWPDDESLRNIRERFESVSGIPNV 285
Query: 193 WKGQFT----------------------RGDHGKPTIMLEAVASQDLWIWHAFFGIAGSN 230
+T R +I ++AV + G GS
Sbjct: 286 VGSMYTTHIPIIAPKISVASYFNKRHTERNQKTSYSITIQAVVNPKGVFTDLCIGWPGSM 345
Query: 231 NDINVLNQSSVFNEVLSGNAPMVNFSVNETMYNMRYYLADGIYPPWATFVKTIPMPQGEK 290
D VL +S ++ +G + + L D + P+ + + Q
Sbjct: 346 PDDKVLEKSLLYQRANNGGLLKGMWVAG----GPGHPLLDWVLVPYTQ--QNLTWTQHAF 399
Query: 291 RQKFMKRQEAARKDVERAFNVLQSRFAIVRGPSRFWHPNEMKSIMYACIILHNM 344
+K + Q A++ AF L+ R+A ++ + ++ +++ AC +LHN+
Sbjct: 400 NEKMSEVQGVAKE----AFGRLKGRWACLQKRTEV-KLQDLPTVLGACCVLHNI 448
>At5g33270 putative protein
Length = 343
Score = 35.0 bits (79), Expect = 0.076
Identities = 33/164 (20%), Positives = 67/164 (40%), Gaps = 24/164 (14%)
Query: 199 RGDHGKPTIMLEAVASQDLWIWHAFFGIAGSNNDINVLNQSSVFNEVLSGNAPMVNFSVN 258
RG + T+ + A+ + + +A+ G+ G +D VL + +E + P
Sbjct: 138 RGRKSEATMNILALCNFSMKFTYAYVGVPGRAHDTKVLTYCAT-HEASFPHPPAG----- 191
Query: 259 ETMYNMRYYLADGIYPPWATFVKTIPM------------PQGEKRQKFMKRQEAARKDVE 306
+YYL D YP + ++ P R+ F +R + R +E
Sbjct: 192 ------KYYLVDSGYPTRSGYLGPHRRTRYHLELFNRGGPPTNSRELFNRRHSSLRSVIE 245
Query: 307 RAFNVLQSRFAIVRGPSRFWHPNEMKSIMYACIILHNMIVEDER 350
R F V ++++ I+ + + I+ + + LHN I + ++
Sbjct: 246 RTFGVWKAKWRILDRKHLKYEVKKWIKIVTSTMALHNYIRDSQQ 289
>At5g27250 putative protein
Length = 348
Score = 34.7 bits (78), Expect = 0.100
Identities = 26/94 (27%), Positives = 42/94 (44%), Gaps = 14/94 (14%)
Query: 265 RYYLADGIYPPWATFVKTIPMPQ-------GEKR------QKFMKRQEAARKDVERAFNV 311
+YYLAD +P F+ + + GE R + F R + R +ER F +
Sbjct: 189 KYYLADCGFPNRRNFLAPLRSTRYHLQDFRGEGRDPTNQNELFNLRHASLRNVIERIFGI 248
Query: 312 LQSRFAIVRGPSRFWHPNEMKSIMYACIILHNMI 345
+SRF I + F + + I+ +C LHN +
Sbjct: 249 FKSRFLIFKSAPPFSFKTQAE-IVLSCAALHNFL 281
>At3g44640 putative protein
Length = 377
Score = 34.3 bits (77), Expect = 0.13
Identities = 74/362 (20%), Positives = 135/362 (36%), Gaps = 63/362 (17%)
Query: 8 DLDIYDVIIDDIMNDTTIEDMMQEEMEFYQQHANTVRPKRTRKVIKRDREAGNERLWKDY 67
DL YDV + ++ T+ +MM ME + + K+ D R+ +Y
Sbjct: 4 DLSFYDVFFNIMLCATS--NMMVFNMENIGDEEENLNDRE--KIEAEDVRRSVSRVGYNY 59
Query: 68 FSENPVYTEELFRRRFRMQKHVFLRIVGALRSHNPYFLMSVDEVGRQGLSPLQKCTAVIR 127
FR+ +RM+ VF ++ + +N + + L K ++
Sbjct: 60 IQTALTVNPAHFRQLYRMEPEVFHKLCDVILLYNGH-------ISEIKLFYKHKFHKALK 112
Query: 128 MLVYGSPADSVDEYVR----IGESTAIECLNFFVEGVCAVFGETYLRRPNQEDITRLLQW 183
L + +P VR I EST+ +F + + A+ G I ++
Sbjct: 113 ALAFVAPNWMAKPEVRVPAKIRESTSF--YPYFKDCIGAIDGT---------HIFAMVPT 161
Query: 184 GESRGFPVAWKGQFTRGDHGKPTIMLEAVASQDLWIWHAFFGIAGSNNDINVLNQSSVFN 243
++ F R G + + A + DL + G GS +D VLN + N
Sbjct: 162 CDAASF---------RNRKGFISQNVLAACNFDLQFIYILSGWKGSAHDSKVLNDALTRN 212
Query: 244 ----EVLSGNAPMVNFSV----------NETMYNMRYYLADGIYPPWATFVKTIPMPQGE 289
+V G +V+ T Y+++ + G P KT
Sbjct: 213 TNRLQVPEGKFYLVDCGYANRRKFLAPFRRTRYHLQDFRGQGRDP------KT------- 259
Query: 290 KRQKFMKRQEAARKDVERAFNVLQSRFAIVRGPSRFWHPNEMKSIMYACIILHNMIVEDE 349
+ + F R + R +ER F + +SRF I + + + + ++ AC LHN + ++
Sbjct: 260 QNELFNLRHASLRNVIERIFGIFKSRFLIFKSAPPYPFKTQTELVL-ACAGLHNFLHQEC 318
Query: 350 RN 351
R+
Sbjct: 319 RS 320
>At1g61510 hypothetical protein
Length = 608
Score = 33.9 bits (76), Expect = 0.17
Identities = 41/208 (19%), Positives = 79/208 (37%), Gaps = 42/208 (20%)
Query: 155 FFVEGVCAVFGETYLRRPNQEDITRLLQWGESRGFPVAWKGQFTRGDHGKPTIMLEAVAS 214
+F++ + A+ G RP D+ R RG + T+ + AV +
Sbjct: 377 YFIDCIGALDGTHVSVRPPSGDVERY------------------RGRKSEATMNILAVCN 418
Query: 215 QDLWIWHAFFGIAGSNNDINVLNQSSVFNEVLSGNAPMVNFSVNETMYNMRYYLADGIYP 274
+ A+ G+ G +D VL + +E + P +YYL D YP
Sbjct: 419 FSMKFNIAYVGVPGRAHDTKVLTYCAT-HEASFPHPPAG-----------KYYLVDSGYP 466
Query: 275 PWATFVKTIPM------------PQGEKRQKFMKRQEAARKDVERAFNVLQSRFAIVRGP 322
+ ++ P R+ F +R + R +ER F V ++++ I+
Sbjct: 467 TRSGYLGPHRRTRYHLELFNRGGPPTNSRELFNRRHSSLRSVIERTFGVWKAKWRILDRK 526
Query: 323 SRFWHPNEMKSIMYACIILHNMIVEDER 350
+ + I+ + + LHN I + ++
Sbjct: 527 HPKYEVKKWIKIVTSTMALHNYIRDSQQ 554
>At3g29620 hypothetical protein
Length = 222
Score = 32.0 bits (71), Expect = 0.65
Identities = 33/148 (22%), Positives = 60/148 (40%), Gaps = 27/148 (18%)
Query: 211 AVASQDLWIWHAFFGIAGSNNDINVLNQSSVFNEVLSGNAPMVNFSVNETMYNMRYYLAD 270
AV + D+ + + G+ GS +D VL+ + + G+ + + +YYL D
Sbjct: 31 AVCNFDMLFTYIYVGVFGSAHDTKVLSLA------MEGDPNFPHPHIG------KYYLVD 78
Query: 271 GIYPPWATFVKTIPM------------PQGEKRQKFMKRQEAARKDVERAFNVLQSRFAI 318
Y ++ P ++KF R R +ER F V + ++ I
Sbjct: 79 SGYALRRGYLGPFRQTWYHHNQFQNQAPPNNHKEKFNWRHSLLRCVIERTFGVWKGKWRI 138
Query: 319 VRGPSRFWHP-NEMKSIMYACIILHNMI 345
++ R W+ + IM A + LHN +
Sbjct: 139 MQ--DRAWYNIVTTRKIMVATMALHNFV 164
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,419,663
Number of Sequences: 26719
Number of extensions: 399398
Number of successful extensions: 925
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 893
Number of HSP's gapped (non-prelim): 31
length of query: 415
length of database: 11,318,596
effective HSP length: 102
effective length of query: 313
effective length of database: 8,593,258
effective search space: 2689689754
effective search space used: 2689689754
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0039.8


