

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0039.7
(754 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
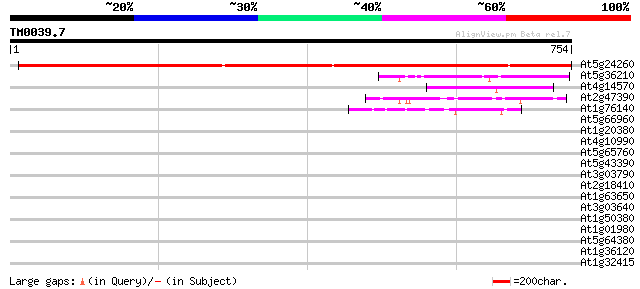
Sequences producing significant alignments: (bits) Value
At5g24260 dipeptidyl peptidase IV-like protein 1068 0.0
At5g36210 acyl-peptide hydrolase-like 68 2e-11
At4g14570 acylamino acid-releasing enzyme (aare) 59 1e-08
At2g47390 unknown protein 59 1e-08
At1g76140 prolyl endopeptidase like protein 44 3e-04
At5g66960 protease-like 41 0.002
At1g20380 hypothetical protein 41 0.003
At4g10990 putative retrotransposon polyprotein 33 0.77
At5g65760 lysosomal Pro-X carboxypeptidase 31 2.2
At5g43390 putative protein 31 2.2
At3g03790 unknown protein 30 3.8
At2g18410 hypothetical protein 30 3.8
At1g63650 putative transcription factor (BHLH2) 30 3.8
At3g03640 beta-glucosidase 30 5.0
At1g50380 oligopeptidase-like protein 30 5.0
At1g01980 hypothetical protein 30 5.0
At5g64380 fructose-bisphosphatase-like protein 29 8.5
At1g36120 putative reverse transcriptase gb|AAD22339.1 29 8.5
At1g32415 unknown protein 29 8.5
>At5g24260 dipeptidyl peptidase IV-like protein
Length = 746
Score = 1068 bits (2761), Expect = 0.0
Identities = 518/748 (69%), Positives = 622/748 (82%), Gaps = 10/748 (1%)
Query: 13 DDNILFPVEEIVQYPLPGYVSPSSINFSPDDSLISYLYSPEQTLNRKVFAFNVKTKAQEL 72
D ++ F VE+IVQ PLPGYV+P++++FSPDDSLI+YL+SPE+ L R+V+AF+V L
Sbjct: 3 DKDVPFGVEDIVQTPLPGYVAPTAVSFSPDDSLITYLFSPEKNLKRRVYAFDVNKGESNL 62
Query: 73 LFSPPDGGLDESNISPEEKLRRERLRERGLGVTRYEWVKTSSKRKAVMVPLPAGIYIQDI 132
+FSPPDGG+DESNISPEEKLRRERLRERGLGVTRYEWVKT+SK + ++VPLPAG+Y++D+
Sbjct: 63 VFSPPDGGVDESNISPEEKLRRERLRERGLGVTRYEWVKTNSKMRFIVVPLPAGVYMKDL 122
Query: 133 SQS-KAELKLSSIPSSPIIDPHLSPDGSMLAYVRDHELHVLNLFTNESKQLTHGAKENGL 191
S S EL + S P+SPIIDP LSP+G LAYVR+ ELHVLNL N+++QLT GA + L
Sbjct: 123 SSSPNPELIVPSSPTSPIIDPRLSPNGLFLAYVRESELHVLNLLKNQTQQLTSGANGSTL 182
Query: 192 TRGLAEYIAQEEMDRKNGYWWSLDSKYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQEDHP 251
+ GLAEYIAQEEMDR+NGYWWSLDSK+IA+TEVDSS+IPLFRIMHQGK SVG EAQEDH
Sbjct: 183 SHGLAEYIAQEEMDRRNGYWWSLDSKFIAYTEVDSSQIPLFRIMHQGKRSVGSEAQEDHA 242
Query: 252 YPFAGASNVKVRLGVVSAAGSS-ITWMDLLCGGKEQQANEEEYLARVNWMHGNTLTAQVL 310
YPFAGA N +RLGVVS+AG TWM+L+CGG+ E+EYL RVNW+ GN L QVL
Sbjct: 243 YPFAGALNSTLRLGVVSSAGGGKTTWMNLVCGGRGN--TEDEYLGRVNWLPGNVLIVQVL 300
Query: 311 NRHHTKIKILKFDIRTGQGKNILVEENSTWINIHDCFTPLDK-GITKFSGGFIWASEKTG 369
NR +K+KI+ FDI TGQG +L EE+ TW+ +HDCFTPL+K ++ SGGFIWASE+TG
Sbjct: 301 NRSQSKLKIISFDINTGQGNVLLTEESDTWVTLHDCFTPLEKVPSSRGSGGFIWASERTG 360
Query: 370 FRHLYLHDANGTCLGPITEGEWMVEQIAGVNEATGLLYFTGTLDGPLESNLYCAKLYVDG 429
FRHLYL++++GTCLG IT GEWMVEQIAGVNE L+YFTGTLDGPLE+NLYCAKL
Sbjct: 361 FRHLYLYESDGTCLGAITSGEWMVEQIAGVNEPMSLVYFTGTLDGPLETNLYCAKLEAGN 420
Query: 430 THPPEAPTRLTLNKGKHIVVLDHHMQSFVDIYDSLGCPPRVLLCSLEDGRVIMPLFEQPF 489
T + P RLT KGKHIVVLDH M++FVDI+DS+ PPRV LCSL DG V+ L+EQ
Sbjct: 421 TSRCQ-PMRLTHGKGKHIVVLDHQMKNFVDIHDSVDSPPRVSLCSLSDGTVLKILYEQTS 479
Query: 490 SVPRFKKLQLEPPEIVEIQANDG-TVLYGALYKPDASRFGPPPYKTMINVYGGPSVQLVS 548
+ K L+LEPPE V+IQANDG T LYGA+YKPD+S+FGPPPYKTMINVYGGPSVQLV
Sbjct: 480 PIQILKSLKLEPPEFVQIQANDGKTTLYGAVYKPDSSKFGPPPYKTMINVYGGPSVQLVY 539
Query: 549 NSWLNTVDMRAQYLRNQGILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLV 608
+SW+NTVDMR QYLR++GILVWKLDNRGTARRGLKFES++KH G VDA+DQ+TGA+WL+
Sbjct: 540 DSWINTVDMRTQYLRSRGILVWKLDNRGTARRGLKFESWMKHNCGYVDAEDQVTGAKWLI 599
Query: 609 KQGLAKAGHIGLYGWSYGGYLSAMTLSRYPDFFKCAVAGAPVTSWDGYDTFYTEKYMGLP 668
+QGLAK HIG+YGWSYGGYLSA L+RYP+ F CAV+GAPVTSWDGYD+FYTEKYMGLP
Sbjct: 600 EQGLAKPDHIGVYGWSYGGYLSATLLTRYPEIFNCAVSGAPVTSWDGYDSFYTEKYMGLP 659
Query: 669 SENQSEYESGSVMNQVHKL--KGRLLLVHGMIDENVHFRHTARLINALVAAGKPYELVLF 726
+E + Y SVM+ V L K +L+LVHGMIDENVHFRHTARL+NALV AGK YEL++F
Sbjct: 660 TE-EERYLKSSVMHHVGNLTDKQKLMLVHGMIDENVHFRHTARLVNALVEAGKRYELLIF 718
Query: 727 PDERHMPRRHSDRVYMEERMWEFIDRNL 754
PDERHMPR+ DR+YME+R+WEFI++NL
Sbjct: 719 PDERHMPRKKKDRIYMEQRIWEFIEKNL 746
>At5g36210 acyl-peptide hydrolase-like
Length = 678
Score = 67.8 bits (164), Expect = 2e-11
Identities = 68/268 (25%), Positives = 116/268 (42%), Gaps = 21/268 (7%)
Query: 496 KLQLEPPEIVEIQAN-DGTVLYGALYKP-----DASRFGPPPYKTMINVYGGPSVQLVSN 549
K PE++E G Y Y P +AS PP ++ +GGP+ + S
Sbjct: 400 KAYFSVPELIEFPTEVPGQNAYAYFYPPTNPLYNASMEEKPPL--LVKSHGGPTAE--SR 455
Query: 550 SWLNTVDMRAQYLRNQGILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVK 609
LN + QY ++G ++ G+ G ++ L + G VD DD A++LV
Sbjct: 456 GSLN---LNIQYWTSRGWAFVDVNYGGSTGYGREYRERLLRQWGIVDVDDCCGCAKYLVS 512
Query: 610 QGLAKAGHIGLYGWSYGGYLSAMTLSRYPDFFKC-----AVAGAPVTSWDGYDTFYTEKY 664
G A + + G S GGY + +L+ + D FK VA + +G+ + +Y
Sbjct: 513 SGKADVKRLCISGGSAGGYTTLASLA-FRDVFKAGASLYGVADLKMLKEEGHK--FESRY 569
Query: 665 MGLPSENQSEYESGSVMNQVHKLKGRLLLVHGMIDENVHFRHTARLINALVAAGKPYELV 724
+ ++ ++ S +N V K ++L G+ D+ V + ++ AL G P LV
Sbjct: 570 IDNLVGDEKDFYERSPINFVDKFSCPIILFQGLEDKVVTPDQSRKIYEALKKKGLPVALV 629
Query: 725 LFPDERHMPRRHSDRVYMEERMWEFIDR 752
+ E+H R+ + Y E+ F R
Sbjct: 630 EYEGEQHGFRKAENIKYTLEQQMVFFAR 657
>At4g14570 acylamino acid-releasing enzyme (aare)
Length = 764
Score = 58.9 bits (141), Expect = 1e-08
Identities = 46/185 (24%), Positives = 82/185 (43%), Gaps = 14/185 (7%)
Query: 561 YLRNQGILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGLAKAGHIGL 620
YL + G ++ RG+ G L K+G D D L + ++ G+A I +
Sbjct: 555 YLSSIGYSQLIINYRGSLGYGEDALQSLPGKVGSQDVKDCLLAVDHAIEMGIADPSRITV 614
Query: 621 YGWSYGGYLSAMTLSRYPDFFKCAVAGAPVTS------------WDGYDTFYTEK-YMGL 667
G S+GG+L+ + + PD F A A PV + W ++ + + Y
Sbjct: 615 LGGSHGGFLTTHLIGQAPDKFVAAAARNPVCNMASMVGITDIPDWCFFEAYGDQSHYTEA 674
Query: 668 PS-ENQSEYESGSVMNQVHKLKGRLLLVHGMIDENVHFRHTARLINALVAAGKPYELVLF 726
PS E+ S + S ++ + K+K L + G D V + + + AL G ++++F
Sbjct: 675 PSAEDLSRFHQMSPISHISKVKTPTLFLLGTKDLRVPISNGFQYVRALKEKGVEVKVLVF 734
Query: 727 PDERH 731
P++ H
Sbjct: 735 PNDNH 739
>At2g47390 unknown protein
Length = 961
Score = 58.5 bits (140), Expect = 1e-08
Identities = 75/296 (25%), Positives = 123/296 (41%), Gaps = 51/296 (17%)
Query: 479 RVIMPLFEQPFSVPRFKKLQLEPPEIVEIQANDGTVLYGALYKP---DASRFGPPP---- 531
R + + P P+ LQ E++ Q DG L LY P D S+ GP P
Sbjct: 633 RKVQQITNFPHPYPQLASLQ---KEMIRYQRKDGVQLTATLYLPPGYDPSKDGPLPCLFW 689
Query: 532 -----YKTMI---NVYGGPS--VQLVSNSWLNTVDMRAQYLRNQGI-LVWKLDNRGTARR 580
+K+ V G P+ + S S L + R L I ++ + D R
Sbjct: 690 SYPGEFKSKDAAGQVRGSPNEFAGIGSTSALLWLARRFAILSGPTIPIIGEGDEEANDR- 748
Query: 581 GLKFESYLKHKLGQVDADDQLTGAEWLVKQGLAKAGHIGLYGWSYGGYLSAMTLSRYPDF 640
Y++ + +A E +V++G+A I + G SYG +++A L+ P
Sbjct: 749 ------YVEQLVASAEA-----AVEEVVRRGVADRSKIAVGGHSYGAFMTANLLAHAPHL 797
Query: 641 FKCAVAGAPVTSWDGYDTFYTEKYMGLPSENQSEYESGSVMNQV------HKLKGRLLLV 694
F C +A + Y+ T G +E+++ +E+ +V ++ +K+K +LL+
Sbjct: 798 FACGIARS-----GAYNRTLTP--FGFQNEDRTLWEATNVYVEMSPFMSANKIKKPILLI 850
Query: 695 HGMIDEN--VHFRHTARLINALVAAGKPYELVLFPDERHMPRRHSDRVYMEERMWE 748
HG D N + R NAL G LV+ P E H +S R + +WE
Sbjct: 851 HGEEDNNPGTLTMQSDRFFNALKGHGALCRLVVLPHESH---GYSARESIMHVLWE 903
>At1g76140 prolyl endopeptidase like protein
Length = 731
Score = 43.9 bits (102), Expect = 3e-04
Identities = 54/244 (22%), Positives = 104/244 (42%), Gaps = 26/244 (10%)
Query: 456 SFVDIYDSLGCPPRVLLCSLEDGRVIMPLFEQPFSVPRFKKLQLEPPEIVEIQANDGTVL 515
+F + S P + C L + + +F + +VP F + + ++ + DGT +
Sbjct: 407 TFFFSFTSFLTPGVIYKCDLANESPEVKVFRE-VTVPGFDREAFQAIQVF-YPSKDGTKI 464
Query: 516 -YGALYKPDASRFGPPPYKTMINVYGGPSVQLVSNSWLNTVDMRAQYLRNQGILVWKLDN 574
+ K D G P ++ YGG ++ + + + + + ++ G++ +
Sbjct: 465 PMFIVAKKDIKLDGSHP--CLLYAYGGFNISITPSFSASRIVLS----KHLGVVFCFANI 518
Query: 575 RGTARRGLKFESYLKHKLGQVDA-----DDQLTGAEWLVKQGLAKAGHIGLYGWSYGGYL 629
RG G ++ HK G + DD ++GAE+LV G + + + G S GG L
Sbjct: 519 RGGGEYGEEW-----HKAGSLAKKQNCFDDFISGAEYLVSAGYTQPSKLCIEGGSNGGLL 573
Query: 630 SAMTLSRYPDFFKCAVAGAPVTSWDGYDTF-----YTEKYMGLPSENQSEYESGSVMNQV 684
+++ PD + CA+A V + F +T Y SEN+ E+ + +
Sbjct: 574 VGACINQRPDLYGCALAHVGVMDMLRFHKFTIGHAWTSDYG--CSENEEEFHWLIKYSPL 631
Query: 685 HKLK 688
H +K
Sbjct: 632 HNVK 635
>At5g66960 protease-like
Length = 792
Score = 41.2 bits (95), Expect = 0.002
Identities = 40/190 (21%), Positives = 85/190 (44%), Gaps = 23/190 (12%)
Query: 506 EIQANDG-----TVLYGALYKPDASRFGPPPYKTMINVYGGPSVQLVSNSWLNTVDMRAQ 560
E+ ++DG +++Y K + + G +++V+G +++ W + + +
Sbjct: 535 EVSSHDGAMVPLSIVYSRAQKEENQKPG------LLHVHGAYG-EMLDKRWRSEL----K 583
Query: 561 YLRNQGILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGLAKAGHIGL 620
L ++G ++ D RG +G K+ + D + A++LV+ + + +
Sbjct: 584 SLLDRGWVLAYADVRGGGGKGKKWHQDGRGAKKLNSIKDYIQCAKYLVENNIVEENKLAG 643
Query: 621 YGWSYGGYLSAMTLSRYPDFFKCAVAGAPVTSWDGYDTFYTEKYMGLP--SENQSEYESG 678
+G+S GG + A ++ PD F+ AV P D +T Y LP +E+ E+
Sbjct: 644 WGYSAGGLVVASAINHCPDLFQAAVLKVPF-----LDPTHTLIYPILPLTAEDYEEFGYP 698
Query: 679 SVMNQVHKLK 688
+N H ++
Sbjct: 699 GDINDFHAIR 708
>At1g20380 hypothetical protein
Length = 731
Score = 40.8 bits (94), Expect = 0.003
Identities = 46/192 (23%), Positives = 81/192 (41%), Gaps = 19/192 (9%)
Query: 461 YDSLGCPPRVLLCSLEDGRVIMPLFEQPFSVPRFKKLQLEPPEIVEIQANDGT-VLYGAL 519
+ S P + +C L + +F + VP F + + ++ + DGT + +
Sbjct: 412 FTSFLTPGVIYICDLSHEAPEVTVFRE-IGVPGFDRTAFQVTQVF-YPSKDGTDIPMFIV 469
Query: 520 YKPDASRFGPPPYKTMINVYGGPSVQLVSNSWLNTVDMRAQYLRNQGILVWKLDNRGTAR 579
+ D G P ++ YGG S+ + + + R+ G + + RG
Sbjct: 470 ARKDIKLDGSHP--CLLYAYGGFSISMTPFFSATRIVLG----RHLGTVFCFANIRGGGE 523
Query: 580 RGLKFESYLKHKLG-----QVDADDQLTGAEWLVKQGLAKAGHIGLYGWSYGGYLSAMTL 634
G ++ HK G Q DD ++GAE+LV G + + + G S GG L +
Sbjct: 524 YGEEW-----HKSGALANKQNCFDDFISGAEYLVSAGYTQPRKLCIEGGSNGGILVGACI 578
Query: 635 SRYPDFFKCAVA 646
++ PD F CA+A
Sbjct: 579 NQRPDLFGCALA 590
>At4g10990 putative retrotransposon polyprotein
Length = 1203
Score = 32.7 bits (73), Expect = 0.77
Identities = 44/180 (24%), Positives = 71/180 (39%), Gaps = 23/180 (12%)
Query: 133 SQSKAELKLSSIPSSPIIDPHLSPDGSMLAYVR----DHELHVLNLFTNESKQLTHGAKE 188
+ + E SSIP I P+ P + ++Y + H E K T K
Sbjct: 477 TSTSIETPSSSIPPKKITTPY--PMSTAISYDKLTPLFHSYICAYNVETEPKAFTQAMKS 534
Query: 189 NGLTRGLAE----------YIAQEEMDRKN--GYWWSLDSKYIAFTEVDSSEIPLFRIMH 236
TR E +I + + KN G W KY ++ + R++
Sbjct: 535 EKWTRAANEELHALEQNKTWIVESLTEGKNVVGCKWVFTIKYNPDGSIERYKA---RLVA 591
Query: 237 QG-KSSVGLEAQEDHPYPFAGASNVKVRLGVVSAAGSSITWMDLLCGGKEQQANEEEYLA 295
QG G++ E P A +VK+ LG+ +A G S+T MD+ + +EE Y++
Sbjct: 592 QGFTQQEGIDYMETFS-PVAKFGSVKLLLGLAAATGWSLTQMDVSNAFLHGELDEEIYMS 650
>At5g65760 lysosomal Pro-X carboxypeptidase
Length = 515
Score = 31.2 bits (69), Expect = 2.2
Identities = 24/73 (32%), Positives = 37/73 (49%), Gaps = 4/73 (5%)
Query: 613 AKAGHIGLYGWSYGGYLSAMTLSRYPDFFKCAVA-GAPVTSWDGYDTFYTEKYMGLPSEN 671
A+A + L+G SYGG L+A +YP A+A AP+ ++ D E + + S N
Sbjct: 181 AEACPVVLFGGSYGGMLAAWMRLKYPHIAIGALASSAPILQFE--DVVPPETFYDIAS-N 237
Query: 672 QSEYESGSVMNQV 684
+ ES S N +
Sbjct: 238 DFKRESSSCFNTI 250
>At5g43390 putative protein
Length = 643
Score = 31.2 bits (69), Expect = 2.2
Identities = 23/75 (30%), Positives = 35/75 (46%), Gaps = 6/75 (8%)
Query: 78 DGGLDESNISPEEKLRRERLRER---GLGVTRYEWVKTSSKRKAVMVPLPAGIYIQDISQ 134
D D++ + E RR LRE G+ Y + RK V+VPL + + ++S
Sbjct: 271 DSSYDKTTLICEAIARRMFLREEYEEGIEEVHYAYRIRDRLRKEVLVPLHKALELPEVSM 330
Query: 135 SKAE---LKLSSIPS 146
S E LK + +PS
Sbjct: 331 SAKEWNLLKYNRVPS 345
>At3g03790 unknown protein
Length = 1078
Score = 30.4 bits (67), Expect = 3.8
Identities = 42/216 (19%), Positives = 75/216 (34%), Gaps = 41/216 (18%)
Query: 211 WWSLDSKYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQEDHPYPFAGASNVKVRLGVVSAA 270
W S + +T VD+ P + L+A+ S VVS
Sbjct: 266 WGSNREGQLGYTSVDTQATP--------RKVTSLKAK------IVAVSAANKHTAVVSDC 311
Query: 271 GSSITWM-----DLLCGGKEQQANEEEYLARVNWMHGNTLTAQVLNRHHTKI-----KIL 320
G TW L G +N L V+++ G TA +++HT + ++
Sbjct: 312 GEVFTWGCNKEGQLGYGTSNSASNYSPRL--VDYLKGKVFTAIASSKYHTLVLRNDGEVY 369
Query: 321 KFDIRTGQGKNILVEEN-----STWINIHDC----FTPLDKG------ITKFSGGFIWAS 365
+ R + +++ N +T +N H T + G + + F W S
Sbjct: 370 TWGHRLVTPRRVIISRNLKKAGNTLLNFHRRRPLRLTAIAAGMVHSLALAEDGAFFYWVS 429
Query: 366 EKTGFRHLYLHDANGTCLGPITEGEWMVEQIAGVNE 401
+ R LH +G + I+ G++ + E
Sbjct: 430 SDSNLRCQQLHSLHGKTVVSISAGKYWASAVTSTGE 465
>At2g18410 hypothetical protein
Length = 392
Score = 30.4 bits (67), Expect = 3.8
Identities = 15/35 (42%), Positives = 20/35 (56%), Gaps = 4/35 (11%)
Query: 330 KNILVEENSTWINIHDCFTP----LDKGITKFSGG 360
K I+V +S WI I DC+T +D+ T FS G
Sbjct: 77 KGIVVSSSSKWIRILDCYTDPLGWIDQSSTSFSEG 111
>At1g63650 putative transcription factor (BHLH2)
Length = 596
Score = 30.4 bits (67), Expect = 3.8
Identities = 25/88 (28%), Positives = 43/88 (48%), Gaps = 5/88 (5%)
Query: 53 EQTLNRKVFAFNVKTKAQELLFSPPDGGLDESNISPEEKLRRERLRERGLGVTRYEWVKT 112
++ + + +F + K +ELL PD + N + EK RRE+L ER + T + +
Sbjct: 376 QKMIKKILFEVPLMNKKEELL---PDTPEETGNHALSEKKRREKLNERFM--TLRSIIPS 430
Query: 113 SSKRKAVMVPLPAGIYIQDISQSKAELK 140
SK V + Y+QD+ + EL+
Sbjct: 431 ISKIDKVSILDDTIEYLQDLQKRVQELE 458
>At3g03640 beta-glucosidase
Length = 531
Score = 30.0 bits (66), Expect = 5.0
Identities = 37/136 (27%), Positives = 57/136 (41%), Gaps = 25/136 (18%)
Query: 574 NRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGLAKAGHIGLYGWSYGGYLSAMT 633
++G + G+KF + L ++L +T +W V Q L YGG+LS
Sbjct: 126 DKGVSETGVKFYNDLINELIANGVTPLVTLFQWDVPQALED---------EYGGFLSDRI 176
Query: 634 LSRYPDFFKCAV--AGAPVTSW-----------DGYDTFYTEKYMGLPSENQSE-YESGS 679
L + DF + A G V W GY+T EK G S+ +E +G
Sbjct: 177 LEDFRDFAQFAFNKYGDRVKHWVTINEPYEFSRGGYET--GEKAPGRCSKYVNEKCVAGK 234
Query: 680 VMNQVHKLKGRLLLVH 695
++V+ + LLL H
Sbjct: 235 SGHEVYTVSHNLLLAH 250
>At1g50380 oligopeptidase-like protein
Length = 710
Score = 30.0 bits (66), Expect = 5.0
Identities = 16/51 (31%), Positives = 24/51 (46%)
Query: 599 DQLTGAEWLVKQGLAKAGHIGLYGWSYGGYLSAMTLSRYPDFFKCAVAGAP 649
D + AE L++ + + G S GG L ++ PD FK +AG P
Sbjct: 534 DFIACAERLIELKYCSKEKLCMEGRSAGGLLMGAVVNMRPDLFKVVIAGVP 584
>At1g01980 hypothetical protein
Length = 541
Score = 30.0 bits (66), Expect = 5.0
Identities = 18/53 (33%), Positives = 29/53 (53%), Gaps = 1/53 (1%)
Query: 586 SYLKHKLGQVDADDQLTGAEWLVKQGLAKAGHIGLYGWSYGGYLSAMTLSRYP 638
S+LK K V+ + G ++L K+ L +AG +GL YGG +S + + P
Sbjct: 375 SFLKRKSDYVEKEISKDGLDFLCKK-LMEAGKLGLVFNPYGGKMSEVATTATP 426
>At5g64380 fructose-bisphosphatase-like protein
Length = 404
Score = 29.3 bits (64), Expect = 8.5
Identities = 14/41 (34%), Positives = 20/41 (48%)
Query: 439 LTLNKGKHIVVLDHHMQSFVDIYDSLGCPPRVLLCSLEDGR 479
+TL G H LDH FV + ++ P R + S+ D R
Sbjct: 241 VTLGSGTHAFTLDHSTGEFVLTHQNIKIPTRGQIYSVNDAR 281
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 29.3 bits (64), Expect = 8.5
Identities = 32/119 (26%), Positives = 50/119 (41%), Gaps = 21/119 (17%)
Query: 454 MQSFVDIYDSL-GCPPR---VLLCSLEDGRVIMPLFEQPFSVPRFKKLQLEPPEIVEIQA 509
+Q F D++ SL G PP LE G PL + P+ ++ P EI E++
Sbjct: 504 VQEFEDVFQSLQGLPPSRSDPFTIELELGTA--PLSKTPY--------RMVPAEIAELKK 553
Query: 510 NDGTVLYGALYKPDASRFGPPPYK--TMINVYGGPSV-----QLVSNSWLNTVDMRAQY 561
+L +P SR+G P T+ N Y P + QL + + +D+ Y
Sbjct: 554 QLEDLLGKGFIRPSTSRWGAPGLNRVTLKNKYPLPRIDELLDQLRGATCFSKIDLTPGY 612
>At1g32415 unknown protein
Length = 945
Score = 29.3 bits (64), Expect = 8.5
Identities = 21/63 (33%), Positives = 29/63 (45%), Gaps = 3/63 (4%)
Query: 175 FTNESKQLTHGAKENGLT--RGLAEYIAQEEMDRKNGYWWSLDSKYIAFTEVDSSEIPLF 232
F+NE + E GL R L + I Q + YW SL SKY +D + + LF
Sbjct: 42 FSNEEALILRRLSEGGLVHARHLLDKIPQRGSINRVVYWTSLLSKYAKTGYLDEARV-LF 100
Query: 233 RIM 235
+M
Sbjct: 101 EVM 103
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.137 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,802,734
Number of Sequences: 26719
Number of extensions: 879622
Number of successful extensions: 1791
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 1770
Number of HSP's gapped (non-prelim): 21
length of query: 754
length of database: 11,318,596
effective HSP length: 107
effective length of query: 647
effective length of database: 8,459,663
effective search space: 5473401961
effective search space used: 5473401961
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0039.7


