

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0039.6
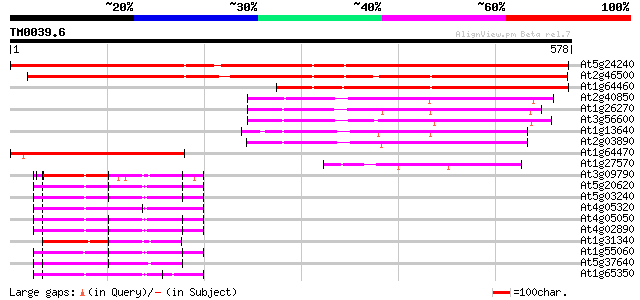
(578 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g24240 ubiquitin 714 0.0
At2g46500 putative ubiquitin 684 0.0
At1g64460 unknown protein 412 e-115
At2g40850 putative protein 211 7e-55
At1g26270 unknown protein 209 3e-54
At3g56600 putative protein 208 6e-54
At1g13640 unknown protein 207 1e-53
At2g03890 unknown protein 207 1e-53
At1g64470 ubiquitin, putative 184 9e-47
At1g27570 hypothetical protein 102 4e-22
At3g09790 polyubiquitin 74 3e-13
At5g20620 polyubiquitin 4 UBQ4 71 1e-12
At5g03240 polyubiquitin (ubq3) 71 1e-12
At4g05320 polyubiquitin (ubq10) 71 1e-12
At4g05050 70 2e-12
At4g02890 polyubiquitin 70 2e-12
At1g31340 putative ubiquitin (AtRUB1) 69 7e-12
At1g55060 ubiquitin-like (UBQ12) 67 2e-11
At5g37640 polyubiquitin 4 67 3e-11
At1g65350 67 3e-11
>At5g24240 ubiquitin
Length = 574
Score = 714 bits (1843), Expect = 0.0
Identities = 360/577 (62%), Positives = 447/577 (77%), Gaps = 11/577 (1%)
Query: 1 MACLALRPMHEEA--FGDFTSCFSHHLRDSILIYLTVGGSVVPMHIMETDSIASMKLRIQ 58
+A +AL P EE F F +L D IL++LT+ GSV+P +ME+DSIAS+KLRIQ
Sbjct: 3 VASVALSPALEELVNFPGIIGRFGFNLDDPILVFLTIAGSVIPKRVMESDSIASVKLRIQ 62
Query: 59 TFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFG 118
+ KGFFV+KQKL+++G+E++R+ +RDYG++DG +LHLV+RLSDLQAI+VRT GKEF
Sbjct: 63 SIKGFFVKKQKLLYDGREVSRNDSQIRDYGLADGKLLHLVIRLSDLQAISVRTVDGKEFE 122
Query: 119 FYVGKKRNVGYVKQQIARKGQGLLDLADQELVWEGEALEDQRLIEDICKDNDAVIHFLVR 178
V + RNVGYVKQQIA K + L D EL +GE L+DQRLI D+C++ D VIH L+
Sbjct: 123 LVVERSRNVGYVKQQIASKEKELGIPRDHELTLDGEELDDQRLITDLCQNGDNVIHLLIS 182
Query: 179 TSDAKVTTKPVGKDFELSIEAYTVPSFSAGQLGSVSVTNRALKRNELTREFLLEPIVVNP 238
S AKV KPVGKDFE+ IE G+ G + + +EF +EP +VNP
Sbjct: 183 KS-AKVRAKPVGKDFEVFIEDVNHKHNVDGRRG------KNISSEAKPKEFFVEPFIVNP 235
Query: 239 KIKIPQVIQELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAI 298
+IK+P +++ELI T EGLEKG PI+SS+GSGGAY MQD G KYVSVFKPIDEEPMA+
Sbjct: 236 EIKLPILLKELISSTLEGLEKGNGPIRSSDGSGGAYFMQDPSGHKYVSVFKPIDEEPMAV 295
Query: 299 NNPRGLPLSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTV 358
NNP G P+SVDGEG LKKGT+VG+GA+REVAAYILD+P GPR++ + D+ GFAGVPPT
Sbjct: 296 NNPHGQPVSVDGEG-LKKGTQVGEGAIREVAAYILDYPMTGPRTFPH-DQTGFAGVPPTT 353
Query: 359 MVKCLHKGFHHPEGYNSASDNVKLGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRL 418
MVKCLHK F+HP GY+ + +N K+GSLQMF+ N+GSCEDMG VFPV++VHKISVLDIRL
Sbjct: 354 MVKCLHKDFNHPNGYSFSPENTKIGSLQMFVSNVGSCEDMGYRVFPVDQVHKISVLDIRL 413
Query: 419 ANADRHAGNILVAKDGENGSTVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTIEY 478
ANADRHAGNILV++DG++G VL PIDHGYC P FEDCTF+WLYWPQAKEPYS +T+EY
Sbjct: 414 ANADRHAGNILVSRDGKDGQMVLTPIDHGYCFPNKFEDCTFEWLYWPQAKEPYSSETLEY 473
Query: 479 IKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRETLKKK 538
IKSLD E+DI+LL+FHGWE+PP C R+LRISTMLLKKG+ KGL PFTIGSIMCRETLK++
Sbjct: 474 IKSLDPEKDIELLRFHGWEIPPSCTRVLRISTMLLKKGSAKGLTPFTIGSIMCRETLKEE 533
Query: 539 SVIEQIIEEAEEGALPGTSEASFLDLVSVIMDSHLEE 575
SVIEQII +AE T+E F+ VS IMD+ L++
Sbjct: 534 SVIEQIIHDAEAIVPTETTEDEFISTVSAIMDNRLDQ 570
>At2g46500 putative ubiquitin
Length = 566
Score = 684 bits (1765), Expect = 0.0
Identities = 343/556 (61%), Positives = 433/556 (77%), Gaps = 19/556 (3%)
Query: 19 SCFSHHLRDSILIYLTVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELA 78
SC + D+I+IYLT+ GSV+PM ++E+DSI S+KLRIQ+++GF VR QKLVF G+ELA
Sbjct: 25 SCLDSYPDDTIMIYLTLPGSVIPMRVLESDSIESVKLRIQSYRGFVVRNQKLVFGGRELA 84
Query: 79 RDKLCVRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKG 138
R +RDYGVS+GN+LHLVL+LSDLQ + V+T CGK F+V + RN+GYVK+QI++K
Sbjct: 85 RSNSNMRDYGVSEGNILHLVLKLSDLQVLDVKTTCGKHCRFHVERGRNIGYVKKQISKKR 144
Query: 139 QGLLDLADQELVWEGEALEDQRLIEDICKDNDAVIHFLVRTSDAKVTTKPVGKDFELSIE 198
+D +QE+++EGE LEDQ LI DIC+++D+V+H LVR S AKV KPV K+FELSI
Sbjct: 145 GDFVDPDEQEILYEGEKLEDQSLINDICRNDDSVLHLLVRRS-AKVRVKPVEKNFELSIV 203
Query: 199 AYTVPSFSAGQLGSVSVTNRALKRNELTREFLLEPIVVNPKIKIPQVIQELIKITSEGLE 258
A + S+ ++ LEP+VVN K K+P V++++I+ S+GL+
Sbjct: 204 APQAKDKKGREAKSIVPP----------KKLSLEPVVVNSKAKVPLVVKDMIQSASDGLK 253
Query: 259 KGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAINNPRGLPLSVDGEGGLKKGT 318
G P++SSEG+GGAY MQ G K+V VFKPIDEEPMA NNP+GLPLS +GEG LKKGT
Sbjct: 254 SGNSPVRSSEGTGGAYFMQGPSGNKFVGVFKPIDEEPMAENNPQGLPLSPNGEG-LKKGT 312
Query: 319 RVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVMVKCLHKGFHHPEGYNSASD 378
+VG+GALREVAAYILDHPK G +S +E GFAGVPPT M++CLH GF+HP+G +
Sbjct: 313 KVGEGALREVAAYILDHPKSGNKSMFG-EEIGFAGVPPTAMIECLHPGFNHPKGIKT--- 368
Query: 379 NVKLGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRLANADRHAGNILVAKDGENGS 438
K+GSLQMF +N GSCEDMGP FPVEEVHKISVLDIRLANADRH GNIL+ KD E+G
Sbjct: 369 --KIGSLQMFTENDGSCEDMGPLSFPVEEVHKISVLDIRLANADRHGGNILMTKD-ESGK 425
Query: 439 TVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTIEYIKSLDAEEDIKLLKFHGWEL 498
VL+PIDHGYCLPESFEDCTF+WLYWPQA++PYS +T EYI+SLDAEEDI LLKFHGW++
Sbjct: 426 LVLVPIDHGYCLPESFEDCTFEWLYWPQARKPYSAETQEYIRSLDAEEDIDLLKFHGWKM 485
Query: 499 PPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRETLKKKSVIEQIIEEAEEGALPGTSE 558
P E A+ LRISTMLLKKG E+GL F IG+IMCRETL KKS++E+++EEA+E LPGTSE
Sbjct: 486 PAETAQTLRISTMLLKKGVERGLTAFEIGTIMCRETLSKKSLVEEMVEEAQEAVLPGTSE 545
Query: 559 ASFLDLVSVIMDSHLE 574
A+FL+ +S +MD HL+
Sbjct: 546 AAFLEALSDVMDYHLD 561
>At1g64460 unknown protein
Length = 301
Score = 412 bits (1059), Expect = e-115
Identities = 201/301 (66%), Positives = 242/301 (79%), Gaps = 4/301 (1%)
Query: 276 MQDSLGLKYVSVFKPIDEEPMAINNPRGLPLSVDGEGGLKKGTRVGQGALREVAAYILDH 335
MQDS GL YVSVFKP+DEEPMA+NNP+ LP+S DG+G LK+GTRVG+GA REVAAY+LDH
Sbjct: 1 MQDSSGLNYVSVFKPMDEEPMAVNNPQQLPVSSDGQG-LKRGTRVGEGATREVAAYLLDH 59
Query: 336 PKKGPRSYHNSDEKGFAGVPPTVMVKCLHKGFHHPEGYNS-ASDNVKLGSLQMFMKNIGS 394
PK G RS + + GFAGVPPT MV+ HK +++P G++S A+ + K+GSLQMFMKN GS
Sbjct: 60 PKSGLRSV-SKEVMGFAGVPPTAMVRSSHKVYNYPNGFSSCATKDAKVGSLQMFMKNNGS 118
Query: 395 CEDMGPSVFPVEEVHKISVLDIRLANADRHAGNILVAKDGENGSTVLIPIDHGYCLPESF 454
CED+GP FPVEEVHKI V DIR+ANADRHAGNIL K E G T+LIPIDHGYCLPE+F
Sbjct: 119 CEDIGPGAFPVEEVHKICVFDIRMANADRHAGNILTGKS-EEGKTLLIPIDHGYCLPENF 177
Query: 455 EDCTFDWLYWPQAKEPYSPDTIEYIKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLK 514
EDCTF+WLYWPQAK P+S DTI+YI SLD+E+DI LL+ HGW +P +R LRISTMLLK
Sbjct: 178 EDCTFEWLYWPQAKLPFSADTIDYINSLDSEQDIALLQLHGWNVPEAVSRTLRISTMLLK 237
Query: 515 KGAEKGLPPFTIGSIMCRETLKKKSVIEQIIEEAEEGALPGTSEASFLDLVSVIMDSHLE 574
KG E+ L P+ IGS+MCRET+ K S IE+I+ EA LP +SEA+FL+ VSV MD L+
Sbjct: 238 KGVERNLTPYQIGSVMCRETVNKDSAIEEIVREAHNSVLPASSEATFLEAVSVAMDRRLD 297
Query: 575 E 575
E
Sbjct: 298 E 298
>At2g40850 putative protein
Length = 561
Score = 211 bits (538), Expect = 7e-55
Identities = 122/324 (37%), Positives = 182/324 (55%), Gaps = 23/324 (7%)
Query: 246 IQELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAINNPRGLP 305
++ L+ + + G +P+ G GGAYL+Q G ++V KP+DEEP+A NNP+
Sbjct: 107 VRALVAEVTMAMVSGAQPLLLPSGMGGAYLLQTGKGHN-IAVAKPVDEEPLAFNNPKKSG 165
Query: 306 LSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVMVKCLHK 365
+ G+ G+K VG+ +RE+AAY+LD+ +GF+GVPPT +V H
Sbjct: 166 NLMLGQPGMKHSIPVGETGIRELAAYLLDY-------------QGFSGVPPTALVSISHV 212
Query: 366 GFHHPEGYNSASDNVKLGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRLANADRHA 425
FH + ++ +S K+ SLQ F+ + ++GP F VH+I +LD+RL N DRHA
Sbjct: 213 PFHVSDAFSFSSMPYKVASLQRFVGHDFDAGELGPGSFTATSVHRIGILDVRLLNLDRHA 272
Query: 426 GNILVA----KDGEN--GSTVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTIEYI 479
GN+LV K+ N G+ L+PIDHG CLPE +D F+WL WPQA P+S ++YI
Sbjct: 273 GNMLVKRCDKKEAYNRLGTAELVPIDHGLCLPECLDDPYFEWLNWPQALVPFSDTELDYI 332
Query: 480 KSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRETLKKK- 538
+LD +D +LL+ LP R+L + T+ LK+ A GL IG M R+ K +
Sbjct: 333 SNLDPFKDAELLRTELHSLPESAIRVLVVCTVFLKQAAAAGLCLAEIGEKMTRDFSKGEE 392
Query: 539 --SVIEQIIEEAEEGALPGTSEAS 560
S++E + +A+ TSE S
Sbjct: 393 SFSLLETLCTKAKASVFGKTSEDS 416
>At1g26270 unknown protein
Length = 630
Score = 209 bits (533), Expect = 3e-54
Identities = 123/314 (39%), Positives = 180/314 (57%), Gaps = 25/314 (7%)
Query: 246 IQELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAINNPRGLP 305
++++ K ++ ++KG P+ + G GGAY ++S G + V++ KP DEEP A NNP+G
Sbjct: 148 VRQMAKDITKAVKKGIDPVAVNSGLGGAYYFKNSRG-ESVAIVKPTDEEPYAPNNPKGFV 206
Query: 306 LSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVMVKCLHK 365
G+ GLK+ RVG+ REVAAY+LD ++ FA VPPT +VK H
Sbjct: 207 GKALGQPGLKRSVRVGETGYREVAAYLLD-------------KEHFANVPPTALVKITHS 253
Query: 366 GFHHPEGYNSASDNVKL-----GSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRLAN 420
F+ +G ++ K+ SLQ F+ + + G S FPV VH+I +LDIR+ N
Sbjct: 254 IFNVNDGVKASKPMEKMLVSKIASLQQFIPHDYDASEHGTSNFPVSAVHRIGILDIRILN 313
Query: 421 ADRHAGNILVAK---DGENGSTVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTIE 477
DRH+GN+LV K DG G L+PIDHG CLPE+ ED F+W++WPQA P+S D ++
Sbjct: 314 TDRHSGNLLVKKLDGDGMFGQVELVPIDHGLCLPETLEDPYFEWIHWPQASIPFSEDELK 373
Query: 478 YIKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRETL-- 535
YI +LD D ++L+ + R+L + T+ LK+ A GL IG +M RE
Sbjct: 374 YIANLDPLGDCEMLRRELPMVREASLRVLVLCTIFLKEAAANGLCLAEIGEMMTREVRPG 433
Query: 536 -KKKSVIEQIIEEA 548
++ S IE + EA
Sbjct: 434 DEEPSEIEVVCLEA 447
>At3g56600 putative protein
Length = 533
Score = 208 bits (530), Expect = 6e-54
Identities = 118/325 (36%), Positives = 181/325 (55%), Gaps = 29/325 (8%)
Query: 246 IQELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAINNPRGLP 305
++ L+ + + G +P+ G GGAYL+Q G ++V KP+DEEP+A NNP+G
Sbjct: 87 VRSLVAEVTIAIVSGAQPLLLPSGLGGAYLLQTEKG-NNIAVAKPVDEEPLAFNNPKGSG 145
Query: 306 LSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVMVKCLHK 365
G+ G+K+ RVG+ +RE+AAY+LDH +GF+ VPPT +V+ H
Sbjct: 146 GLTLGQPGMKRSIRVGESGIRELAAYLLDH-------------QGFSSVPPTALVRISHV 192
Query: 366 GFHHPEGYNSASDNVKLGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRLANADRHA 425
FH ++A K+ SLQ F+ + ++GP F V VH+I +LD+R+ N DRHA
Sbjct: 193 PFHDRGSDHAA---YKVASLQRFVGHDFDAGELGPGSFTVVSVHRIGILDVRVLNLDRHA 249
Query: 426 GNILVAKDGEN---------GSTVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTI 476
GN+LV K + G+ L+PIDHG CLPE +D F+WL WPQA P++ +
Sbjct: 250 GNMLVKKIHDQDETTCSNGVGAAELVPIDHGLCLPECLDDPYFEWLNWPQASVPFTDIEL 309
Query: 477 EYIKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRETLK 536
+YI +LD +D +LL+ + R+L + T+ LK+ A GL IG M R+ +
Sbjct: 310 QYISNLDPFKDAELLRTELDSIQESSLRVLIVCTIFLKEAAAAGLSLAEIGEKMTRDICR 369
Query: 537 ---KKSVIEQIIEEAEEGALPGTSE 558
SV+E + +A+ A+ G+ +
Sbjct: 370 GEESSSVLEILCNKAKASAVSGSDD 394
>At1g13640 unknown protein
Length = 622
Score = 207 bits (528), Expect = 1e-53
Identities = 121/306 (39%), Positives = 168/306 (54%), Gaps = 30/306 (9%)
Query: 240 IKIPQVIQELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAIN 299
+ + Q +++K G+E PI + G GGAY +D G + V++ KP DEEP A N
Sbjct: 142 LSLKQTANDIVKAMKMGVE----PIPVNGGLGGAYYFRDEKG-QSVAIVKPTDEEPFAPN 196
Query: 300 NPRGLPLSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVM 359
NP+G G+ GLK RVG+ REVAAY+LD+ FA VPPT +
Sbjct: 197 NPKGFVGKALGQPGLKPSVRVGETGFREVAAYLLDYDH-------------FANVPPTAL 243
Query: 360 VKCLHKGFHHPEGYNSASD-------NVKLGSLQMFMKNIGSCEDMGPSVFPVEEVHKIS 412
VK H F+ +G + + K+ S Q F+ + D G S FPV VH+I
Sbjct: 244 VKITHSVFNVNDGMDGNKSREKKKLVSSKIASFQKFVPHDFDASDHGTSSFPVASVHRIG 303
Query: 413 VLDIRLANADRHAGNILVAK-----DGENGSTVLIPIDHGYCLPESFEDCTFDWLYWPQA 467
+LDIR+ N DRH GN+LV K G G LIPIDHG CLPE+ ED F+W++WPQA
Sbjct: 304 ILDIRILNTDRHGGNLLVKKLDDGGVGRFGQVELIPIDHGLCLPETLEDPYFEWIHWPQA 363
Query: 468 KEPYSPDTIEYIKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIG 527
P+S + ++YI+SLD +D ++L+ + C R+L + T+ LK+ A GL IG
Sbjct: 364 SIPFSEEELDYIQSLDPVKDCEMLRRELPMIREACLRVLVLCTVFLKEAAVFGLCLAEIG 423
Query: 528 SIMCRE 533
+M RE
Sbjct: 424 EMMTRE 429
>At2g03890 unknown protein
Length = 650
Score = 207 bits (527), Expect = 1e-53
Identities = 117/297 (39%), Positives = 167/297 (55%), Gaps = 22/297 (7%)
Query: 245 VIQELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAINNPRGL 304
+++ ++K + ++ G +P+ G GGAY ++ G + V++ KP DEEP A NNP+G
Sbjct: 151 IVKHMVKDIVKAMKMGVEPLPVHSGLGGAYYFRNKRG-ESVAIVKPTDEEPFAPNNPKGF 209
Query: 305 PLSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVMVKCLH 364
G+ GLK RVG+ REVAAY+LD+ + FA VPPT +VK H
Sbjct: 210 VGKALGQPGLKSSVRVGETGFREVAAYLLDYGR-------------FANVPPTALVKITH 256
Query: 365 KGFHHPEGYNSASDNVK-----LGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRLA 419
F+ +G K + S Q F+ + D G S FPV VH+I +LDIR+
Sbjct: 257 SVFNVNDGVKGNKPREKKLVSKIASFQKFVAHDFDASDHGTSSFPVTSVHRIGILDIRIF 316
Query: 420 NADRHAGNILVAK-DGEN--GSTVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTI 476
N DRH GN+LV K DG G LIPIDHG CLPE+ ED F+W++WPQA P+S + +
Sbjct: 317 NTDRHGGNLLVKKLDGVGMFGQVELIPIDHGLCLPETLEDPYFEWIHWPQASLPFSDEEV 376
Query: 477 EYIKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRE 533
+YI+SLD +D +L+ + C R+L + T+ LK+ + GL IG +M RE
Sbjct: 377 DYIQSLDPVKDCDMLRRELPMIREACLRVLVLCTIFLKEASAYGLCLAEIGEMMTRE 433
>At1g64470 ubiquitin, putative
Length = 213
Score = 184 bits (468), Expect = 9e-47
Identities = 95/184 (51%), Positives = 138/184 (74%), Gaps = 4/184 (2%)
Query: 1 MACLALRPMHEE---AFGDF-TSCFSHHLRDSILIYLTVGGSVVPMHIMETDSIASMKLR 56
+A +AL P+H A G F S +H+ S+L++L+V GS +PM I+E+DSIA +KLR
Sbjct: 3 VADVALSPIHRGSAFAVGGFGQSTTTHYSVKSVLVFLSVSGSTMPMLILESDSIAEVKLR 62
Query: 57 IQTFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGNVLHLVLRLSDLQAITVRTECGKE 116
IQT GF VR+QKLVF G+ELAR+ V+DYGV+ G+VLHLVL+L D +TV T CGK
Sbjct: 63 IQTCNGFRVRRQKLVFSGRELARNASRVKDYGVTGGSVLHLVLKLYDPLLVTVITTCGKV 122
Query: 117 FGFYVGKKRNVGYVKQQIARKGQGLLDLADQELVWEGEALEDQRLIEDICKDNDAVIHFL 176
F F+V ++RNVGY+K++I+++G+G ++ DQE++++GE L+D R+I+ ICKD ++VIH L
Sbjct: 123 FQFHVDRRRNVGYLKKRISKEGKGFPEVDDQEILFKGEKLDDNRIIDGICKDGNSVIHLL 182
Query: 177 VRTS 180
V+ S
Sbjct: 183 VKKS 186
>At1g27570 hypothetical protein
Length = 649
Score = 102 bits (255), Expect = 4e-22
Identities = 69/215 (32%), Positives = 101/215 (46%), Gaps = 24/215 (11%)
Query: 324 ALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVMVKCLHKGFHHPEGYNSASDNVKLG 383
A E AY+LDHP+ G RS + GF+ V V V+C K + +K+G
Sbjct: 396 ARNEAIAYLLDHPEDGHRS-QSEQIYGFSRVRRAVWVRCRIK------------NQMKMG 442
Query: 384 SLQMFMKNIGSCEDMG------PSVFPVEEVHKISVLDIRLANADRHAGNILVAKDGENG 437
L F+++ + + +G P EE+HKI VLDIR N DR+ GN+LV + NG
Sbjct: 443 VLIEFLESSVTVQSLGTVYSELPDDVGAEEIHKIVVLDIRFGNIDRNLGNLLVQAEPRNG 502
Query: 438 STV-LIPIDHGYCL---PESFEDCTFDWLYW-PQAKEPYSPDTIEYIKSLDAEEDIKLLK 492
S L+PIDH + C W+ W Q + +S + Y+ +LD + D++ L+
Sbjct: 503 SAAHLVPIDHELSFFNDAHPYITCGACWIKWLEQIDKDFSSQLVNYVAALDPDRDLEFLR 562
Query: 493 FHGWELPPECARLLRISTMLLKKGAEKGLPPFTIG 527
GWE + LKK +GL IG
Sbjct: 563 HCGWEPNQRYIENFTVFATFLKKAVSQGLTALQIG 597
>At3g09790 polyubiquitin
Length = 631
Score = 73.6 bits (179), Expect = 3e-13
Identities = 50/152 (32%), Positives = 82/152 (53%), Gaps = 4/152 (2%)
Query: 28 SILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLCVRD 86
+I IY T+ + + + +DSI ++K +IQ +G + +Q+L+F GK+L D L + D
Sbjct: 2 TIQIYAKTLTEKTITLDVETSDSIHNVKAKIQNKEGIPLDQQRLIFAGKQL-EDGLTLAD 60
Query: 87 YGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLDLAD 146
Y + + LHLVLRL I V+T GK V + VK +I K +G+L
Sbjct: 61 YNIQKESTLHLVLRLRGGMQIFVQTLTGKTITLEVKSSDTIDNVKAKIQDK-EGILP-RQ 118
Query: 147 QELVWEGEALEDQRLIEDICKDNDAVIHFLVR 178
Q L++ G+ LED R + D ++ +H ++R
Sbjct: 119 QRLIFAGKQLEDGRTLADYNIQKESTLHLVLR 150
Score = 63.2 bits (152), Expect = 4e-10
Identities = 58/191 (30%), Positives = 93/191 (48%), Gaps = 20/191 (10%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G R+Q+L+F GK+L D
Sbjct: 75 LRGGMQIFVQTLTGKTITLEVKSSDTIDNVKAKIQDKEGILPRQQRLIFAGKQL-EDGRT 133
Query: 84 VRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEF-----GFYVGKKRNVGYVKQQIA-RK 137
+ DY + + LHLVLRL I V T GK F V + VK +I R+
Sbjct: 134 LADYNIQKESTLHLVLRLCGGMQIFVSTFSGKNFTSDTLTLKVESSDTIENVKAKIQDRE 193
Query: 138 GQGLLDLADQELVWEGEAL--EDQRLIEDICKDNDAVIHFLVR-TSDAKVTTKPV----- 189
G L Q L++ GE L ED R + D N + + +R D + K +
Sbjct: 194 G---LRPDHQRLIFHGEELFTEDNRTLADYGIRNRSTLCLALRLRGDMYIFVKNLPYNSF 250
Query: 190 -GKDFELSIEA 199
G++F L +E+
Sbjct: 251 TGENFILEVES 261
Score = 46.2 bits (108), Expect = 5e-05
Identities = 45/173 (26%), Positives = 73/173 (42%), Gaps = 12/173 (6%)
Query: 36 GGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGNVL 95
GG ++ + ++ +D+I S+K +IQ G +Q L+F G +L +D + DY + + + L
Sbjct: 401 GGKIITLEVLSSDTIKSVKAKIQDKVGSPPDQQILLFRGGQL-QDGRTLGDYNIRNESTL 459
Query: 96 HLVLRLSDLQAITVR-------TECGKEFGFYVGKKRNVGYVKQQIARKGQGLLDLADQE 148
HL + I V+ T K V + VK +I K + L Q
Sbjct: 460 HLFFHIRHGMQIFVKTFSFSGETPTCKTITLEVESSDTIDNVKVKIQHKVG--IPLDRQR 517
Query: 149 LVWEGEALEDQRLIEDICKDNDAVIH--FLVRTSDAKVTTKPVGKDFELSIEA 199
L++ G L R + D + IH FL R GK L +E+
Sbjct: 518 LIFGGRVLVGSRTLLDYNIQKGSTIHQLFLQRGGMQIFIKTLTGKTIILEVES 570
Score = 42.7 bits (99), Expect = 5e-04
Identities = 23/68 (33%), Positives = 42/68 (60%), Gaps = 1/68 (1%)
Query: 34 TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGN 93
T+ G + + + +D+IA++K +IQ +G +Q L+F G++L D + + DY + +
Sbjct: 558 TLTGKTIILEVESSDTIANVKEKIQVKEGIKPDQQMLIFFGQQL-EDGVTLGDYDIHKKS 616
Query: 94 VLHLVLRL 101
L+LVLRL
Sbjct: 617 TLYLVLRL 624
Score = 42.0 bits (97), Expect = 0.001
Identities = 37/167 (22%), Positives = 75/167 (44%), Gaps = 9/167 (5%)
Query: 18 TSCFSHHLRDSILIYL------TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLV 71
T C + LR + I++ + G + + +D+I ++K ++Q + + +L+
Sbjct: 227 TLCLALRLRGDMYIFVKNLPYNSFTGENFILEVESSDTIDNVKAKLQDKERIPMDLHRLI 286
Query: 72 FEGKELARDKLCVRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVK 131
F GK L + + Y + G+ L+LV R I V+T K V + VK
Sbjct: 287 FAGKPLEGGR-TLAHYNIQKGSTLYLVTRFRCGMQIFVKTLTRKRINLEVESWDTIENVK 345
Query: 132 QQIARKGQGLLDLAD-QELVWEGEALEDQRLIEDICKDNDAVIHFLV 177
+ K +G+ + Q L++ G+ L+D + D ++ +H ++
Sbjct: 346 AMVQDK-EGIQPQPNLQRLIFLGKELKDGCTLADYSIQKESTLHLVL 391
>At5g20620 polyubiquitin 4 UBQ4
Length = 382
Score = 71.2 bits (173), Expect = 1e-12
Identities = 53/178 (29%), Positives = 92/178 (50%), Gaps = 6/178 (3%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D
Sbjct: 149 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRT 207
Query: 84 VRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLD 143
+ DY + + LHLVLRL I V+T GK V + VK +I K +G +
Sbjct: 208 LADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK-EG-IP 265
Query: 144 LADQELVWEGEALEDQRLIEDICKDNDAVIHFLVR-TSDAKVTTKPV-GKDFELSIEA 199
Q L++ G+ LED R + D ++ +H ++R ++ K + GK L +E+
Sbjct: 266 PDQQRLIFAGKQLEDGRTLADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVES 323
Score = 71.2 bits (173), Expect = 1e-12
Identities = 53/178 (29%), Positives = 92/178 (50%), Gaps = 6/178 (3%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D
Sbjct: 73 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRT 131
Query: 84 VRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLD 143
+ DY + + LHLVLRL I V+T GK V + VK +I K +G +
Sbjct: 132 LADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK-EG-IP 189
Query: 144 LADQELVWEGEALEDQRLIEDICKDNDAVIHFLVR-TSDAKVTTKPV-GKDFELSIEA 199
Q L++ G+ LED R + D ++ +H ++R ++ K + GK L +E+
Sbjct: 190 PDQQRLIFAGKQLEDGRTLADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVES 247
Score = 70.5 bits (171), Expect = 2e-12
Identities = 48/155 (30%), Positives = 82/155 (51%), Gaps = 4/155 (2%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D
Sbjct: 225 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRT 283
Query: 84 VRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLD 143
+ DY + + LHLVLRL I V+T GK V + VK +I K +G +
Sbjct: 284 LADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK-EG-IP 341
Query: 144 LADQELVWEGEALEDQRLIEDICKDNDAVIHFLVR 178
Q L++ G+ LED R + D ++ +H ++R
Sbjct: 342 PDQQRLIFAGKQLEDGRTLADYNIQKESTLHLVLR 376
Score = 70.1 bits (170), Expect = 3e-12
Identities = 50/168 (29%), Positives = 86/168 (50%), Gaps = 5/168 (2%)
Query: 34 TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGN 93
T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D + DY + +
Sbjct: 7 TLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRTLADYNIQKES 65
Query: 94 VLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLDLADQELVWEG 153
LHLVLRL I V+T GK V + VK +I K +G + Q L++ G
Sbjct: 66 TLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK-EG-IPPDQQRLIFAG 123
Query: 154 EALEDQRLIEDICKDNDAVIHFLVR-TSDAKVTTKPV-GKDFELSIEA 199
+ LED R + D ++ +H ++R ++ K + GK L +E+
Sbjct: 124 KQLEDGRTLADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVES 171
Score = 48.1 bits (113), Expect = 1e-05
Identities = 27/78 (34%), Positives = 47/78 (59%), Gaps = 2/78 (2%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D
Sbjct: 301 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRT 359
Query: 84 VRDYGVSDGNVLHLVLRL 101
+ DY + + LHLVLRL
Sbjct: 360 LADYNIQKESTLHLVLRL 377
>At5g03240 polyubiquitin (ubq3)
Length = 306
Score = 71.2 bits (173), Expect = 1e-12
Identities = 53/178 (29%), Positives = 92/178 (50%), Gaps = 6/178 (3%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D
Sbjct: 73 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRT 131
Query: 84 VRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLD 143
+ DY + + LHLVLRL I V+T GK V + VK +I K +G +
Sbjct: 132 LADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK-EG-IP 189
Query: 144 LADQELVWEGEALEDQRLIEDICKDNDAVIHFLVR-TSDAKVTTKPV-GKDFELSIEA 199
Q L++ G+ LED R + D ++ +H ++R ++ K + GK L +E+
Sbjct: 190 PDQQRLIFAGKQLEDGRTLADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVES 247
Score = 70.5 bits (171), Expect = 2e-12
Identities = 48/155 (30%), Positives = 82/155 (51%), Gaps = 4/155 (2%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D
Sbjct: 149 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRT 207
Query: 84 VRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLD 143
+ DY + + LHLVLRL I V+T GK V + VK +I K +G +
Sbjct: 208 LADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK-EG-IP 265
Query: 144 LADQELVWEGEALEDQRLIEDICKDNDAVIHFLVR 178
Q L++ G+ LED R + D ++ +H ++R
Sbjct: 266 PDQQRLIFAGKQLEDGRTLADYNIQKESTLHLVLR 300
Score = 70.1 bits (170), Expect = 3e-12
Identities = 50/168 (29%), Positives = 86/168 (50%), Gaps = 5/168 (2%)
Query: 34 TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGN 93
T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D + DY + +
Sbjct: 7 TLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRTLADYNIQKES 65
Query: 94 VLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLDLADQELVWEG 153
LHLVLRL I V+T GK V + VK +I K +G + Q L++ G
Sbjct: 66 TLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK-EG-IPPDQQRLIFAG 123
Query: 154 EALEDQRLIEDICKDNDAVIHFLVR-TSDAKVTTKPV-GKDFELSIEA 199
+ LED R + D ++ +H ++R ++ K + GK L +E+
Sbjct: 124 KQLEDGRTLADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVES 171
Score = 48.1 bits (113), Expect = 1e-05
Identities = 27/78 (34%), Positives = 47/78 (59%), Gaps = 2/78 (2%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D
Sbjct: 225 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRT 283
Query: 84 VRDYGVSDGNVLHLVLRL 101
+ DY + + LHLVLRL
Sbjct: 284 LADYNIQKESTLHLVLRL 301
>At4g05320 polyubiquitin (ubq10)
Length = 464
Score = 71.2 bits (173), Expect = 1e-12
Identities = 53/178 (29%), Positives = 92/178 (50%), Gaps = 6/178 (3%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D
Sbjct: 225 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRT 283
Query: 84 VRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLD 143
+ DY + + LHLVLRL I V+T GK V + VK +I K +G +
Sbjct: 284 LADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK-EG-IP 341
Query: 144 LADQELVWEGEALEDQRLIEDICKDNDAVIHFLVR-TSDAKVTTKPV-GKDFELSIEA 199
Q L++ G+ LED R + D ++ +H ++R ++ K + GK L +E+
Sbjct: 342 PDQQRLIFAGKQLEDGRTLADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVES 399
Score = 71.2 bits (173), Expect = 1e-12
Identities = 53/178 (29%), Positives = 92/178 (50%), Gaps = 6/178 (3%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D
Sbjct: 149 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRT 207
Query: 84 VRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLD 143
+ DY + + LHLVLRL I V+T GK V + VK +I K +G +
Sbjct: 208 LADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK-EG-IP 265
Query: 144 LADQELVWEGEALEDQRLIEDICKDNDAVIHFLVR-TSDAKVTTKPV-GKDFELSIEA 199
Q L++ G+ LED R + D ++ +H ++R ++ K + GK L +E+
Sbjct: 266 PDQQRLIFAGKQLEDGRTLADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVES 323
Score = 71.2 bits (173), Expect = 1e-12
Identities = 53/178 (29%), Positives = 92/178 (50%), Gaps = 6/178 (3%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D
Sbjct: 73 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRT 131
Query: 84 VRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLD 143
+ DY + + LHLVLRL I V+T GK V + VK +I K +G +
Sbjct: 132 LADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK-EG-IP 189
Query: 144 LADQELVWEGEALEDQRLIEDICKDNDAVIHFLVR-TSDAKVTTKPV-GKDFELSIEA 199
Q L++ G+ LED R + D ++ +H ++R ++ K + GK L +E+
Sbjct: 190 PDQQRLIFAGKQLEDGRTLADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVES 247
Score = 70.1 bits (170), Expect = 3e-12
Identities = 50/168 (29%), Positives = 86/168 (50%), Gaps = 5/168 (2%)
Query: 34 TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGN 93
T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D + DY + +
Sbjct: 7 TLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRTLADYNIQKES 65
Query: 94 VLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLDLADQELVWEG 153
LHLVLRL I V+T GK V + VK +I K +G + Q L++ G
Sbjct: 66 TLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK-EG-IPPDQQRLIFAG 123
Query: 154 EALEDQRLIEDICKDNDAVIHFLVR-TSDAKVTTKPV-GKDFELSIEA 199
+ LED R + D ++ +H ++R ++ K + GK L +E+
Sbjct: 124 KQLEDGRTLADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVES 171
Score = 57.0 bits (136), Expect = 3e-08
Identities = 37/114 (32%), Positives = 60/114 (52%), Gaps = 2/114 (1%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D
Sbjct: 301 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRT 359
Query: 84 VRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARK 137
+ DY + + LHLVLRL I V+T GK V + VK +I K
Sbjct: 360 LADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK 413
>At4g05050
Length = 229
Score = 70.5 bits (171), Expect = 2e-12
Identities = 48/155 (30%), Positives = 82/155 (51%), Gaps = 4/155 (2%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D
Sbjct: 73 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRT 131
Query: 84 VRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLD 143
+ DY + + LHLVLRL I V+T GK V + VK +I K +G +
Sbjct: 132 LADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK-EG-IP 189
Query: 144 LADQELVWEGEALEDQRLIEDICKDNDAVIHFLVR 178
Q L++ G+ LED R + D ++ +H ++R
Sbjct: 190 PDQQRLIFAGKQLEDGRTLADYNIQKESTLHLVLR 224
Score = 70.1 bits (170), Expect = 3e-12
Identities = 50/168 (29%), Positives = 86/168 (50%), Gaps = 5/168 (2%)
Query: 34 TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGN 93
T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D + DY + +
Sbjct: 7 TLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRTLADYNIQKES 65
Query: 94 VLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLDLADQELVWEG 153
LHLVLRL I V+T GK V + VK +I K +G + Q L++ G
Sbjct: 66 TLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK-EG-IPPDQQRLIFAG 123
Query: 154 EALEDQRLIEDICKDNDAVIHFLVR-TSDAKVTTKPV-GKDFELSIEA 199
+ LED R + D ++ +H ++R ++ K + GK L +E+
Sbjct: 124 KQLEDGRTLADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVES 171
Score = 48.1 bits (113), Expect = 1e-05
Identities = 27/78 (34%), Positives = 47/78 (59%), Gaps = 2/78 (2%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D
Sbjct: 149 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRT 207
Query: 84 VRDYGVSDGNVLHLVLRL 101
+ DY + + LHLVLRL
Sbjct: 208 LADYNIQKESTLHLVLRL 225
>At4g02890 polyubiquitin
Length = 229
Score = 70.5 bits (171), Expect = 2e-12
Identities = 48/155 (30%), Positives = 82/155 (51%), Gaps = 4/155 (2%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D
Sbjct: 73 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRT 131
Query: 84 VRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLD 143
+ DY + + LHLVLRL I V+T GK V + VK +I K +G +
Sbjct: 132 LADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK-EG-IP 189
Query: 144 LADQELVWEGEALEDQRLIEDICKDNDAVIHFLVR 178
Q L++ G+ LED R + D ++ +H ++R
Sbjct: 190 PDQQRLIFAGKQLEDGRTLADYNIQKESTLHLVLR 224
Score = 70.1 bits (170), Expect = 3e-12
Identities = 50/168 (29%), Positives = 86/168 (50%), Gaps = 5/168 (2%)
Query: 34 TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGN 93
T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D + DY + +
Sbjct: 7 TLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRTLADYNIQKES 65
Query: 94 VLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLDLADQELVWEG 153
LHLVLRL I V+T GK V + VK +I K +G + Q L++ G
Sbjct: 66 TLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK-EG-IPPDQQRLIFAG 123
Query: 154 EALEDQRLIEDICKDNDAVIHFLVR-TSDAKVTTKPV-GKDFELSIEA 199
+ LED R + D ++ +H ++R ++ K + GK L +E+
Sbjct: 124 KQLEDGRTLADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVES 171
Score = 48.1 bits (113), Expect = 1e-05
Identities = 27/78 (34%), Positives = 47/78 (59%), Gaps = 2/78 (2%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D
Sbjct: 149 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRT 207
Query: 84 VRDYGVSDGNVLHLVLRL 101
+ DY + + LHLVLRL
Sbjct: 208 LADYNIQKESTLHLVLRL 225
>At1g31340 putative ubiquitin (AtRUB1)
Length = 156
Score = 68.9 bits (167), Expect = 7e-12
Identities = 41/144 (28%), Positives = 78/144 (53%), Gaps = 3/144 (2%)
Query: 34 TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGN 93
T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D + DY + +
Sbjct: 7 TLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRTLADYNIQKES 65
Query: 94 VLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLDLADQELVWEG 153
LHLVLRL I V+T GKE + + +K+++ K +G+ + Q L++ G
Sbjct: 66 TLHLVLRLRGGTMIKVKTLTGKEIEIDIEPTDTIDRIKERVEEK-EGIPPV-QQRLIYAG 123
Query: 154 EALEDQRLIEDICKDNDAVIHFLV 177
+ L D + +D + +V+H ++
Sbjct: 124 KQLADDKTAKDYNIEGGSVLHLVL 147
Score = 50.8 bits (120), Expect = 2e-06
Identities = 27/68 (39%), Positives = 43/68 (62%), Gaps = 1/68 (1%)
Query: 34 TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGN 93
T+ G + + I TD+I +K R++ +G +Q+L++ GK+LA DK +DY + G+
Sbjct: 83 TLTGKEIEIDIEPTDTIDRIKERVEEKEGIPPVQQRLIYAGKQLADDK-TAKDYNIEGGS 141
Query: 94 VLHLVLRL 101
VLHLVL L
Sbjct: 142 VLHLVLAL 149
>At1g55060 ubiquitin-like (UBQ12)
Length = 230
Score = 67.4 bits (163), Expect = 2e-11
Identities = 44/145 (30%), Positives = 75/145 (51%), Gaps = 3/145 (2%)
Query: 34 TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGN 93
T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D + DY + +
Sbjct: 83 TLTGKTITLEVESSDTIDNLKAKIQDKEGIPPDQQRLIFAGKQL-EDGRTLADYNIQKES 141
Query: 94 VLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLDLADQELVWEG 153
LHLVLRL I V+T GK V + VK +I K +G + Q L++ G
Sbjct: 142 TLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK-EG-ISPDQQRLIFAG 199
Query: 154 EALEDQRLIEDICKDNDAVIHFLVR 178
+ ED R + D ++ +H ++R
Sbjct: 200 KQHEDGRTLADYNIQKESTLHLVLR 224
Score = 65.1 bits (157), Expect = 1e-10
Identities = 47/168 (27%), Positives = 84/168 (49%), Gaps = 5/168 (2%)
Query: 34 TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGN 93
T+ G + + +D+I ++K +IQ +G + +L+F GK+L D + DY V + +
Sbjct: 7 TLTGKTKVLEVESSDTIDNVKAKIQDIEGIPPDQHRLIFAGKQL-EDGRTLADYNVQEDS 65
Query: 94 VLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLDLADQELVWEG 153
LHL+LR I V+T GK V + +K +I K +G + Q L++ G
Sbjct: 66 TLHLLLRFRGGMQIFVKTLTGKTITLEVESSDTIDNLKAKIQDK-EG-IPPDQQRLIFAG 123
Query: 154 EALEDQRLIEDICKDNDAVIHFLVR-TSDAKVTTKPV-GKDFELSIEA 199
+ LED R + D ++ +H ++R ++ K + GK L +E+
Sbjct: 124 KQLEDGRTLADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVES 171
Score = 46.6 bits (109), Expect = 4e-05
Identities = 26/78 (33%), Positives = 46/78 (58%), Gaps = 2/78 (2%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+ D
Sbjct: 149 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGISPDQQRLIFAGKQ-HEDGRT 207
Query: 84 VRDYGVSDGNVLHLVLRL 101
+ DY + + LHLVLRL
Sbjct: 208 LADYNIQKESTLHLVLRL 225
>At5g37640 polyubiquitin 4
Length = 322
Score = 67.0 bits (162), Expect = 3e-11
Identities = 48/155 (30%), Positives = 80/155 (50%), Gaps = 4/155 (2%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D
Sbjct: 75 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGVPPDQQRLIFAGKQL-DDGRT 133
Query: 84 VRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLD 143
+ DY + + LHLVLRL I VRT K V VK +I K +G +
Sbjct: 134 LADYNIQKESTLHLVLRLRGGMQIFVRTLTRKTIALEVESSDTTDNVKAKIQDK-EG-IP 191
Query: 144 LADQELVWEGEALEDQRLIEDICKDNDAVIHFLVR 178
Q L++ G+ LED R + D ++ +H ++R
Sbjct: 192 PDQQRLIFAGKQLEDGRTLADYNIQKESTLHLVLR 226
Score = 66.2 bits (160), Expect = 5e-11
Identities = 42/145 (28%), Positives = 74/145 (50%), Gaps = 3/145 (2%)
Query: 34 TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGN 93
T+ + + ++ +D+I ++K +IQ +G + +Q+L+F GK L D + DY + +
Sbjct: 9 TLTEKTITIDVVSSDTINNVKAKIQDIEGIPLDQQRLIFSGK-LLDDGRTLADYSIQKDS 67
Query: 94 VLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLDLADQELVWEG 153
+LHL LRL I V+T GK V + VK +I K D Q L++ G
Sbjct: 68 ILHLALRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGVPPD--QQRLIFAG 125
Query: 154 EALEDQRLIEDICKDNDAVIHFLVR 178
+ L+D R + D ++ +H ++R
Sbjct: 126 KQLDDGRTLADYNIQKESTLHLVLR 150
Score = 60.5 bits (145), Expect = 3e-09
Identities = 45/156 (28%), Positives = 77/156 (48%), Gaps = 5/156 (3%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ + + + +D+ ++K +IQ +G +Q+L+F GK+L D
Sbjct: 151 LRGGMQIFVRTLTRKTIALEVESSDTTDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRT 209
Query: 84 VRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLD 143
+ DY + + LHLVLRL I V T GK V + VK +I K + +
Sbjct: 210 LADYNIQKESTLHLVLRLCGGMQIFVNTLTGKTITLEVESSDTIDNVKAKIQDKER--IQ 267
Query: 144 LADQELVWEGEALED-QRLIEDICKDNDAVIHFLVR 178
Q L++ GE LED + D ++ +H ++R
Sbjct: 268 PDQQRLIFAGEQLEDGYYTLADYNIQKESTLHLVLR 303
Score = 43.9 bits (102), Expect = 2e-04
Identities = 21/68 (30%), Positives = 39/68 (56%)
Query: 34 TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGN 93
T+ G + + + +D+I ++K +IQ + +Q+L+F G++L + DY + +
Sbjct: 237 TLTGKTITLEVESSDTIDNVKAKIQDKERIQPDQQRLIFAGEQLEDGYYTLADYNIQKES 296
Query: 94 VLHLVLRL 101
LHLVLRL
Sbjct: 297 TLHLVLRL 304
>At1g65350
Length = 294
Score = 67.0 bits (162), Expect = 3e-11
Identities = 50/168 (29%), Positives = 87/168 (51%), Gaps = 6/168 (3%)
Query: 34 TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGN 93
T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D + DY + +
Sbjct: 7 TLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRTLADYNIQKES 65
Query: 94 VLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLDLADQELVWEG 153
LHLVLRL I V+T GK V + VK +I K +G + Q L++ G
Sbjct: 66 TLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK-EG-IPPDQQRLIFAG 123
Query: 154 EALEDQRLIEDICKDNDAVIHFLVR-TSDAKVTTKPV-GKDFELSIEA 199
+ LED R + D + ++ +H ++R ++ K + GK L +E+
Sbjct: 124 KQLEDGRTLADNIQ-KESTLHLVLRLRGGMQIFVKTLTGKTITLEVES 170
Score = 63.5 bits (153), Expect = 3e-10
Identities = 51/178 (28%), Positives = 89/178 (49%), Gaps = 7/178 (3%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ +G +Q+L+F GK+L D
Sbjct: 73 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPDQQRLIFAGKQL-EDGRT 131
Query: 84 VRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLD 143
+ D + + LHLVLRL I V+T GK V + VK +I K +
Sbjct: 132 LAD-NIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDK--EWIP 188
Query: 144 LADQELVWEGEALEDQRLIEDICKDNDAVIHFLVR-TSDAKVTTKPV-GKDFELSIEA 199
Q L++ G+ LED R + D ++ +H ++R ++ K + GK L +E+
Sbjct: 189 PDQQRLIFAGKQLEDGRTLADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVES 246
Score = 56.6 bits (135), Expect = 4e-08
Identities = 42/134 (31%), Positives = 70/134 (51%), Gaps = 4/134 (2%)
Query: 25 LRDSILIYL-TVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDKLC 83
LR + I++ T+ G + + + +D+I ++K +IQ + +Q+L+F GK+L D
Sbjct: 148 LRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEWIPPDQQRLIFAGKQL-EDGRT 206
Query: 84 VRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGLLD 143
+ DY + + LHLVLRL I V+T GK V + VK +I K +G +
Sbjct: 207 LADYNIQKESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSGTIDNVKAKIQDK-EG-IP 264
Query: 144 LADQELVWEGEALE 157
Q L++ G+ LE
Sbjct: 265 PDQQRLIFAGKQLE 278
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.138 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,542,466
Number of Sequences: 26719
Number of extensions: 616626
Number of successful extensions: 1546
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 35
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 1392
Number of HSP's gapped (non-prelim): 80
length of query: 578
length of database: 11,318,596
effective HSP length: 105
effective length of query: 473
effective length of database: 8,513,101
effective search space: 4026696773
effective search space used: 4026696773
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0039.6


