

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
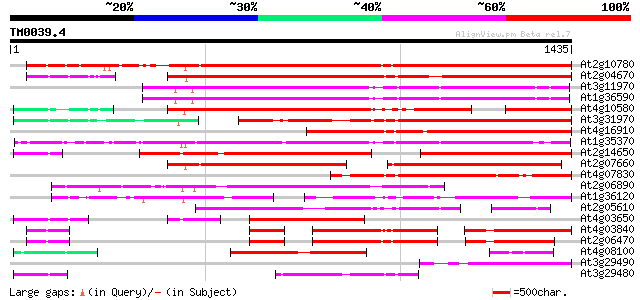
Query= TM0039.4
(1435 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g10780 pseudogene 1159 0.0
At2g04670 putative retroelement pol polyprotein 1038 0.0
At3g11970 hypothetical protein 754 0.0
At1g36590 hypothetical protein 749 0.0
At4g10580 putative reverse-transcriptase -like protein 748 0.0
At3g31970 hypothetical protein 724 0.0
At4g16910 retrotransposon like protein 679 0.0
At1g35370 hypothetical protein 664 0.0
At2g14650 putative retroelement pol polyprotein 600 e-171
At2g07660 putative retroelement pol polyprotein 533 e-151
At4g07830 putative reverse transcriptase 521 e-148
At2g06890 putative retroelement integrase 503 e-142
At1g36120 putative reverse transcriptase gb|AAD22339.1 495 e-140
At2g05610 putative retroelement pol polyprotein 484 e-136
At4g03650 putative reverse transcriptase 401 e-111
At4g03840 putative transposon protein 318 1e-86
At2g06470 putative retroelement pol polyprotein 318 2e-86
At4g08100 putative polyprotein 312 1e-84
At3g29490 hypothetical protein 308 1e-83
At3g29480 hypothetical protein 280 3e-75
>At2g10780 pseudogene
Length = 1611
Score = 1159 bits (2998), Expect = 0.0
Identities = 646/1466 (44%), Positives = 888/1466 (60%), Gaps = 137/1466 (9%)
Query: 42 FDGGYDPEGANRWLRKIEQIYESLPTSEDRMIAYASYLFHEEARNWWVHAKSRITPPDGV 101
F G DP A+ W ++++ ++S ED A + +A NWW+ + R D V
Sbjct: 182 FMGSTDPIVADEWRSRLKRNFKSTRCPEDYQRDIAVHFLEGDAHNWWLTVEKR--RGDEV 239
Query: 102 LTWSIFKEAFLEKYFPADVKGKKETEFLELKQGEMFVGQYAARFEELSQFHPYYGTTADD 161
+++ F++ F +KYFP + + E +L+L QG V +Y EE ++ Y G ++
Sbjct: 240 RSFADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTVREYD---EEFNRLRRYVGRELEE 296
Query: 162 ASKCIR-FECGLRPDIRAAIGHQQIRTF---TVLVEKCRIFEENDRARREYYKSS---KF 214
+R F GLR +IR H +RTF + LVE+ + EE R + +
Sbjct: 297 EQAQLRRFIRGLRIEIR---NHCLVRTFNSVSELVERAAMIEEGIEEERYLNREKAPIRN 353
Query: 215 NKTSKRREERKKPYSPRNYKPELQN---------------RNYGGARPTNPNSHV----- 254
N+++K ++++K N K + + + G H
Sbjct: 354 NQSTKPADKKRKFDKVDNTKSDAKTGECVTCGKNHSGTCWKAIGACGRCGSKDHAIQSCP 413
Query: 255 ---------------TCYRCGKEGHKSWSC---TATTTSGQTNLKPPVVGGTNTTSAPRN 296
TC+ CGK GH C TA +GQ + + GG P+
Sbjct: 414 KMEPGQSKVLGEETRTCFYCGKTGHLKRECPKLTAEKQAGQRDNR----GGNGLPPPPKR 469
Query: 297 SAGASA--QKPGRPVNKGKVFAMTGA---EANTSEDFIQGTCFLCDISLVVLYDSGATHS 351
A AS + + G A+TG E NT ++ +Y +G
Sbjct: 470 QAVASRVYELSEEANDAGNFRAITGGFRKEPNTDYGMVRAA------GGQAMYPTGLV-- 521
Query: 352 FISHERAKSLKLVITQLPYDLVVTTPTKESAVTSSVCKKCPLVIEDREYITNLVCLPLEG 411
R S+ + +P DL++ +PL+
Sbjct: 522 -----RGISVVVNGVNMPADLII--------------------------------VPLKK 544
Query: 412 LDIILGMNWLSINNVLLDC-RLRVPIF----LQKYKEKHTAS------------LPEKEP 454
D+ILGM+WL +DC R R+ + K++ T S + K
Sbjct: 545 HDVILGMDWLGKYKAHIDCHRGRIQFERDEGMLKFQGIRTTSGSLVISAIQAERMLGKGC 604
Query: 455 SAYLILFSSEGTKRPA-MEDIPVVREFPEVFPEDMTELPPEREVEFAIDVIPGTTPISAA 513
AYL +++ A ++DIP+V EF +VF ++ +PP+R F I++ PGTTPIS A
Sbjct: 605 EAYLATITTKEVGASAELKDIPIVNEFSDVFAA-VSGVPPDRSDPFTIELEPGTTPISKA 663
Query: 514 PYRISPLELAELQKQVEELLSKGFIRPSVSPWGAPVLLVKKKDGSMRLCVDYRQLNKVTI 573
PYR++P E+A+L+KQ+EELL KGFIRPS SPWGAPVL VKKKDGS RLC+DYR LNKVT+
Sbjct: 664 PYRMAPAEMAKLKKQLEELLDKGFIRPSSSPWGAPVLFVKKKDGSFRLCIDYRGLNKVTV 723
Query: 574 KNRYPLPRIDDLMDQLKGARVFSKIDLRSGYHQIRVKSDDVQKTAFRTRYGHYEYLVMPF 633
KN+YPLPRID+LMDQL GA+ FSKIDL SGYHQI ++ DV+KTAFRTRY H+E++VMPF
Sbjct: 724 KNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYDHFEFVVMPF 783
Query: 634 GVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDILIYSKSKEEHVEHMQVVLKVLKDRKLYA 693
G+TNAPA FM MN +F +LD+FVI+FI+DIL+YSKS E H EH++ VL+ L++ +L+A
Sbjct: 784 GLTNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWEAHQEHLRAVLERLREHELFA 843
Query: 694 KLSKCEFWLEQVQFLGHVVSEDGIAVDPAKVEAVNSWKVPETVTGVRSFLGLAGYYRRFI 753
KLSKC FW V FLGHV+S+ G++VDP K+ ++ W P T +RSFLGLAGYYRRF+
Sbjct: 844 KLSKCSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLGLAGYYRRFV 903
Query: 754 EGFSKIATPLTQLTKKDHPFVWTEKCE*SFQTLKERLTKAPVLTLPDPSKDYDVYCDASK 813
F+ +A PLT+LT KD F W+++CE SF LK LT APVL LP+ + Y VY DAS
Sbjct: 904 MSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLTNAPVLVLPEEGEPYTVYTDASI 963
Query: 814 SGLGCVLMQERKVIAYASQQLRPHEQNYPTHDMELAAVVFALKIWRHYLYGVKFTIYSDH 873
GLGCVLMQ+ VIAYAS+QLR HE+NYPTHD+E+AAVVF LKIWR YLYG K IY+DH
Sbjct: 964 VGLGCVLMQKGSVIAYASRQLRKHEKNYPTHDLEMAAVVFFLKIWRSYLYGAKVQIYTDH 1023
Query: 874 QSLKYLFDQKTLNMRQRRWVEFLEDYDFKLQYHPGKANVVADALSRK--SLHAARLMIEE 931
+SLKY+F Q LN+RQRRW+E + DY+ + YHPGKAN VADALSR+ + A R ++
Sbjct: 1024 KSLKYIFTQPELNLRQRRWMELVADYNLDIAYHPGKANQVADALSRRRSEVEAERSQVDL 1083
Query: 932 TELIEKFRDMNLIMETLPQGTRLGTLTLTNEFIEEVKKEQARDENLQKEAHGRDSMSRPD 991
++ L E P G LG + + ++ Q RDE E G ++ +
Sbjct: 1084 VNMMGTLHVNALSKEVEPLG--LGAADQA-DLLSRIRLAQERDE----EIKGWAQNNKTE 1136
Query: 992 FLKGPDGLWRYQGRLCVPEGGELRQKILEEGHKSDFSIHPGTTKMYQDLKKMFWWPGMKK 1051
+ +G GR+CVP L+++IL E H+S FSIHPG+ KMY+DLK+ + W GMKK
Sbjct: 1137 YQTSNNGTIVVNGRVCVPNDRALKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMKK 1196
Query: 1052 DIMKKVTSCLTCQKVKGEHQKPSGSLQPLSIPEWKWEGISMDFVSGLPR-TTTGHDAIWV 1110
D+ + V C TCQ VK EHQ PSG LQ L IPEWKW+ I+MDFV+GLP + H+A+WV
Sbjct: 1197 DVARWVAKCPTCQLVKAEHQVPSGLLQNLPIPEWKWDHITMDFVTGLPTGIKSKHNAVWV 1256
Query: 1111 IVDRLTKSAHFIAVNMTFPSEKLARIYVKEIVRLHGVPANIVSDRDPRFVSKFWGSLHEA 1170
+VDRLTKSAHF+A++ +E +A Y+ EIVRLHG+P +IVSDRD RF SKFW + +A
Sbjct: 1257 VVDRLTKSAHFMAISDKDGAEIIAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKAFQKA 1316
Query: 1171 LGTRLSLSSAYHPQSDGQSERTIQTLEDMLRACVLDYKGSWEDFLPLAEFSYNNSYHSSL 1230
LGTR++LS+AYHPQ+D QSERTIQTLEDMLRACVLD+ G+WE +L L EF+YNNS+ +S+
Sbjct: 1317 LGTRVNLSTAYHPQTDEQSERTIQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASI 1376
Query: 1231 GMAPFEALYGRRCKTPLCWLSGEDKITLGPELLQEMTEKVRSIREKLRIAQDRQKSYYDK 1290
GM+P+EALYGR C+TPLCW ++ GP ++ E TE+++ ++ KL+ AQDRQKSY +K
Sbjct: 1377 GMSPYEALYGRACRTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKEAQDRQKSYANK 1436
Query: 1291 RHKPLEFQEGDHVFLRVTPITGVGRSIHSKKLTPKYLGPYQILDRIGAVAYRIALPPSLS 1350
R K LEFQ GD V+L+ G GR KKL+P+Y+GPY++++R+GAVAY++ LPP L+
Sbjct: 1437 RRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDLPPKLN 1496
Query: 1351 NLHDVFHISQLRKYLPDSSHVIEPDNIELEENLTYPTQPVKILERREKQLRKRTVPLVKL 1410
H+VFH+SQLRK L D +E L+EN+T PV+I++R K R + L+K+
Sbjct: 1497 AFHNVFHVSQLRKCLSDQEESVEDIPPGLKENMTVEAWPVRIMDRMTKGTRGKARDLLKV 1556
Query: 1411 AWS-DDNQDATWELEESARKRYPSLF 1435
W+ ++ TWE E + +P F
Sbjct: 1557 LWNCRGREEYTWETENKMKANFPEWF 1582
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 1038 bits (2683), Expect = 0.0
Identities = 531/1053 (50%), Positives = 703/1053 (66%), Gaps = 61/1053 (5%)
Query: 403 NLVCLPLEGLDIILGMNWLSINNVLLDC-RLRVPIFLQKYKEKHTASLP----------- 450
+L+ P+E D+ILGM+WL V LDC R RV + + + P
Sbjct: 394 DLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEGRLVYQGVRPTSGSLVISAVQ 453
Query: 451 -----EKEPSAYLILFSS-EGTKRPAMEDIPVVREFPEVFPEDMTELPPEREVEFAIDVI 504
EK AYL+ S E + A+ DI V++EF +VF + + LPP R F I++
Sbjct: 454 AKKMIEKGCEAYLVTISMPESLGQVAVSDIRVIQEFEDVF-QSLQGLPPSRSDPFTIELE 512
Query: 505 PGTTPISAAPYRISPLELAELQKQVEELLSKGFIRPSVSPWGAPVLLVKKKDGSMRLCVD 564
PGT P+S APYR++P E+ EL+KQ+E+LL KGFIRPS SPWGAPVL VKKKDGS RLC+D
Sbjct: 513 PGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRPSTSPWGAPVLFVKKKDGSFRLCID 572
Query: 565 YRQLNKVTIKNRYPLPRIDDLMDQLKGARVFSKIDLRSGYHQIRVKSDDVQKTAFRTRYG 624
YR LN VT+KN+YPLPRID+L+DQL+GA FSKIDL SGYHQI + DV+KTAFRTRYG
Sbjct: 573 YRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYG 632
Query: 625 HYEYLVMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDILIYSKSKEEHVEHMQVVLK 684
H+E++VMPF +TNAPA FM MN +F +LD+FVI+FIDDIL+YSKS EEH H++ V++
Sbjct: 633 HFEFVVMPFALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEHEVHLRRVME 692
Query: 685 VLKDRKLYAKLSKCEFWLEQVQFLGHVVSEDGIAVDPAKVEAVNSWKVPETVTGVRSFLG 744
L+++KL+AKLSKC FW ++ FLGH+VS +G++VDP K+EA+ W P T +RSFL
Sbjct: 693 KLREQKLFAKLSKCSFWQREIGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFLR 752
Query: 745 LAGYYRRFIEGFSKIATPLTQLTKKDHPFVWTEKCE*SFQTLKERLTKAPVLTLPDPSKD 804
L GYYRRF++GF+ +A P+T+LT KD PFVW+ +CE F +LKE LT PVL LP+ +
Sbjct: 753 LTGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALPEHGQP 812
Query: 805 YDVYCDASKSGLGCVLMQERKVIAYASQQLRPHEQNYPTHDMELAAVVFALKIWRHYLYG 864
Y VY DAS+ GLGCVLMQ KVIAYAS+QLR HE NYPTHD+E+A V+FALKIWR YLYG
Sbjct: 813 YMVYTDASRVGLGCVLMQRGKVIAYASRQLRKHEGNYPTHDLEMAVVIFALKIWRSYLYG 872
Query: 865 VKFTIYSDHQSLKYLFDQKTLNMRQRRWVEFLEDYDFKLQYHPGKANVVADALSRKSLHA 924
K +++DH+SLKY+F+Q LN+RQ RW+E + DYD ++ YHPGKANVVADALSRK + A
Sbjct: 873 GKVQVFTDHKSLKYIFNQPELNLRQMRWMELVADYDLEIAYHPGKANVVADALSRKRVGA 932
Query: 925 ARLMIEETELIEKFRDMNL-IMETLPQGTRLGTLTLTNEFIEEVKKEQARDENLQKEAHG 983
A E L+ + + L ++ P G + + + Q +DE L +
Sbjct: 933 APGQSVEA-LVSEIGALRLCVVAREPLGLEAVDRA---DLLTRARLAQEKDEGLIAASKA 988
Query: 984 RDSMSRPDFLKGPDGLWRYQGRLCVPEGGELRQKILEEGHKSDFSIHPGTTKMYQDLKKM 1043
S ++ +G GR+CVP+ ELR++IL E H S FSIHPG TKMY+DLK+
Sbjct: 989 EGS----EYQFAANGTIFVYGRVCVPKDEELRREILSEAHASMFSIHPGATKMYRDLKRY 1044
Query: 1044 FWWPGMKKDIMKKVTSCLTCQKVKGEHQKPSGSLQPLSIPEWKWEGISMDFVSGLPRTTT 1103
+ W GMK+D+ V C CQ VK EHQ P
Sbjct: 1045 YQWVGMKRDVANWVAECDVCQLVKAEHQVP------------------------------ 1074
Query: 1104 GHDAIWVIVDRLTKSAHFIAVNMTFPSEKLARIYVKEIVRLHGVPANIVSDRDPRFVSKF 1163
DAIWVI+DRLTKSAHF+A+ T + LA+ YV EIV+LHGVP +IVSDRD +F F
Sbjct: 1075 --DAIWVIMDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTFAF 1132
Query: 1164 WGSLHEALGTRLSLSSAYHPQSDGQSERTIQTLEDMLRACVLDYKGSWEDFLPLAEFSYN 1223
W + +GT++ +S+AYHPQ+DGQSERTIQTLEDMLR CVLD+ G W D L L EF+YN
Sbjct: 1133 WRAFQAKMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYN 1192
Query: 1224 NSYHSSLGMAPFEALYGRRCKTPLCWLSGEDKITLGPELLQEMTEKVRSIREKLRIAQDR 1283
NSY +S+GMAPFEALYGR C TPL W E++ G + +QE TE++R ++ ++ AQ R
Sbjct: 1193 NSYQASIGMAPFEALYGRPCWTPLRWTQVEERSIYGADYVQETTERIRVLKLNMKEAQAR 1252
Query: 1284 QKSYYDKRHKPLEFQEGDHVFLRVTPITGVGRSIHSKKLTPKYLGPYQILDRIGAVAYRI 1343
Q+SY DKR + LEF+ GD V+L++ + G RSI KL+P+Y+GP++I++R+G VAYR+
Sbjct: 1253 QRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSILETKLSPRYMGPFRIVERVGPVAYRL 1312
Query: 1344 ALPPSLSNLHDVFHISQLRKYLPDSSHVIEPDNIELEENLTYPTQPVKILERREKQLRKR 1403
LP + H VFH+ LRK L V+ +L+ N+T +PV++LERR K+LR++
Sbjct: 1313 ELPDVMRAFHKVFHVLMLRKCLHKDDEVLVKIPEDLQPNMTLEARPVRVLERRIKELRRK 1372
Query: 1404 TVPLVKLAWSDDN-QDATWELEESARKRYPSLF 1435
+PL+K+ W D + TWE E + R+ F
Sbjct: 1373 KIPLIKVLWDCDGVTEETWEPEARMKARFKKWF 1405
Score = 78.6 bits (192), Expect = 2e-14
Identities = 64/230 (27%), Positives = 94/230 (40%), Gaps = 10/230 (4%)
Query: 42 FDGGYDPEGANRWLRKIEQIYESLPTSEDRMIAYASYLFHEEARNWWVHAKSRITPPDGV 101
F G PE A+ W ++++ + S + + A + +A WW +R D
Sbjct: 138 FSSGTSPEEADSWRSRVQRNFGSSRCPAEYRVDLAVHFLEGDAHLWWRSVTARRRQAD-- 195
Query: 102 LTWSIFKEAFLEKYFPADVKGKKETEFLELKQGEMFVGQYAARFEELSQFHPYYGT-TAD 160
++W+ F F KYFP + + E FLEL QGE V +Y F L Y G D
Sbjct: 196 MSWADFVAEFNAKYFPQEALDRMEARFLELTQGEWSVREYDREFNRLL---AYAGRGMED 252
Query: 161 DASKCIRFECGLRPDIRAAIGHQQIRTFTVLVEKCRIFEENDRARREYYKSSKFNKTSKR 220
D ++ RF GLRPD+R Q T LVE EE+ +R+ S +T K
Sbjct: 253 DQAQMRRFLRGLRPDLRVQCRVSQYATKAALVETAAEVEED--LQRQVVGVSPAVQT-KN 309
Query: 221 REERKKPYSPRNYKPELQNRNYGGARPTNPNSHVTCYRCGKEGHKSWSCT 270
+++ P S + Q R + + CG H CT
Sbjct: 310 TQQQVTP-SKGGKPAQGQKRKWDHPSRAGQGGRAGYFSCGSLDHTVADCT 358
>At3g11970 hypothetical protein
Length = 1499
Score = 754 bits (1946), Expect = 0.0
Identities = 437/1130 (38%), Positives = 648/1130 (56%), Gaps = 72/1130 (6%)
Query: 340 LVVLYDSGATHSFISHERAKSLKLVITQLPYDLVVTTPTKESAVTSSVCKKCPLVIEDRE 399
+ +L DSG+TH+F+ A L + V ++ V V ++
Sbjct: 401 IFILIDSGSTHNFLDPNTAAKLGCKVDTAGLTRVSVADGRKLRVEGKVTD-FSWKLQTTT 459
Query: 400 YITNLVCLPLEGLDIILGMNWLS-----------------INNVLLDCRLRVPIFLQKYK 442
+ ++++ +PL+G+D++LG+ WL NN + +++ K
Sbjct: 460 FQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFNNQKVLLHGLTSGSVREVK 519
Query: 443 EKHTASLPEKEPSAYLILF-----SSEG--------TKRPAMEDI--PVVREFPEVFPED 487
+ L E + ++ S+EG T E + V+ E+P++F E
Sbjct: 520 AQKLQKLQEDQVQLAMLCVQEVSESTEGELCTINALTSELGEESVVEEVLNEYPDIFIEP 579
Query: 488 MTELPPEREVE-FAIDVIPGTTPISAAPYRISPLELAELQKQVEELLSKGFIRPSVSPWG 546
T LPP RE I ++ G+ P++ PYR S + E+ K VE+LL+ G ++ S SP+
Sbjct: 580 -TALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSPYA 638
Query: 547 APVLLVKKKDGSMRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGARVFSKIDLRSGYHQ 606
+PV+LVKKKDG+ RLCVDYR+LN +T+K+ +P+P I+DLMD+L GA +FSKIDLR+GYHQ
Sbjct: 639 SPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQ 698
Query: 607 IRVKSDDVQKTAFRTRYGHYEYLVMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDIL 666
+R+ DD+QKTAF+T GH+EYLVMPFG+TNAPA F MN IF P+L KFV+VF DDIL
Sbjct: 699 VRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDIL 758
Query: 667 IYSKSKEEHVEHMQVVLKVLKDRKLYAKLSKCEFWLEQVQFLGHVVSEDGIAVDPAKVEA 726
+YS S EEH +H++ V +V++ KL+AKLSKC F + +V++LGH +S GI DPAK++A
Sbjct: 759 VYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKA 818
Query: 727 VNSWKVPETVTGVRSFLGLAGYYRRFIEGFSKIATPLTQLTKKDHPFVWTEKCE*SFQTL 786
V W P T+ +R FLGLAGYYRRF+ F IA PL LTK D F WT + +F+ L
Sbjct: 819 VKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTD-AFEWTAVAQQAFEDL 877
Query: 787 KERLTKAPVLTLPDPSKDYDVYCDASKSGLGCVLMQERKVIAYASQQLRPHEQNYPTHDM 846
K L +APVL+LP K + V DA G+G VLMQE +AY S+QL+ + + ++
Sbjct: 878 KAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEGHPLAYISRQLKGKQLHLSIYEK 937
Query: 847 ELAAVVFALKIWRHYLYGVKFTIYSDHQSLKYLFDQKTLNMRQRRWVEFLEDYDFKLQYH 906
EL AV+FA++ WRHYL F I +D +SLKYL +Q+ Q++W+ L ++D+++QY
Sbjct: 938 ELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEFDYEIQYR 997
Query: 907 PGKANVVADALSRKSLHAARLMIEETELIEKFRDMNLIMETLPQGTRLGTLTLTNEFIEE 966
GK NVVADALSR +E +E++ + + + +++
Sbjct: 998 QGKENVVADALSR---------VEGSEVLH-----------------MAMTVVECDLLKD 1031
Query: 967 VKKEQARDENLQK--EAHGRDSMSRPDFLKGPDGLWRYQGRLCVPEGGELRQKILEEGHK 1024
++ A D LQ A RD S+ F + R + ++ VP ++ IL H
Sbjct: 1032 IQAGYANDSQLQDIITALQRDPDSKKYF-SWSQNILRRKSKIVVPANDNIKNTILLWLHG 1090
Query: 1025 SDFSIHPGTTKMYQDLKKMFWWPGMKKDIMKKVTSCLTCQKVKGEHQKPSGSLQPLSIPE 1084
S H G +Q +K +F+W GM KDI + SC TCQ+ K + G LQPL IP+
Sbjct: 1091 SGVGGHSGRDVTHQRVKGLFYWKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPD 1150
Query: 1085 WKWEGISMDFVSGLPRTTTGHDAIWVIVDRLTKSAHFIAVNMTFPSEKLARIYVKEIVRL 1144
W +SMDF+ GLP + G I V+VDRL+K+AHFIA++ + + +A Y+ + +L
Sbjct: 1151 TIWSEVSMDFIEGLP-VSGGKTVIMVVVDRLSKAAHFIALSHPYSALTVAHAYLDNVFKL 1209
Query: 1145 HGVPANIVSDRDPRFVSKFWGSLHEALGTRLSLSSAYHPQSDGQSERTIQTLEDMLRACV 1204
HG P +IVSDRD F S+FW G L L+SAYHPQSDGQ+E + LE LR
Sbjct: 1210 HGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCMC 1269
Query: 1205 LDYKGSWEDFLPLAEFSYNNSYHSSLGMAPFEALYGRRCKTPLCWLSGEDKITLGPELLQ 1264
D W +L LAE+ YN +YHSS M PFE +YG+ L +L GE K+ + LQ
Sbjct: 1270 HDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAVVARSLQ 1329
Query: 1265 EMTEKVRSIREKLRIAQDRQKSYYDKRHKPLEFQEGDHVFLRVTPITGVGRSIH-SKKLT 1323
E + + ++ L AQ R K + D+ EF+ GD+V++++ P + ++KL+
Sbjct: 1330 EREDMLLFLKFHLMRAQHRMKQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQKLS 1389
Query: 1324 PKYLGPYQILDRIGAVAYRIALPPSLSNLHDVFHISQLRKYLPDSSHVIEPDNIELEENL 1383
PKY GPY+I+DR G VAY++AL PS S +H VFH+SQL+ + + S + ++ +
Sbjct: 1390 PKYFGPYKIIDRCGEVAYKLAL-PSYSQVHPVFHVSQLKVLVGNVSTTVHLPSVMQD--- 1445
Query: 1384 TYPTQPVKILERREKQLRKRTVPLVKLAWSDDN-QDATWELEESARKRYP 1432
+ P K++ER+ + + V V + WS++ ++ATWE +K +P
Sbjct: 1446 VFEKVPEKVVERKMVNRQGKAVTKVLVKWSNEPLEEATWEFLFDLQKTFP 1495
Score = 30.4 bits (67), Expect = 7.7
Identities = 20/70 (28%), Positives = 30/70 (42%), Gaps = 4/70 (5%)
Query: 18 QQNQPAEDDVYKGIDKFLKRNPPLFDGGYDPEGANRWLRKIEQIYESLPTSEDRMIAYAS 77
QQ + + Y + + K + P FDG E WL K+E+ + T ED + A+
Sbjct: 93 QQVRSDHFNAYNNLTRLGKIDFPRFDGTRLKE----WLFKVEEFFGVDSTPEDMKVKMAA 148
Query: 78 YLFHEEARNW 87
F A W
Sbjct: 149 IHFDSHASTW 158
>At1g36590 hypothetical protein
Length = 1499
Score = 749 bits (1933), Expect = 0.0
Identities = 436/1130 (38%), Positives = 648/1130 (56%), Gaps = 72/1130 (6%)
Query: 340 LVVLYDSGATHSFISHERAKSLKLVITQLPYDLVVTTPTKESAVTSSVCKKCPLVIEDRE 399
+ +L DSG+TH+F+ A L + V ++ V V ++
Sbjct: 401 IFILIDSGSTHNFLDPNTAAKLGCKVDTAGLTRVSVADGRKLRVEGKVTD-FSWKLQTTT 459
Query: 400 YITNLVCLPLEGLDIILGMNWLS-----------------INNVLLDCRLRVPIFLQKYK 442
+ ++++ +PL+G+D++LG+ WL NN + +++ K
Sbjct: 460 FQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFNNQKVLLHGLTSGSVREVK 519
Query: 443 EKHTASLPEKEPSAYLILF-----SSEG--------TKRPAMEDI--PVVREFPEVFPED 487
+ L E + ++ S+EG T E + V+ E+P++F E
Sbjct: 520 AQKLQKLQEDQVQLAMLCVQEVSESTEGELCTINALTSELGEESVVEEVLNEYPDIFIEP 579
Query: 488 MTELPPEREVE-FAIDVIPGTTPISAAPYRISPLELAELQKQVEELLSKGFIRPSVSPWG 546
T LPP RE I ++ G+ P++ PYR S + E+ K VE+LL+ G ++ S SP+
Sbjct: 580 -TALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSPYA 638
Query: 547 APVLLVKKKDGSMRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGARVFSKIDLRSGYHQ 606
+PV+LVKKKDG+ RLCVDYR+LN +T+K+ +P+P I+DLMD+L GA +FSKIDLR+GYHQ
Sbjct: 639 SPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQ 698
Query: 607 IRVKSDDVQKTAFRTRYGHYEYLVMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDIL 666
+R+ DD+QKTAF+T GH+EYLVMPFG+TNAPA F MN IF P+L KFV+VF DDIL
Sbjct: 699 VRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDIL 758
Query: 667 IYSKSKEEHVEHMQVVLKVLKDRKLYAKLSKCEFWLEQVQFLGHVVSEDGIAVDPAKVEA 726
+YS S EEH +H++ V +V++ KL+AKLSKC F + +V++LGH +S GI DPAK++A
Sbjct: 759 VYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKA 818
Query: 727 VNSWKVPETVTGVRSFLGLAGYYRRFIEGFSKIATPLTQLTKKDHPFVWTEKCE*SFQTL 786
V W P T+ +R FLGLAGYYRRF+ F IA PL LTK D F WT + +F+ L
Sbjct: 819 VKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTD-AFEWTAVAQQAFEDL 877
Query: 787 KERLTKAPVLTLPDPSKDYDVYCDASKSGLGCVLMQERKVIAYASQQLRPHEQNYPTHDM 846
K L +APVL+LP K + V DA G+G VLMQE +AY S+QL+ + + ++
Sbjct: 878 KAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEGHPLAYISRQLKGKQLHLSIYEK 937
Query: 847 ELAAVVFALKIWRHYLYGVKFTIYSDHQSLKYLFDQKTLNMRQRRWVEFLEDYDFKLQYH 906
EL AV+FA++ WRHYL F I +D +SLKYL +Q+ Q++W+ L ++D+++QY
Sbjct: 938 ELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEFDYEIQYR 997
Query: 907 PGKANVVADALSRKSLHAARLMIEETELIEKFRDMNLIMETLPQGTRLGTLTLTNEFIEE 966
GK NVVADALSR +E +E++ + + + +++
Sbjct: 998 QGKENVVADALSR---------VEGSEVLH-----------------MAMTVVECDLLKD 1031
Query: 967 VKKEQARDENLQK--EAHGRDSMSRPDFLKGPDGLWRYQGRLCVPEGGELRQKILEEGHK 1024
++ A D LQ A RD S+ F + R + ++ VP ++ IL H
Sbjct: 1032 IQAGYANDSQLQDIITALQRDPDSKKYF-SWSQNILRRKSKIVVPANDNIKNTILLWLHG 1090
Query: 1025 SDFSIHPGTTKMYQDLKKMFWWPGMKKDIMKKVTSCLTCQKVKGEHQKPSGSLQPLSIPE 1084
S H G +Q +K +F+ GM KDI + SC TCQ+ K + G LQPL IP+
Sbjct: 1091 SGVGGHSGRDVTHQRVKGLFYSKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPD 1150
Query: 1085 WKWEGISMDFVSGLPRTTTGHDAIWVIVDRLTKSAHFIAVNMTFPSEKLARIYVKEIVRL 1144
W +SMDF+ GLP + G I V+VDRL+K+AHFIA++ + + +A+ Y+ + +L
Sbjct: 1151 TIWSEVSMDFIEGLP-VSGGKTVIMVVVDRLSKAAHFIALSHPYSALTVAQAYLDNVFKL 1209
Query: 1145 HGVPANIVSDRDPRFVSKFWGSLHEALGTRLSLSSAYHPQSDGQSERTIQTLEDMLRACV 1204
HG P +IVSDRD F S+FW G L L+SAYHPQSDGQ+E + LE LR
Sbjct: 1210 HGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCMC 1269
Query: 1205 LDYKGSWEDFLPLAEFSYNNSYHSSLGMAPFEALYGRRCKTPLCWLSGEDKITLGPELLQ 1264
D W +L LAE+ YN +YHSS M PFE +YG+ L +L GE K+ + LQ
Sbjct: 1270 HDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAVVARSLQ 1329
Query: 1265 EMTEKVRSIREKLRIAQDRQKSYYDKRHKPLEFQEGDHVFLRVTPITGVGRSIH-SKKLT 1323
E + + ++ L AQ R K + D+ EF+ GD+V++++ P + ++KL+
Sbjct: 1330 EREDMLLFLKFHLMRAQHRMKQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQKLS 1389
Query: 1324 PKYLGPYQILDRIGAVAYRIALPPSLSNLHDVFHISQLRKYLPDSSHVIEPDNIELEENL 1383
PKY GPY+I+DR G VAY++AL PS S +H VFH+SQL+ + + S + ++ +
Sbjct: 1390 PKYFGPYKIIDRCGEVAYKLAL-PSYSQVHPVFHVSQLKVLVGNVSTTVHLPSVMQD--- 1445
Query: 1384 TYPTQPVKILERREKQLRKRTVPLVKLAWSDDN-QDATWELEESARKRYP 1432
+ P K++ER+ + + V V + WS++ ++ATWE +K +P
Sbjct: 1446 VFEKVPEKVVERKMVNRQGKAVTKVLVKWSNEPLEEATWEFLFDLQKTFP 1495
Score = 30.4 bits (67), Expect = 7.7
Identities = 20/70 (28%), Positives = 30/70 (42%), Gaps = 4/70 (5%)
Query: 18 QQNQPAEDDVYKGIDKFLKRNPPLFDGGYDPEGANRWLRKIEQIYESLPTSEDRMIAYAS 77
QQ + + Y + + K + P FDG E WL K+E+ + T ED + A+
Sbjct: 93 QQVRSDHFNAYNNLTRLGKIDFPRFDGTRLKE----WLFKVEEFFGVDSTPEDMKVKMAA 148
Query: 78 YLFHEEARNW 87
F A W
Sbjct: 149 IHFDSHASTW 158
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 748 bits (1930), Expect = 0.0
Identities = 398/799 (49%), Positives = 521/799 (64%), Gaps = 81/799 (10%)
Query: 403 NLVCLPLEGLDIILGMNWLSINNVLLDCRLRVPIFLQKYKEK------------------ 444
+L+ P+E D+ILGM+WL V LD R +F ++ + +
Sbjct: 368 DLIISPVELYDVILGMDWLDYYRVHLDWH-RGRVFFERPEGRLVYQGVRPISGSLVISAV 426
Query: 445 HTASLPEKEPSAYLILFSS-EGTKRPAMEDIPVVREFPEVFPEDMTELPPEREVEFAIDV 503
+ EK AYL+ S E + A+ DI VV+EF +VF + + LPP + F I++
Sbjct: 427 QAEKMIEKGCEAYLVTISMPESVGQVAVSDIRVVQEFQDVF-QSLQGLPPSQSDPFTIEL 485
Query: 504 IPGTTPISAAPYRISPLELAELQKQVEELLSKGFIRPSVSPWGAPVLLVKKKDGSMRLCV 563
PGT P+S APYR++P E+AEL+KQ+++LL KGFIRPS SPWGAPVL VKKKDGS RLC+
Sbjct: 486 EPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPSTSPWGAPVLFVKKKDGSFRLCI 545
Query: 564 DYRQLNKVTIKNRYPLPRIDDLMDQLKGARVFSKIDLRSGYHQIRVKSDDVQKTAFRTRY 623
DYR+LN+VT+KNRYPLPRID+L+DQL+GA FSKIDL SGYHQI + DV+KTAFRTRY
Sbjct: 546 DYRELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRY 605
Query: 624 GHYEYLVMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDILIYSKSKEEHVEHMQVVL 683
GH+E++VMPFG+TNAPA+FM MN +F +LD+FVI+FIDDIL+YSKS EE H++ V+
Sbjct: 606 GHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEQEVHLRRVM 665
Query: 684 KVLKDRKLYAKLSKCEFWLEQVQFLGHVVSEDGIAVDPAKVEAVNSWKVPETVTGVRSFL 743
+ L+++KL+AKLSKC FW ++ FLGH+VS +G++VDP K+EA+ W P T +RSFL
Sbjct: 666 EKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFL 725
Query: 744 GLAGYYRRFIEGFSKIATPLTQLTKKDHPFVWTEKCE*SFQTLKERLTKAPVLTLPDPSK 803
G AGYYRRF++GF+ +A P+T+LT KD PFVW+++CE F +LKE LT PVL LP+ +
Sbjct: 726 GWAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSQECEEGFVSLKEMLTSTPVLALPEHGQ 785
Query: 804 DYDVYCDASKSGLGCVLMQERKVIAYASQQLRPHEQNYPTHDMELAAVVFALKIWRHYLY 863
Y VY DAS+ GLGCVLMQ KVIAYAS+QL HE NYPTHD+E+AAV+FALKIWR YLY
Sbjct: 786 PYMVYTDASRVGLGCVLMQHGKVIAYASRQLMKHEGNYPTHDLEMAAVIFALKIWRSYLY 845
Query: 864 GVKFTIYSDHQSLKYLFDQKTLNMRQRRWVEFLEDYDFKLQYHPGKANVVADALSRKSLH 923
G K +++DH+SLKY+F Q LN+RQRRW+E + DYD ++ YHPGKANVV DALSRK +
Sbjct: 846 GGKVQVFTDHKSLKYIFTQPELNLRQRRWMELVADYDLEIAYHPGKANVVVDALSRKRVG 905
Query: 924 AARLMIEETELIEKFRDMNLIMETLPQGTRLGTLTLTNEFIEEVKKEQARDENLQKEAHG 983
AA + + E++ + +G L L + +E G
Sbjct: 906 AA--LGQSVEVLV---------------SEIGALRLC---------------AVAREPLG 933
Query: 984 RDSMSRPDFLKGPDGLWRYQGRLCVPEGGELRQKILEEGHKSDFSIHPGTTKMYQDLKKM 1043
+++ R D L + RL + LR TKMY+DLK+
Sbjct: 934 LEAVDRADLLT--------RVRLAQKKDEGLR-----------------ATKMYRDLKRY 968
Query: 1044 FWWPGMKKDIMKKVTSCLTCQKVKGEHQKPSGSLQPLSIPEWKWEGISMDFVSGLPRTTT 1103
+ W GMK D+ V C CQ VK EHQ G LQ L IPEWKW+ I+MD V GL R +
Sbjct: 969 YQWVGMKMDVANWVAECDVCQLVKAEHQVLGGMLQSLPIPEWKWDFITMDLVVGL-RVSR 1027
Query: 1104 GHDAIWVIVDRLTKSAHFIAVNMTFPSEKLARIYVKEIVRLHGVPANI--VSDRDPRFVS 1161
DAIWVIVDRLTKSAHF+A+ T + LA+ +V EIV+LHGVP N+ DR +
Sbjct: 1028 TKDAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKFVSEIVKLHGVPLNMKEAQDRQRSYAD 1087
Query: 1162 KFWGSLHEALGTRLSLSSA 1180
K L +G R+ L A
Sbjct: 1088 KRRRELEFEVGDRVYLKMA 1106
Score = 143 bits (360), Expect = 8e-34
Identities = 68/168 (40%), Positives = 106/168 (62%), Gaps = 1/168 (0%)
Query: 1269 KVRSIREKLRIAQDRQKSYYDKRHKPLEFQEGDHVFLRVTPITGVGRSIHSKKLTPKYLG 1328
K+ + ++ AQDRQ+SY DKR + LEF+ GD V+L++ + G RSI KL+P+Y+G
Sbjct: 1067 KLHGVPLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLSPRYMG 1126
Query: 1329 PYQILDRIGAVAYRIALPPSLSNLHDVFHISQLRKYLPDSSHVIEPDNIELEENLTYPTQ 1388
P++I++R+ VAYR+ LP + H VFH+S LRK L + +L+ N+T +
Sbjct: 1127 PFKIVERVEPVAYRLELPDVMRAFHKVFHVSMLRKCLHKDDEALAKIPEDLQPNMTLEAR 1186
Query: 1389 PVKILERREKQLRKRTVPLVKLAWSDDN-QDATWELEESARKRYPSLF 1435
PV++LERR K+LR++ +PL+K+ W D + TWE E + R+ F
Sbjct: 1187 PVRVLERRIKELRQKKIPLIKVLWDCDGVTEETWEPEARMKARFKKWF 1234
Score = 56.2 bits (134), Expect = 1e-07
Identities = 57/260 (21%), Positives = 95/260 (35%), Gaps = 39/260 (15%)
Query: 9 LANQAARAEQQNQPAED--DVYKGIDKFLKRNPPLFDGGYDPEGANRWLRKIEQIYESLP 66
+A + A +Q+ AE+ + +++ + + F GG PE A+ W ++ + + S
Sbjct: 117 VAPRVAEVQQRAAVAEEVLSYLRMMEQLQRIDTGYFSGGTSPEEADSWRSRVGRNFGSSR 176
Query: 67 TSEDRMIAYASYLFHEEARNWWVHAKSRITPPDGVLTWSIFKEAFLEKYFPADVKGKKET 126
+ + A + +A WW +R D ++W+ F F KYFP +
Sbjct: 177 CPAEYRVDLAVHFLEGDAHLWWRSVTARRRQTD--MSWADFVAEFKAKYFPQEA------ 228
Query: 127 EFLELKQGEMFVGQYAARFEELSQFHPYYGT-TADDASKCIRFECGLRPDIRAAIGHQQI 185
PY G DD ++ RF GLRPD+R Q
Sbjct: 229 ------------------------LDPYAGQGMEDDQAQMRRFLRGLRPDLRVRCRVSQY 264
Query: 186 RTFTVLVEKCRIFEENDRARREYYKSSKFNKTSKRREERKKPYSPRNYKPELQNRNYGGA 245
T LVE EE+ +R+ S + K +++ S + Q R +
Sbjct: 265 ATKAALVETAAEVEED--FQRQVVGVSPVVQPKKTQQQVTP--SKGGKPAQGQKRKWDHP 320
Query: 246 RPTNPNSHVTCYRCGKEGHK 265
C+ CG HK
Sbjct: 321 SRAGQGGRARCFSCGSLDHK 340
>At3g31970 hypothetical protein
Length = 1329
Score = 724 bits (1869), Expect = 0.0
Identities = 388/852 (45%), Positives = 522/852 (60%), Gaps = 82/852 (9%)
Query: 586 MDQLKGARVFSKIDLRSGYHQIRVKSDDVQKTAFRTRYGHYEYLVMPFGVTNAPAIFMDY 645
M + G S I + + I + +V+KTAFRTRYGH+E++VMPFG+TN P FM
Sbjct: 552 MPESVGQVAVSDIRVVHEFEDIPIAEANVRKTAFRTRYGHFEFVVMPFGLTNIPTAFMRL 611
Query: 646 MNRIFHPYLDKFVIVFIDDILIYSKSKEEHVEHMQVVLKVLKDRKLYAKLSKCEFWLEQV 705
MN +F +LD+FVI+FIDDIL+YSKS EEH ++ ++ S E
Sbjct: 612 MNSVFQEFLDEFVIIFIDDILVYSKSPEEH--------------EVQSEESDGEA----- 652
Query: 706 QFLGHVVSEDGIAVDPAKVEAVNSWKVPETVTGVRSFLGLAGYYRRFIEGFSKIATPLTQ 765
+E G++VD K+EA+ W P T +RSFLGLAGYY+RF++ F+ +A P+T+
Sbjct: 653 -----AGAEVGVSVDLEKIEAIRDWPRPTNATEIRSFLGLAGYYKRFVKVFASMAQPMTK 707
Query: 766 LTKKDHPFVWTEKCE*SFQTLKERLTKAPVLTLPDPSKDYDVYCDASKSGLGCVLMQERK 825
LT KD PFVW+ +C+ F +LKE LT PVL LP+ + Y VY DAS+ GL CVLMQ K
Sbjct: 708 LTGKDVPFVWSPECDEGFMSLKEMLTSTPVLALPEHGEPYMVYTDASRVGLDCVLMQRGK 767
Query: 826 VIAYASQQLRPHEQNYPTHDMELAAVVFALKIWRHYLYGVKFTIYSDHQSLKYLFDQKTL 885
V ++DH+SLKY+F Q L
Sbjct: 768 V-------------------------------------------FTDHKSLKYIFTQPEL 784
Query: 886 NMRQRRWVEFLEDYDFKLQYHPGKANVVADALSRKSLHAARLMIEETELIEKFRDMNL-I 944
N+RQRRW+E + DYD ++ YH GKANVVADALSRK + + L+ + + L +
Sbjct: 785 NLRQRRWMELVADYDLEIAYHSGKANVVADALSRKRVGGS-----VEALVSEIGALRLCV 839
Query: 945 METLPQGTRLGTLTLTNEFIEEVKKEQARDENLQKEAHGRDSMSRPDFLKGPDGLWRYQG 1004
M P G + + V+ Q +DE L + S ++ +G G
Sbjct: 840 MAQEPLGLEAVDRA---DLLTRVRLAQEKDEGLIAASKADGS----EYQFAANGTILVHG 892
Query: 1005 RLCVPEGGELRQKILEEGHKSDFSIHPGTTKMYQDLKKMFWWPGMKKDIMKKVTSCLTCQ 1064
R+CVP+ ELR++IL E H S FSIHP TKMY+DLK+ + W GMK+D+ VT C CQ
Sbjct: 893 RVCVPKDEELRREILSEAHASMFSIHPRATKMYRDLKRYYQWVGMKRDVANWVTECDVCQ 952
Query: 1065 KVKGEHQKPSGSLQPLSIPEWKWEGISMDFVSGLPRTTTGHDAIWVIVDRLTKSAHFIAV 1124
VK EHQ P G LQ L I EWKW+ I+MDFV GLP + T DAIWVIVDRLTKSAHF+A+
Sbjct: 953 LVKAEHQVPGGLLQSLPILEWKWDFITMDFVVGLPVSRT-KDAIWVIVDRLTKSAHFLAI 1011
Query: 1125 NMTFPSEKLARIYVKEIVRLHGVPANIVSDRDPRFVSKFWGSLHEALGTRLSLSSAYHPQ 1184
T + LA+ YV EIV LHGVP +IVSDRD +F S FW + +GT++ +S+AYHPQ
Sbjct: 1012 RKTDGAVLLAKKYVSEIVELHGVPVSIVSDRDSKFTSAFWRAFQGEMGTKVQMSTAYHPQ 1071
Query: 1185 SDGQSERTIQTLEDMLRACVLDYKGSWEDFLPLAEFSYNNSYHSSLGMAPFEALYGRRCK 1244
+DGQSERTIQTLEDMLR CVLD G W D L L EF+YNNSY +S+ MAPFEALYGR C+
Sbjct: 1072 TDGQSERTIQTLEDMLRMCVLDRGGHWADHLSLVEFAYNNSYQASIRMAPFEALYGRPCR 1131
Query: 1245 TPLCWLSGEDKITLGPELLQEMTEKVRSIREKLRIAQDRQKSYYDKRHKPLEFQEGDHVF 1304
TPLCW ++ G + + E TE++R ++ ++ AQDRQ+SY DKR + LEF+ GD V+
Sbjct: 1132 TPLCWTQVGERSIYGADYVLETTERIRVLKLNMKEAQDRQRSYADKRRRELEFEVGDRVY 1191
Query: 1305 LRVTPITGVGRSIHSKKLTPKYLGPYQILDRIGAVAYRIALPPSLSNLHDVFHISQLRKY 1364
L++ + G RSI KLTP+Y+GP++I++R+G VAYR+ LP + H VFH+S LRK
Sbjct: 1192 LKMAMLRGPNRSISETKLTPRYMGPFRIVERVGPVAYRLELPDVMRAFHKVFHVSMLRKC 1251
Query: 1365 LPDSSHVIEPDNIELEENLTYPTQPVKILERREKQLRKRTVPLVKLAWSDDN-QDATWEL 1423
L V+ +L+ N+T +PV+ILERR K+LR++ +PL+K+ W+ D + TWE
Sbjct: 1252 LHKDDEVLAKILEDLQPNMTLEARPVRILERRIKELRRKKIPLIKVLWNCDGVTEETWEP 1311
Query: 1424 EESARKRYPSLF 1435
E + + F
Sbjct: 1312 EARMKASFKKWF 1323
Score = 99.8 bits (247), Expect = 1e-20
Identities = 119/499 (23%), Positives = 181/499 (35%), Gaps = 72/499 (14%)
Query: 9 LANQAARAEQQNQPAED--DVYKGIDKFLKRNPPLFDGGYDPEGANRWLRKIEQIYESLP 66
+A + A +Q+ AE+ + +++ + F GG PE A+ W ++E + S
Sbjct: 123 VAPRVAEVQQRAAVAEEVPSYLRMMEQLQRIGTRYFFGGTSPEEADSWKSRVEHNFGSSR 182
Query: 67 TSEDRMIAYASYLFHEEARNWWVHAKSRITPPDGVLTWSIFKEAFLEKYFPADVKGKKET 126
+ + A + +A WW +R ++W+ F F KYFP + + E
Sbjct: 183 CPAEYRVDLAVHFLEGDAHLWWRSVTARRRQAH--MSWADFVAEFNAKYFPQEALDRMEA 240
Query: 127 EFLELKQGEMFVGQYAARFEELSQFHPYYGT-TADDASKCIRFECGLRPDIRAAIGHQQI 185
FLEL QGE V +Y F L Y G DD ++ RF GLRPD+R Q
Sbjct: 241 RFLELTQGERSVREYEREFNRLL---VYAGRGMQDDQAQMRRFLRGLRPDLRVRCRVLQY 297
Query: 186 RTFTVLVEKCRIFEENDRARREYYKSSKFNKTSKRREERKKPYSPRNYKPELQNRNYGGA 245
T LVE EE+ +R+ S K K +++
Sbjct: 298 ATKAALVETAAEVEED--LQRQVVGVSPAVKPKKTQQQ---------------------V 334
Query: 246 RPTNPNSHVTCYRCGKEGHKSWSCTATTTSGQTNLKPPVVGGTNTTSAPRNSAGASAQKP 305
P+ + + GH +C ++P V G + A +A
Sbjct: 335 APSKGGKPAQGQKRKERGHLRPNCPKLQRMAVAVVQPAVQHGAQVQQRVQQLAHIAAAPQ 394
Query: 306 G---RPVNKGKVFAMTGAEANTSEDFIQGTCFLCDISLVVLYDSGATHSFISHERAKSLK 362
G R + A+T A VL+DSGA+H FI+ E A
Sbjct: 395 GYTTREIGSTSKRAITEAH--------------------VLFDSGASHCFITPESASRGN 434
Query: 363 LVITQLPYDLVVTTPTKESAVTSSVCKKCPLVIEDREYITNLVCLPLEGLDIILGMNWLS 422
+ V + K + I +L+ P+E D+ILGM+WL
Sbjct: 435 IRGDPGEQLGAVKVAGGQFLAVLGRAKGVDIQIAGESMPVDLIISPVELYDVILGMDWLD 494
Query: 423 INNVLLDC-----------------RLRVPIFLQKYKEKHTASLPEKEPSAYLILFS-SE 464
V LDC R+R + EK AYL+ S E
Sbjct: 495 HYRVHLDCHRGRVSFERPEGRLVYQRVRPTSRSLVISAVQAEKMIEKGCEAYLVTISMPE 554
Query: 465 GTKRPAMEDIPVVREFPEV 483
+ A+ DI VV EF ++
Sbjct: 555 SVGQVAVSDIRVVHEFEDI 573
>At4g16910 retrotransposon like protein
Length = 687
Score = 679 bits (1751), Expect = 0.0
Identities = 341/681 (50%), Positives = 462/681 (67%), Gaps = 19/681 (2%)
Query: 759 IATPLTQLTKKDHPFVWTEKCE*SFQTLKERLTKAPVLTLPDPSKDYDVYCDASKSGLGC 818
+A P+TQLT KD F W+E+CE SF LK LT APVL LP+ + Y VY DAS GLGC
Sbjct: 1 MAQPMTQLTGKDTAFNWSEECERSFLELKAMLTNAPVLVLPEEGEPYTVYTDASIVGLGC 60
Query: 819 VLMQERKVIAYASQQLRPHEQNYPTHDMELAAVVFALKIWRHYLYGVKFTIYSDHQSLKY 878
VL+Q+ VIAYAS+QLR HE+NYPT+D+E+AAVVFALKIWR YLYG K I++DH+SLKY
Sbjct: 61 VLIQKGSVIAYASRQLRKHEKNYPTNDLEMAAVVFALKIWRSYLYGAKVQIFTDHKSLKY 120
Query: 879 LFDQKTLNMRQRRWVEFLEDYDFKLQYHPGKANVVADALSRKS--LHAARLMIEETELIE 936
+F Q LN+RQRRW++ + DYD + YHPGKAN V DALSR + A R + ++
Sbjct: 121 IFTQPELNLRQRRWIKLVADYDLNIAYHPGKANQVVDALSRLRTVVEAERNQVNLVNMMG 180
Query: 937 KFRDMNLIMETLPQGTRLGTLTLTNEFIEEVKKEQARDENLQKEAHGRDSMSRPDFLKGP 996
L E P G R + + ++ Q RDE ++ A ++ ++
Sbjct: 181 TLHLNALSKEVEPLGLRAANQA---DLLSRIRSAQERDEEIKGWAQN----NKTEYQSSN 233
Query: 997 DGLWRYQGRLCVPEGGELRQKILEEGHKSDFSIHPGTTKMYQDLKKMFWWPGMKKDIMKK 1056
+G GR+C P L+++IL+E H+S FSIHPG+ KMY+DLK+ + W GMKKD+ +
Sbjct: 234 NGTIVVNGRVCGPNDKALKEEILKEAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARW 293
Query: 1057 VTSCLTCQKVKGEHQKPSGSLQPLSIPEWKWEGISMDFVSGLPR-TTTGHDAIWVIVDRL 1115
V K EHQ PSG LQ L IPEWKW+ I MDFV+GLP + H+A+WV+VDRL
Sbjct: 294 VA--------KEEHQVPSGMLQNLPIPEWKWDHIMMDFVTGLPTGIKSKHNAVWVVVDRL 345
Query: 1116 TKSAHFIAVNMTFPSEKLARIYVKEIVRLHGVPANIVSDRDPRFVSKFWGSLHEALGTRL 1175
TKSAHF+A++ +E +A Y+ EIVRLHG+P +IVSDRD RF SKFW + LGTR+
Sbjct: 346 TKSAHFMAISDKDAAEIIAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKPFQKVLGTRV 405
Query: 1176 SLSSAYHPQSDGQSERTIQTLEDMLRACVLDYKGSWEDFLPLAEFSYNNSYHSSLGMAPF 1235
+LS+AYHPQ+DGQSERTIQTLEDMLRACVLD+ G+WE +L L EF+YNNS+ +S+GM+P+
Sbjct: 406 NLSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPY 465
Query: 1236 EALYGRRCKTPLCWLSGEDKITLGPELLQEMTEKVRSIREKLRIAQDRQKSYYDKRHKPL 1295
EALYGR +TPLCW ++ GP ++ E T+K++ ++ KL+ AQDRQKSY +KR K L
Sbjct: 466 EALYGRAGRTPLCWTPVGERRLFGPAVVDETTKKMKFLKIKLKEAQDRQKSYANKRRKEL 525
Query: 1296 EFQEGDHVFLRVTPITGVGRSIHSKKLTPKYLGPYQILDRIGAVAYRIALPPSLSNLHDV 1355
EFQ GD V+L+ G GR KKL P+Y+GPY++++R+GAVAY++ LPP L H+V
Sbjct: 526 EFQVGDLVYLKAMTYKGAGRFTSRKKLRPRYVGPYKVIERVGAVAYKLDLPPKLDAFHNV 585
Query: 1356 FHISQLRKYLPDSSHVIEPDNIELEENLTYPTQPVKILERREKQLRKRTVPLVKLAWS-D 1414
FH+SQLRK L + +E L+EN+T PV+I+++ +K R +++ L+K+ W+
Sbjct: 586 FHVSQLRKCLSEQEESMEDVPPGLKENMTVEAWPVRIMDQMKKGTRGKSMDLLKILWNCG 645
Query: 1415 DNQDATWELEESARKRYPSLF 1435
++ TWE E + +P F
Sbjct: 646 GREEYTWETETKMKANFPEWF 666
>At1g35370 hypothetical protein
Length = 1447
Score = 664 bits (1712), Expect = 0.0
Identities = 469/1470 (31%), Positives = 732/1470 (48%), Gaps = 169/1470 (11%)
Query: 12 QAARAEQQNQPAEDDVYKGIDKFLKRNPPLFDGGYDPEGANRWLRKIEQIYESLPTSEDR 71
+ R E NQ Y + + K + P FDG N WL K+E+ + T E+
Sbjct: 98 EMVRQEVNNQ------YGNLTRLGKIDFPRFDGSR----INEWLFKVEEFFGVDFTPEEM 147
Query: 72 MIAYASYLFHEEARNWWVHAKSRITPPDGVLTWSIFKEAFLEKYFPADVKGKKETEFLEL 131
+ + F A W D W + + +++ A E + L+
Sbjct: 148 KVKMVAIHFDSHAATWHHSFIQSGIGLDVFFNWPEYVKLLKDRFEDACDDPMAELKKLQE 207
Query: 132 KQGEMFVGQYAARFEELSQFHPYYGTTADDASKCIRFECGLRPDIRAAIGHQQIRTFTVL 191
G + +Y +FE I+ L + ++ +RT T +
Sbjct: 208 TDG---IVEYHQQFE------------------LIKVRLNLSEEYLVSVYLAGLRTDTQM 246
Query: 192 VEKCRIFEEN---DRARR-EYYKSSKFNKTSKRREERKKPYSPRNYKP--ELQNRNYGGA 245
R+FE D R +YY+ + KT +K S +Y+P E++ ++
Sbjct: 247 --HVRMFEPKTVRDCLRLGKYYERAHPKKTVSSTWSQKGTRSGGSYRPVKEVEQKS---- 300
Query: 246 RPTNPNSHV-TCYRCGKEGHKSWSCTATTTSGQTNLKPPVVGGTNTTSAPRNSAGASAQK 304
H+ CY C ++ ++ +T L V + S QK
Sbjct: 301 ------DHLGLCYFCDEK----FTPEHYLVHKKTQLFRMDVDEEFEDAVEVLSDDDHEQK 350
Query: 305 PGRPVNKGKVFAMTGAEANTSEDFIQGTCFLCDISLVVLYDSGATHSFISHERAKSLKLV 364
P ++ V ++G + ++GT D L +L DSG+TH+FI A L
Sbjct: 351 PMPQISVNAVSGISGYKTMG----VKGTVDKRD--LFILIDSGSTHNFIDSTVAAKLGCH 404
Query: 365 ITQLPYDLVVTTPTKESAVTSSVCKKCPLVIEDREYITNLVCLPLEGLDIILGMNWL-SI 423
+ V ++ V + K ++ + ++++ +PL+G+D++LG+ WL ++
Sbjct: 405 VESAGLTKVAVADGRKLNVDGQI-KGFTWKLQSTTFQSDILLIPLQGVDMVLGVQWLETL 463
Query: 424 NNVLLDCR-LRVPIF------------------LQKYKEKHTAS--------------LP 450
+ + + L + F ++ +K + T +
Sbjct: 464 GRISWEFKKLEMQFFYKNQRVWLHGIITGSVRDIKAHKLQKTQADQIQLAMVCVREVVSD 523
Query: 451 EKEPSAYLILFSSEGTKRPAMEDIPVVREFPEVFPEDMTELPPEREV-EFAIDVIPGTTP 509
E++ + +S+ + +++I V EFP+VF E T+LPP RE + I ++ G P
Sbjct: 524 EEQEIGSISALTSDVVEESVVQNI--VEEFPDVFAEP-TDLPPFREKHDHKIKLLEGANP 580
Query: 510 ISAAPYRISPLELAELQKQVEELLSKGFIRPSVSPWGAPVLLVKKKDGSMRLCVDYRQLN 569
++ PYR + E+ K V++++ G I+ S SP+ +PV+LVKKKDG+ RLCVDY +LN
Sbjct: 581 VNQRPYRYVVHQKDEIDKIVQDMIKSGTIQVSSSPFASPVVLVKKKDGTWRLCVDYTELN 640
Query: 570 KVTIKNRYPLPRIDDLMDQLKGARVFSKIDLRSGYHQIRVKSDDVQKTAFRTRYGHYEYL 629
+T+K+R+ +P I+DLMD+L G+ VFSKIDLR+GYHQ+R+ DD+QKTAF+T GH+EYL
Sbjct: 641 GMTVKDRFLIPLIEDLMDELGGSVVFSKIDLRAGYHQVRMDPDDIQKTAFKTHNGHFEYL 700
Query: 630 VMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDILIYSKSKEEHVEHMQVVLKVLKDR 689
VM FG+TNAPA F MN +F +L KFV+VF DDILIYS S EEH EH+++V +V++
Sbjct: 701 VMLFGLTNAPATFQSLMNSVFRDFLRKFVLVFFDDILIYSSSIEEHKEHLRLVFEVMRLH 760
Query: 690 KLYAKLSKCEFWLEQVQFLGHVVSEDGIAVDPAKVEAVNSWKVPETVTGVRSFLGLAGYY 749
KL+AK SK + LGH +S I DPAK++AV W P TV VR FLG AGYY
Sbjct: 761 KLFAKGSK--------EHLGHFISAREIETDPAKIQAVKEWPTPTTVKQVRGFLGFAGYY 812
Query: 750 RRFIEGFSKIATPLTQLTKKDHPFVWTEKCE*SFQTLKERLTKAPVLTLPDPSKDYDVYC 809
RRF+ F IA PL LTK D F W+ + + +F TLK L APVL LP K + V
Sbjct: 813 RRFVRNFGVIAGPLHALTKTD-GFCWSLEAQSAFDTLKAVLCNAPVLALPVFDKQFMVET 871
Query: 810 DASKSGLGCVLMQERKVIAYASQQLRPHEQNYPTHDMELAAVVFALKIWRHYLYGVKFTI 869
DA G+ VLMQ+ +AY S+QL+ + + ++ EL A +FA++ WRHYL F I
Sbjct: 872 DACGQGIRAVLMQKGHPLAYISRQLKGKQLHLSIYEKELLAFIFAVRKWRHYLLPSHFII 931
Query: 870 YSDHQSLKYLFDQKTLNMRQRRWVEFLEDYDFKLQYHPGKANVVADALSRKSLHAARLMI 929
+D +SLKYL +Q+ Q++W+ L ++D+++QY GK N+VADALSR +
Sbjct: 932 KTDQRSLKYLLEQRLNTPVQQQWLPKLLEFDYEIQYRQGKENLVADALSR---------V 982
Query: 930 EETELIEKFRDMNLIMETLPQGTRLGTLTLTNEFIEEVKKEQARDENLQKEAHGRDSMSR 989
E +E++ + + +F++E++ D L+
Sbjct: 983 EGSEVL-----------------HMALSIVECDFLKEIQVAYESDGVLKDIISA--LQQH 1023
Query: 990 PDFLK---GPDGLWRYQGRLCVPEGGELRQKILEEGHKSDFSIHPGTTKMYQDLKKMFWW 1046
PD K + R + ++ VP E+ K+L+ H S G +Q +K +F+W
Sbjct: 1024 PDAKKHYSWSQDILRRKSKIVVPNDVEITNKLLQWLHCSGMGGRSGRDASHQRVKSLFYW 1083
Query: 1047 PGMKKDIMKKVTSCLTCQKVKGEHQKPSGSLQPLSIPEWKWEGISMDFVSGLPRTTTGHD 1106
GM KDI + SC TCQ+ K ++ G LQPL IP+ W +SMDF+ GLP + G
Sbjct: 1084 KGMVKDIQAFIRSCGTCQQCKSDNAAYPGLLQPLPIPDKIWCDVSMDFIEGLP-NSGGKS 1142
Query: 1107 AIWVIVDRLTKSAHFIAVNMTFPSEKLARIYVKEIVRLHGVPANIVSDRDPRFVSKFWGS 1166
I V+VDRL+K+AHF+A+ + + +A+ ++ + + HG P +IVSDRD F S FW
Sbjct: 1143 VIMVVVDRLSKAAHFVALAHPYSALTVAQAFLDNVYKHHGCPTSIVSDRDVLFTSDFWKE 1202
Query: 1167 LHEALGTRLSLSSAYHPQSDGQSERTIQTLEDMLRACVLDYKGSWEDFLPLAEFSYNNSY 1226
+ G L +SSAYHPQSDGQ+E + LE+ LR W +LPLAE+ YN +Y
Sbjct: 1203 FFKLQGVELRMSSAYHPQSDGQTEVVNRCLENYLRCMCHARPHLWNKWLPLAEYWYNTNY 1262
Query: 1227 HSSLGMAPFEALYGRRCKTPLCWLSGEDKITLGPELLQEMTEKVRSIREKLRIAQDRQKS 1286
HSS M PFE +YG+ L +L G+ K+ + LQE + ++ L AQ R K
Sbjct: 1263 HSSSQMTPFELVYGQAPPIHLPYLPGKSKVAVVARSLQERENMLLFLKFHLMRAQHRMKQ 1322
Query: 1287 YYDKRHKPLEFQEGDHVFLRVTPITGVGRSIH-SKKLTPKYLGPYQILDRIGAVAYRIAL 1345
+ D+ F GD V++++ P + ++KL+PKY GPY+I+++ G V
Sbjct: 1323 FADQHRTERTFDIGDFVYVKLQPYRQQSVVLRVNQKLSPKYFGPYKIIEKCGEVM----- 1377
Query: 1346 PPSLSNLHDVFHISQLRKYLPDSSHVIEPDNIELEENLTYPTQPVKILERREKQLRKRTV 1405
+ +V +QL LPD + P ILER+ + + R
Sbjct: 1378 ------VGNVTTSTQLPSVLPD----------------IFEKAPEYILERKLVKRQGRAA 1415
Query: 1406 PLVKLAW-SDDNQDATWELEESARKRYPSL 1434
+V + W + ++ATW+ ++++ L
Sbjct: 1416 TMVLVKWIGEPVEEATWKFLFDRQQKFLGL 1445
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 600 bits (1548), Expect = e-171
Identities = 311/598 (52%), Positives = 406/598 (67%), Gaps = 22/598 (3%)
Query: 331 GTCFLCDISLVVLYDSGATHSFISHERAKSLKLVITQLPYDLVVTTPTKESAVTSSVCKK 390
GT + + VL+DSGA+H FI+ E A + V + K
Sbjct: 304 GTLLVGGVEAHVLFDSGASHCFITPESASRGNIRGDPGEQLGAVKVAGGQFVAVLGRTKG 363
Query: 391 CPLVIEDREYITNLVCLPLEGLDIILGMNWLSINNVLLDC-RLRVPIFLQKYKEKHTASL 449
+ I +L+ P+E D+ILGM+WL V LDC R RV E+ SL
Sbjct: 364 VDIQIAGESMPADLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSF------ERPEGSL 417
Query: 450 PEK--EPSAYLILFSSEGTKRPAMEDIPVVREFPEVFPEDMTELPPEREVEFAIDVIPGT 507
+ P++ ++ S+ ++ + EF +VF + + LPP R F I++ PGT
Sbjct: 418 VYQGVRPTSGSLVISAVQAEKMIEKGCEAYLEFEDVF-QSLQGLPPSRSDPFTIELEPGT 476
Query: 508 TPISAAPYRISPLELAELQKQVEELLSKGFIRPSVSPWGAPVLLVKKKDGSMRLCVDYRQ 567
P+S APYR++P E+AEL+KQ+E+LL GAPVL VKKKDGS RLC+DYR
Sbjct: 477 APLSKAPYRMAPAEMAELKKQLEDLL------------GAPVLFVKKKDGSFRLCIDYRG 524
Query: 568 LNKVTIKNRYPLPRIDDLMDQLKGARVFSKIDLRSGYHQIRVKSDDVQKTAFRTRYGHYE 627
LN VT+KN+YPLPRID+L+DQL+GA FSKIDL SGYH I + DV+KTAFRTRYGH+E
Sbjct: 525 LNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPIAEADVRKTAFRTRYGHFE 584
Query: 628 YLVMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDILIYSKSKEEHVEHMQVVLKVLK 687
++VMPFG+TNAPA FM MN +F LD+FVI+FIDDIL+YSKS EEH H++ V++ L+
Sbjct: 585 FVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYSKSLEEHEVHLRRVMEKLR 644
Query: 688 DRKLYAKLSKCEFWLEQVQFLGHVVSEDGIAVDPAKVEAVNSWKVPETVTGVRSFLGLAG 747
++KL+AKLSKC FW ++ FLGH+VS +G++VDP K+EA+ W P T +RSFLGLAG
Sbjct: 645 EQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWHTPTNATEIRSFLGLAG 704
Query: 748 YYRRFIEGFSKIATPLTQLTKKDHPFVWTEKCE*SFQTLKERLTKAPVLTLPDPSKDYDV 807
YYRRF++GF+ +A P+T+LT KD PFVW+ +CE F +LKE LT PVL LP+ + Y V
Sbjct: 705 YYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALPEHGEPYMV 764
Query: 808 YCDASKSGLGCVLMQERKVIAYASQQLRPHEQNYPTHDMELAAVVFALKIWRHYLYGVKF 867
Y DAS GLGCVLMQ KVIAYAS+QLR HE NYPTHD+E+AAV+FALKIWR YLYG
Sbjct: 765 YTDASGVGLGCVLMQRGKVIAYASRQLRKHEGNYPTHDLEMAAVIFALKIWRSYLYGGNV 824
Query: 868 TIYSDHQSLKYLFDQKTLNMRQRRWVEFLEDYDFKLQYHPGKANVVADALSRKSLHAA 925
+++DH+SLKY+F Q LN+RQR+W+E + DYD ++ YHPGKANVVADALS K + AA
Sbjct: 825 QVFTDHKSLKYIFTQPELNLRQRQWMELVADYDLEIAYHPGKANVVADALSHKRVGAA 882
Score = 408 bits (1048), Expect = e-113
Identities = 199/386 (51%), Positives = 267/386 (68%), Gaps = 2/386 (0%)
Query: 1051 KDIMKKVTSCLTCQKVKGEHQKPSGSLQPLSIPEWKWEGISMDFVSGLPRTTTGHDAIWV 1110
+D+ V C CQ VK EHQ P G LQ L IPEWKW+ I++DFV GLP + T DAIWV
Sbjct: 941 EDVANWVAECDVCQLVKAEHQVPGGMLQSLPIPEWKWDFITIDFVVGLPVSRT-KDAIWV 999
Query: 1111 IVDRLTKSAHFIAVNMTFPSEKLARIYVKEIVRLHGVPANIVSDRDPRFVSKFWGSLHEA 1170
IVDRLTKSAHF+A+ T + LA+ YV EIV+LHGVP +IVSDRD +F S FW +
Sbjct: 1000 IVDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAE 1059
Query: 1171 LGTRLSLSSAYHPQSDGQSERTIQTLEDMLRACVLDYKGSWEDFLPLAEFSYNNSYHSSL 1230
+GT++ +S+AYHPQ+ GQSERTIQTLEDMLR CVLD+ G W D L L EF+YNNSY +S+
Sbjct: 1060 MGTKVQMSTAYHPQTYGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYPASI 1119
Query: 1231 GMAPFEALYGRRCKTPLCWLSGEDKITLGPELLQEMTEKVRSIREKLRIAQDRQKSYYDK 1290
GMAPFEALY R C+TPLC ++ G + +QE TE++R ++ ++ AQDRQ+SY DK
Sbjct: 1120 GMAPFEALYERPCRTPLCLTQVGERSIYGADYVQETTERIRVLKLNMKEAQDRQRSYADK 1179
Query: 1291 RHKPLEFQEGDHVFLRVTPITGVGRSIHSKKLTPKYLGPYQILDRIGAVAYRIALPPSLS 1350
R + LEF+ GD V+L++ + G RSI KL+P+Y+GP++I++R+G VAYR+ LP +
Sbjct: 1180 RRRELEFEVGDRVYLKMAMLRGPNRSISETKLSPRYMGPFRIVERVGPVAYRLELPDVMR 1239
Query: 1351 NLHDVFHISQLRKYLPDSSHVIEPDNIELEENLTYPTQPVKILERREKQLRKRTVPLVKL 1410
H VFH+S LRK L V+ +L+ N+T +PV++LERR K+LR++ +PL+K+
Sbjct: 1240 AFHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARPVRVLERRIKELRRKKIPLIKV 1299
Query: 1411 AWSDDN-QDATWELEESARKRYPSLF 1435
W D TWE E + R+ F
Sbjct: 1300 LWDCDGVTKETWEPEARMKARFKKWF 1325
Score = 56.6 bits (135), Expect = 1e-07
Identities = 35/128 (27%), Positives = 59/128 (45%), Gaps = 4/128 (3%)
Query: 9 LANQAARAEQQNQPAED--DVYKGIDKFLKRNPPLFDGGYDPEGANRWLRKIEQIYESLP 66
+A + A +Q+ AE+ + +++ + F GG PE A+ W ++E+ + S
Sbjct: 121 VAPRVAEVQQRAAVAEEVPSYLRMMEQLQRIGTGYFSGGTSPEVADSWRSRVERNFGSSR 180
Query: 67 TSEDRMIAYASYLFHEEARNWWVHAKSRITPPDGVLTWSIFKEAFLEKYFPADVKGKKET 126
+ I A + +A WW +R D ++W+ F F KYFP + + E
Sbjct: 181 CPAEYRIDLAVHFLEGDAHLWWRSVTARRRQAD--ISWADFVAEFNAKYFPQEALDRMEA 238
Query: 127 EFLELKQG 134
FLEL QG
Sbjct: 239 HFLELTQG 246
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 533 bits (1372), Expect = e-151
Identities = 270/478 (56%), Positives = 347/478 (72%), Gaps = 19/478 (3%)
Query: 403 NLVCLPLEGLDIILGMNWLSINNVLLDCRLRVPIF-----LQKYKEKHTAS--------- 448
+L+ +PL+ D+ILGM+WL +DC F + K++ T S
Sbjct: 31 DLIIVPLKKHDVILGMDWLGKYKGHIDCHRERIQFERDEGMLKFQGIRTTSGSLVISAIQ 90
Query: 449 ---LPEKEPSAYLILFSSEGTKRPA-MEDIPVVREFPEVFPEDMTELPPEREVEFAIDVI 504
+ K AYL +++ A ++DI +V EF +VF ++ +PP+R F I++
Sbjct: 91 AERMLGKGCEAYLATITTKEVGASAELKDILIVNEFSDVFAA-VSGVPPDRSDPFTIELE 149
Query: 505 PGTTPISAAPYRISPLELAELQKQVEELLSKGFIRPSVSPWGAPVLLVKKKDGSMRLCVD 564
PGTTPIS APYR++P E+AEL+KQ+EELL+KGFIRPS SPWGAPVL VKKKDGS RLC+D
Sbjct: 150 PGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSPWGAPVLFVKKKDGSFRLCID 209
Query: 565 YRQLNKVTIKNRYPLPRIDDLMDQLKGARVFSKIDLRSGYHQIRVKSDDVQKTAFRTRYG 624
YR LNKVT+KN+YPLPRID+LMDQL GA+ FSKIDL SGYHQI ++ DV+KTAFRTRYG
Sbjct: 210 YRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYG 269
Query: 625 HYEYLVMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDILIYSKSKEEHVEHMQVVLK 684
H+E++VMPFG+TNAPA FM MN +F +LD+FVI+FIDDIL++SKS E H EH++ VL+
Sbjct: 270 HFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWEAHQEHLRAVLE 329
Query: 685 VLKDRKLYAKLSKCEFWLEQVQFLGHVVSEDGIAVDPAKVEAVNSWKVPETVTGVRSFLG 744
L++ +L+AKLSK FW V FLGHV+S+ G++VDP K+ ++ W P T +RSFLG
Sbjct: 330 RLREHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLG 389
Query: 745 LAGYYRRFIEGFSKIATPLTQLTKKDHPFVWTEKCE*SFQTLKERLTKAPVLTLPDPSKD 804
LAGYYRRF+ F+ +A PLT+LT KD F W+++CE SF LK L APVL LP+ +
Sbjct: 390 LAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLINAPVLVLPEEGEP 449
Query: 805 YDVYCDASKSGLGCVLMQERKVIAYASQQLRPHEQNYPTHDMELAAVVFALKIWRHYL 862
Y VY DAS GLGCVLMQ+ VIAYAS+QLR HE+NYPTHD+E+AAVVFALKIWR YL
Sbjct: 450 YTVYTDASIVGLGCVLMQKGSVIAYASRQLRKHEKNYPTHDLEMAAVVFALKIWRSYL 507
Score = 482 bits (1240), Expect = e-136
Identities = 231/445 (51%), Positives = 320/445 (71%), Gaps = 5/445 (1%)
Query: 967 VKKEQARDENLQKEAHGRDSMSRPDFLKGPDGLWRYQGRLCVPEGGELRQKILEEGHKSD 1026
++ Q RDE ++ G ++ ++ +G GR+CVP L+++IL E H+S
Sbjct: 508 IRLAQERDEEIK----GWTLNNKTEYQTSNNGTIVVNGRVCVPNDRALKEEILREAHQSK 563
Query: 1027 FSIHPGTTKMYQDLKKMFWWPGMKKDIMKKVTSCLTCQKVKGEHQKPSGSLQPLSIPEWK 1086
FSIHPG+ KMY+DLK+ + W GM+KD+ + V C TCQ VK EHQ PSG LQ L I EWK
Sbjct: 564 FSIHPGSNKMYRDLKRYYHWVGMRKDVARWVAKCPTCQLVKAEHQVPSGLLQNLPISEWK 623
Query: 1087 WEGISMDFVSGLPR-TTTGHDAIWVIVDRLTKSAHFIAVNMTFPSEKLARIYVKEIVRLH 1145
W+ I+MDFV+ LP + H+A+WV+VDRLTKSAHF+A++ +E +A Y+ EI+RLH
Sbjct: 624 WDHITMDFVTRLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDGAEIIAEKYIDEIMRLH 683
Query: 1146 GVPANIVSDRDPRFVSKFWGSLHEALGTRLSLSSAYHPQSDGQSERTIQTLEDMLRACVL 1205
G+P +IVSDRD RF SKFW + +ALGTR++LS+AYHPQ+DGQSERTIQTLEDMLRACVL
Sbjct: 684 GIPVSIVSDRDTRFTSKFWNAFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACVL 743
Query: 1206 DYKGSWEDFLPLAEFSYNNSYHSSLGMAPFEALYGRRCKTPLCWLSGEDKITLGPELLQE 1265
D+ G+WE +L L EF+YNNS+ +S+GM+P+EALYGR C+TPLCW ++ GP ++ E
Sbjct: 744 DWGGNWEKYLRLIEFAYNNSFQASIGMSPYEALYGRACRTPLCWTPVGERRLFGPTIVDE 803
Query: 1266 MTEKVRSIREKLRIAQDRQKSYYDKRHKPLEFQEGDHVFLRVTPITGVGRSIHSKKLTPK 1325
TE+++ ++ KL+ AQDRQKSY +KR K LEFQ GD V+L+ G GR KKL+P+
Sbjct: 804 TTERMKFLKIKLKEAQDRQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPR 863
Query: 1326 YLGPYQILDRIGAVAYRIALPPSLSNLHDVFHISQLRKYLPDSSHVIEPDNIELEENLTY 1385
Y+GPY++++R+GAVAY++ LPP L+ H+VFH+SQLRKYL D +E L+EN+T
Sbjct: 864 YVGPYKVIERVGAVAYKLDLPPKLNVFHNVFHVSQLRKYLSDQEESVEDIPPGLKENMTV 923
Query: 1386 PTQPVKILERREKQLRKRTVPLVKL 1410
PV+I++R K R ++ L+K+
Sbjct: 924 EAWPVRIMDRMSKGTRGKSRDLLKV 948
>At4g07830 putative reverse transcriptase
Length = 611
Score = 521 bits (1343), Expect = e-148
Identities = 286/616 (46%), Positives = 381/616 (61%), Gaps = 48/616 (7%)
Query: 821 MQERKVIAYASQQLRPHEQNYPTHDMELAAVVFALKIWRHYLYGVKFTIYSDHQSLKYLF 880
MQ KVIAY S+QL+ HE NYPTHD+E+AAV ++DH+SLKY+F
Sbjct: 1 MQRGKVIAYGSRQLKKHEGNYPTHDLEMAAV------------------FTDHKSLKYIF 42
Query: 881 DQKTLNMRQRRWVEFLEDYDFKLQYHPGKANVVADALSRKSLHAARLMIEETELIEKFRD 940
Q LN+R RRW++ + DYD ++ YHPGKANVV DALSRK + AA ET +IE
Sbjct: 43 TQLELNLRLRRWMKLVADYDLEIAYHPGKANVVTDALSRKRVGAAPGQSVETLVIEIGAL 102
Query: 941 MNLIMETLPQGTRLGTLTLTNEFIEEVKKEQARDENLQKEAHGRDSMSRPDFLKGPDGLW 1000
+ P G T + + V+ Q +DE L + ++ +G
Sbjct: 103 RLCAVAREPLGLEAVDQT---DLLSRVQLAQEKDEGLIAASKAEGF----EYQFAANGTI 155
Query: 1001 RYQGRLCVPEGGELRQKILEEGHKSDFSIHPGTTKMYQDLKKMFWWPGMKKDIMKKVTSC 1060
GR+CVP+ ELRQ+IL E H S FSIHPG TKMY+DLK+ + W GMK+D+ V C
Sbjct: 156 LVHGRVCVPKDKELRQEILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVGNWVEEC 215
Query: 1061 LTCQKVKGEHQKPSGSLQPLSIPEWKWEGISMDFVSGLPRTTTGHDAIWVIVDRLTKSAH 1120
CQ VK EHQ LQ L IPEWKW+ I+MDFV GLP + T DAIWVIVDRLTKSAH
Sbjct: 216 DVCQLVKIEHQVSGSLLQSLPIPEWKWDFITMDFVVGLPVSRT-KDAIWVIVDRLTKSAH 274
Query: 1121 FIAVNMTFPSEKLARIYVKEIVRLHGVPANIVSDRDPRFVSKFWGSLHEALGTRLSLSSA 1180
F+A+ T + LA+ YV EIV+LHGVP +IVSDRD +F S FW + +GT++ +S+A
Sbjct: 275 FLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAEMGTKVQMSTA 334
Query: 1181 YHPQSDGQSERTIQTLEDMLRACVLDYKGSWEDFLPLAEFSYNNSYHSSLGMAPFEALYG 1240
YHPQ+DGQSERTIQTLEDMLR CVLD++G W D L L EF+YNNSY +S+GMAPFE LYG
Sbjct: 335 YHPQTDGQSERTIQTLEDMLRMCVLDWRGHWADHLSLVEFAYNNSYQASIGMAPFEVLYG 394
Query: 1241 RRCKTPLCWLSGEDKITLGPELLQEMTEKVRSIREKLRIAQDRQKSYYDKRHKPLEFQEG 1300
R C+T LCW ++ G + +QE+TE++R ++ ++ AQ+RQ+SY DKR K LEF+ G
Sbjct: 395 RPCRT-LCWTQVGERSIYGADYVQEITERIRVLKLNMKEAQNRQRSYADKRRKELEFEVG 453
Query: 1301 DHVFLRVTPITGVGRSIHSKKLTPKYLGPYQILDRIGAVAYRIALPPSLSNLHDVFHISQ 1360
D V + G S++ R+G VA+R+ L + H VFH+S
Sbjct: 454 DSV-------SQDGHVARSEQ-------------RVGPVAFRLELSDVMRAFHKVFHVSM 493
Query: 1361 LRKYLPDSSHVIEPDNIELEENLTYPTQPVKILERREKQLRKRTVPLVKLAWSDDN-QDA 1419
LRK L V+ +L+ N+T +PV++LERR K+LR++ +PL+K+ + D +
Sbjct: 494 LRKCLHKDDEVLAKIPEDLQPNMTLEARPVRVLERRIKELRRKKIPLIKVLRNCDGVTEE 553
Query: 1420 TWELEESARKRYPSLF 1435
TWE E + R+ F
Sbjct: 554 TWEPEARLKARFKKWF 569
>At2g06890 putative retroelement integrase
Length = 1215
Score = 503 bits (1296), Expect = e-142
Identities = 351/1065 (32%), Positives = 530/1065 (48%), Gaps = 141/1065 (13%)
Query: 108 KEAFLEKYFPADVKGKKETEFLELKQGEMFVGQYAARFEELSQFHPYYGTTADDASKCIR 167
K +++ P+ + + L QG V +Y ++E+ + D + R
Sbjct: 2 KAIMRKRFVPSHYHRELHLKLRNLTQGNRSVEEY---YKEMETLMLRADISEDREATLSR 58
Query: 168 FECGLRPDIRAAIGHQQIRTFTVLVEKCRIFEENDRAR---REYYKSSKFNKTSKRREER 224
F L DI+ + Q ++ K +FE+ + + R Y S K + +REER
Sbjct: 59 FLGDLNRDIQDRLETQYYVQIEEMLHKAILFEQQVKRKSSSRSSYGSGTIAKPTYQREER 118
Query: 225 KKPY------SPR-NYKPELQNRNYGGARP--TNPNSHVTCYRCGKEGHKSWSCTATTT- 274
Y SPR KP +++ G T+ V CY+C +GH + C
Sbjct: 119 TSSYHNKPIVSPRAESKPYAAVQDHKGKAEISTSRVRDVRCYKCQGKGHYANECPNKRVM 178
Query: 275 ----SGQTNLKPPVVGGTNTTSAPRNSAGASAQKPGRPVNKGKVFAMTGAEANTSEDFIQ 330
+G+ + + ++ S+ + + AQ + E ++
Sbjct: 179 ILLDNGEIEPEEEI---PDSPSSLKENEELPAQGELLVARRTLSVQTKTDEQEQRKNLFH 235
Query: 331 GTCFLCDISLVVLYDSGATHSFISHERAKSLKLVITQLPYDLVVTTPTKESAVTSSVCKK 390
C + ++ D G+ + S K L L + + V + V
Sbjct: 236 TRCHVHGKVCSLIIDGGSCTNVASETMVKKL---------GLKWLNDSGKMRVKNQVV-- 284
Query: 391 CPLVIEDREYITNLVC--LPLEGLDIILGMNWLSINNVLLDCRLRVPIFLQK-------- 440
P+VI +Y ++C LP+E I+LG W S V+ D F K
Sbjct: 285 VPIVIG--KYEDEILCDVLPMEAGHILLGRPWQSDRKVMHDGFTNRHSFEFKGGKTILVS 342
Query: 441 ------YKEK-HTASLPE---KEPSAYLI---LFSSEGTKRPAM---------------- 471
Y+++ H E K+P+ + + S+ +K+P +
Sbjct: 343 MTPHEVYQDQIHLKQKKEQVVKQPNFFAKSGEVKSAYSSKQPMLLFVFKEALTSLTNFAP 402
Query: 472 ----EDIPVVREFPEVFPEDMTE-LPPEREVEFAIDVIPGTTPISAAPYRISPLELAELQ 526
E +++++ +VFPED + LPP R +E ID +PG + + YR +P+E ELQ
Sbjct: 403 VLPSEMTSLLQDYKDVFPEDNPKGLPPIRGIEHQIDFVPGASLPNRPAYRTNPVETKELQ 462
Query: 527 KQVEELLSKGFIRPSVSPWGAPVLLVKKKDGSMRLCVDYRQLNKVTIKNRYPLPRIDDLM 586
+Q KDGS R+C D R +N VT+K +P+PR+DD++
Sbjct: 463 RQ--------------------------KDGSWRMCFDCRAINNVTVKYCHPIPRLDDML 496
Query: 587 DQLKGARVFSKIDLRSGYHQIRVKSDDVQKTAFRTRYGHYEYLVMPFGVTNAPAIFMDYM 646
D+L G+ +FSKIDL+SGYHQIR+ D KTAF+T++G YE+LVMPFG+T+AP+ FM M
Sbjct: 497 DELHGSSIFSKIDLKSGYHQIRMNEGDEWKTAFKTKHGLYEWLVMPFGLTHAPSTFMRLM 556
Query: 647 NRIFHPYLDKFVIVFIDDILIYSKSKEEHVEHMQVVLKVLKDRKLYAKLSKCEFWLEQVQ 706
N + ++ FVIV+ DDIL+YS+S EH+EH+ VL VL+ +LYA L KC F + +
Sbjct: 557 NHVLRAFIGIFVIVYFDDILVYSESLREHIEHLDSVLNVLRKEELYANLKKCTFCTDNLV 616
Query: 707 FLGHVVSEDGIAVDPAKVEAVNSWKVPETVTGVRSFLGLAGYYRRFIEGFSKIATPLTQL 766
FLG VVS DG+ VD KV+A+ W P+TV VRSF GLAG+YRRF + FS I PLT++
Sbjct: 617 FLGFVVSADGVKVDEEKVKAIRDWPSPKTVGEVRSFHGLAGFYRRFFKDFSTIVAPLTEV 676
Query: 767 TKKDHPFVWTEKCE*SFQTLKERLTKAPVLTLPDPSKDYDVYCDASKSGLGCVLMQERKV 826
KKD F W + E +FQ+LK++LT APVL L + K +++ CDAS G+G VLMQ++K+
Sbjct: 677 MKKDVGFKWEKAQEEAFQSLKDKLTNAPVLILSEFLKTFEIECDASGIGIGAVLMQDQKL 736
Query: 827 IAYASQQLRPHEQNYPTHDMELAAVVFALKIWRHYLYGVKFTIYSDHQSLKYLFDQKTLN 886
IA+ S++L NYPT+D EL A+V AL+ W+HYL+ F I++DH+SLK+L Q+ LN
Sbjct: 737 IAFFSEKLGGATLNYPTYDKELYALVRALQRWQHYLWPKVFVIHTDHESLKHLKGQQKLN 796
Query: 887 MRQRRWVEFLEDYDFKLQYHPGKANVVADALSRKSLHAARLMIEETELIEKFRDMNLIME 946
R RWVEF+E + + ++Y GK NVVADALS++
Sbjct: 797 KRHARWVEFIETFAYVIKYKKGKDNVVADALSQRY------------------------- 831
Query: 947 TLPQGTRLGTLTLTNEFIEEVKKEQARDENLQKEAHGRDSMSRPDFLKGPDGLWRYQGRL 1006
T L TL + E++K+ D + Q+ + + + + D Y+ RL
Sbjct: 832 -----TLLSTLNVKLMGFEQIKEVYETDHDFQEVYKACEKFASGRYFR-QDKFLFYENRL 885
Query: 1007 CVPEGGELRQKILEEGHKSDFSIHPGTTKMYQDLKKMFWWPGMKKDIMKKVTSCLTCQKV 1066
CVP LR + E H H G K + + + F WP MK D+ + C TC++
Sbjct: 886 CVP-NCSLRDLFVREAHGGGLMGHFGIAKTLEVMTEHFRWPHMKCDVKRICGRCNTCKQA 944
Query: 1067 KGEHQKPSGSLQPLSIPEWKWEGISMDFVSGLPRTTTGHDAIWVI 1111
K + Q P+G PL IP+ W ISMDFV GLPR TG D+I+V+
Sbjct: 945 KSKIQ-PNGLYTPLPIPKHPWNDISMDFVMGLPR--TGKDSIFVV 986
Score = 34.7 bits (78), Expect = 0.41
Identities = 39/176 (22%), Positives = 80/176 (45%), Gaps = 31/176 (17%)
Query: 1233 APFEALYGRRCKTP--LCWLSGEDKITLG----PELLQEMTEKVR-SIREKLRIAQDRQK 1285
+PF+ +YG +P L L +++++ EL+Q++ E R +I EK ++ +
Sbjct: 988 SPFQIVYGFNPISPFDLIPLPLSERVSIDGKKKAELVQQIHENARRNIEEKTKLYAKQA- 1046
Query: 1286 SYYDKRHKPLEFQEGDHVFLRVTPITGVGRSIHSKKLTPKYLGPYQILDRIGAVAYRIAL 1345
+K + F+ GD V++ + + KL P+ GP++I+ RI AY++ L
Sbjct: 1047 ---NKGRREQIFEVGDMVWIHLRKERFPAQ--RKSKLMPRIDGPFKIIKRINDNAYQLDL 1101
Query: 1346 PPS----------------LSNLHDVFHISQLRKYL--PDSSHVIEPDNIELEENL 1383
+ +++D+ H +L + L + + ++ D + EE L
Sbjct: 1102 QDEPDLRSNPFQEGGDDMIMDSINDMEHEPELERELVAEEGAKLVAEDELVAEEKL 1157
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 495 bits (1274), Expect = e-140
Identities = 277/683 (40%), Positives = 378/683 (54%), Gaps = 145/683 (21%)
Query: 754 EGFSKIATPLTQLTKKDHPFVWTEKCE*SFQTLKERLTKAPVLTLPDPSKDYDVYCDASK 813
+GF+ +A P+T+LT+KD PFVW+ +CE F+
Sbjct: 691 KGFASMAQPMTKLTEKDIPFVWSPECEEGFRG---------------------------- 722
Query: 814 SGLGCVLMQERKVIAYASQQLRPHEQNYPTHDMELAAVVFALKIWRHYLYGVKFTIYSDH 873
KVI YAS+QLR HE NYPTHD+E+AA++FALKIW YLYG K +++DH
Sbjct: 723 -----------KVIVYASRQLRKHEGNYPTHDLEMAAIIFALKIWGSYLYGGKVQVFTDH 771
Query: 874 QSLKYLFDQKTLNMRQRRWVEFLEDYDFKLQYHPGKANVVADALSRKSLHAARLMIEETE 933
+SLK +F Q LN+RQRRW+E + DYD ++ YHPGK NVVADALSRK + AA E
Sbjct: 772 KSLKDIFTQPELNLRQRRWMELVADYDLEIAYHPGKTNVVADALSRKRVGAAPGQSVEAL 831
Query: 934 LIEKFRDMNLIMETLPQGTRLGTLTLTNEFIEEVKKEQARDENLQKEAHGRDSMSRPDFL 993
+ E +G L L V +E + E + + A+G +
Sbjct: 832 VSE-----------------IGALRLC-----VVAREPLKLEAVDRAANGTILVHE---- 865
Query: 994 KGPDGLWRYQGRLCVPEGGELRQKILEEGHKSDFSIHPGTTKMYQDLKKMFWWPGMKKDI 1053
R+C+P+ ELR++IL E H S FSIHPG TKMY+DLK+ + W GMK+D+
Sbjct: 866 -----------RVCLPKDEELRREILSEAHASMFSIHPGATKMYRDLKRHYQWVGMKRDV 914
Query: 1054 MKKVTSCLTCQKVKGEHQKPSGSLQPLSIPEWKWEGISMDFVSGLPRTTTGHDAIWVIVD 1113
VT C CQ VK EHQ P G LQ L I EWK D + LP+
Sbjct: 915 ANWVTECDVCQLVKAEHQVPGGLLQSLPISEWK----KTDGAAVLPKK------------ 958
Query: 1114 RLTKSAHFIAVNMTFPSEKLARIYVKEIVRLHGVPANIVSDRDPRFVSKFWGSLHEALGT 1173
YV EIV+LHGVP +I+S RD +F S FW + +GT
Sbjct: 959 -----------------------YVSEIVKLHGVPVSILSHRDSKFTSAFWRAFQVEMGT 995
Query: 1174 RLSLSSAYHPQSDGQSERTIQTLEDMLRACVLDYKGSWEDFLPLAEFSYNNSYHSSLGMA 1233
++ +S+AYHPQ+DGQSERTIQTLEDML+ CVLD+ G W D L L +F+YNNSY +S+GMA
Sbjct: 996 KVQMSTAYHPQTDGQSERTIQTLEDMLQMCVLDWGGHWADHLSLVKFAYNNSYQASIGMA 1055
Query: 1234 PFEALYGRRCKTPLCWLSGEDKITLGPELLQEMTEKVRSIREKLRIAQDRQKSYYDKRHK 1293
PFEALYGR C+T LCW +K G + +QE TE++R ++ ++ AQDRQ+SY DKR +
Sbjct: 1056 PFEALYGRPCRTLLCWTQVGEKSIYGADYVQETTERIRVLKLNMKEAQDRQRSYADKRRR 1115
Query: 1294 PLEFQEGDHVFLRVTPITGVGRSIHSKKLTPKYLGPYQILDRIGAVAYRIALPPSLSNLH 1353
LEF+ G +I++R+G VAYR+ LP + H
Sbjct: 1116 ELEFEVGT-----------------------------EIVERVGPVAYRLELPDVMRAFH 1146
Query: 1354 DVFHISQLRKYLPDSSHVIEPDNIELEENLTYPTQPVKILERREKQLRKRTVPLVKLAWS 1413
+VFH+S LRK L V+ +L+ N+T +PV++LERR K++R++ +P++K+ W
Sbjct: 1147 NVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARPVRVLERRIKEVRRKKIPMIKVLWD 1206
Query: 1414 DDN-QDATWELEESARKRYPSLF 1435
D + TWE E + R+ F
Sbjct: 1207 CDGVTEETWEPEARIKARFKKWF 1229
Score = 292 bits (748), Expect = 8e-79
Identities = 209/596 (35%), Positives = 289/596 (48%), Gaps = 96/596 (16%)
Query: 107 FKEAFLEKYFPADVKGKKETEFLELKQGEMFVGQYAARFEELSQFHPYYGT-TADDASKC 165
F F KYFP + + E FLEL +GE+ V +Y F+ L Y G DD ++
Sbjct: 157 FVAEFNAKYFPQEALDRMEARFLELTKGELSVREYGREFKRLLV---YAGRGMEDDQAQM 213
Query: 166 IRFECGLRPDIRAAIGHQQIRTFTVLVEKCRIFEENDRARREYYKSSKFNKTSKRREERK 225
RF GLRPD+R +CR+ + A + +++ + +R+
Sbjct: 214 RRFLRGLRPDLRV---------------RCRV---SQYATKAALTAAEVEEDLQRQVVAV 255
Query: 226 KP-YSPRNYKPELQNRNYGGARPTNPNSHVTCYRCGKEGHKSWSCTATTTSGQTNLKPPV 284
P P+ + ++ G +R G+ G + ++TS
Sbjct: 256 SPSVQPKKIQQQVAPSKGGKPAQVQKRKWDHPFRAGQGGRAGSAADDSSTS--------- 306
Query: 285 VGGTNTTSAPRNSAGASAQKPGRPVNKGKVFAMTGAEANTSEDFIQGTCFLCDISLV--- 341
+AG +AQ G A + AE + GT +L L+
Sbjct: 307 ------------TAGGAAQSAGA--------ARSAAECAAVSTYCSGTTWLYYADLISGR 346
Query: 342 ----VLYDSGATHSFISHERAKSLKLVITQLPYDLVVTTPTKESAVTSSVCKKCPLVIED 397
VL+DSGA+H FI+ E A + V + K + IE+
Sbjct: 347 CRGHVLFDSGASHCFITPESASRDNIRGEPGEQFGAVKIAGGQFLTVLGRAKGVNIQIEE 406
Query: 398 REYITNLVCLPLEGLDIILGMNWLSINNVLLDC-RLRVPIFLQKYK-------------- 442
+L+ P+ D ILGM+WL V LDC R RV + +
Sbjct: 407 ESMPADLIINPVVLYDAILGMDWLDHYRVHLDCHRGRVSFERPEGRLVYQGVRPTSRSLV 466
Query: 443 --EKHTASLPEKEPSAYLILFS-SEGTKRPAMEDIPVVREFPEVFPEDMTELPPEREVEF 499
E + EK AYL+ S SE + A+ DI VV+EF +VF + + LPP R F
Sbjct: 467 ISEVQVEKMIEKGCEAYLVTISMSESVGQVAVSDIWVVQEFEDVF-QSLQGLPPSRSDPF 525
Query: 500 AIDVIPGTTPISAAPYRISPLELAELQKQVEELLSKGFIRPSVSPWGAPVLLVKKKDGSM 559
I++ GT P+S PYR+ P E+AEL+KQ+E+LL KGFIRPS S WGAP
Sbjct: 526 TIELELGTAPLSKTPYRMVPAEIAELKKQLEDLLGKGFIRPSTSRWGAP----------- 574
Query: 560 RLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGARVFSKIDLRSGYHQIRVKSDDVQKTAF 619
LN+VT+KN+YPLPRID+L+DQL+GA FSKIDL GYHQ + DV+KTAF
Sbjct: 575 -------GLNRVTLKNKYPLPRIDELLDQLRGATCFSKIDLTPGYHQFPIAEADVRKTAF 627
Query: 620 RTRYGHYEYLVMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDILIYSKSKEEH 675
RTRYGH+E++VMPFG+TNAP M MN +F +LD+FVI+FIDDIL+Y KS EEH
Sbjct: 628 RTRYGHFEFVVMPFGLTNAPTALMRLMNSVFQEFLDEFVIIFIDDILVYFKSPEEH 683
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 484 bits (1246), Expect = e-136
Identities = 270/682 (39%), Positives = 398/682 (57%), Gaps = 88/682 (12%)
Query: 476 VVREFPEVFPEDMTELPPEREV-EFAIDVIPGTTPISAAPYRISPLELAELQKQVEELLS 534
VV EFP++F E T+LPP R + I+++ + P++ PYR + E+ K V+++L+
Sbjct: 10 VVTEFPDIFVEP-TDLPPFRAPHDHKIELLEDSNPVNQRPYRYVVHQKNEIDKIVDDMLA 68
Query: 535 KGFIRPSVSPWGAPVLLVKKKDGSMRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGARV 594
G I+ S SP+ +PV+LVKKKDG+ RLCVDYR+LN +T+K+R+P+P I+DLMD+L G+ V
Sbjct: 69 SGTIQASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDRFPIPLIEDLMDELGGSNV 128
Query: 595 FSKIDLRSGYHQIRVKSDDVQKTAFRTRYGHYEYLVMPFGVTNAPAIFMDYMNRIFHPYL 654
+SKIDLR+GYHQ+R+ D+ KTAF+T GHYEYLVMPFG+TNAPA F MN F P+L
Sbjct: 129 YSKIDLRAGYHQVRMDPLDIHKTAFKTHNGHYEYLVMPFGLTNAPASFQSLMNSFFKPFL 188
Query: 655 DKFVIVFIDDILIYSKSKEEHVEHMQVVLKVLKDRKLYAKLSKCEFWLEQVQFLGHVVSE 714
KFV+VF DDILIYS S EEH +H++ V +V++ L+AK+SKC F + +V++LGH +S
Sbjct: 189 RKFVLVFFDDILIYSTSMEEHKKHLEAVFEVMRVHHLFAKMSKCAFAVPRVEYLGHFISG 248
Query: 715 DGIAVDPAKVEAVNSWKVPETVTGVRSFLGLAGYYRRFIEGFSKIATPLTQLTKKDHPFV 774
+GIA DPAK++AV W VP + + FLGL GYYRRF
Sbjct: 249 EGIATDPAKIKAVQDWPVPVNLKQLCGFLGLTGYYRRF---------------------- 286
Query: 775 WTEKCE*SFQTLKERLTKAPVLTLPDPSKDYDVYCDASKSGLGCVLMQERKVIAYASQQL 834
+ V DA G+G VLMQE +A+ S+QL
Sbjct: 287 ------------------------------FVVEMDACGHGIGAVLMQEGHPLAFISRQL 316
Query: 835 RPHEQNYPTHDMELAAVVFALKIWRHYLYGVKFTIYSDHQSLKYLFDQKTLNMRQRRWVE 894
+ + + ++ EL AV+F ++ WRHYL F I +D +SLKYL +Q+ Q++W+
Sbjct: 317 KGKQLHLSIYEKELLAVIFVVRKWRHYLISSHFIIKTDQRSLKYLLEQRLNTPIQQQWLP 376
Query: 895 FLEDYDFKLQYHPGKANVVADALSR----KSLHAARLMIEETELIEKFRDMNLIMETLPQ 950
L ++D+++QY GK N+VADALSR + LH A L + E +L+++ + +
Sbjct: 377 KLLEFDYEIQYKQGKENLVADALSRVEGSEVLHMA-LSVVECDLLKEIQ-----AGYVTD 430
Query: 951 GTRLGTLTLTNEFIEEVKKEQARDENLQKEAHGRDSMSRPDFLKGPDGLWRYQGRLCVPE 1010
G G +T+ LQ++A + + G+ R + ++ VP
Sbjct: 431 GDIQGIITI-----------------LQQQADSKKHYT------WSQGVLRRKNKIVVPN 467
Query: 1011 GGELRQKILEEGHKSDFSIHPGTTKMYQDLKKMFWWPGMKKDIMKKVTSCLTCQKVKGEH 1070
++ IL H S H G +Q +K +F+W M KDI + SC TCQ+ K ++
Sbjct: 468 NSGIKDTILRWLHCSGMGGHSGKEVTHQRVKGLFYWKSMVKDIQAFIRSCGTCQQCKSDN 527
Query: 1071 QKPSGSLQPLSIPEWKWEGISMDFVSGLPRTTTGHDAIWVIVDRLTKSAHFIAVNMTFPS 1130
G LQPL IP+ W +SMDF+ GLP + G I V+VDRL+K+AHFIA+ + +
Sbjct: 528 AASPGLLQPLPIPDRIWSDVSMDFIDGLP-LSNGKTVIMVVVDRLSKAAHFIALAHPYSA 586
Query: 1131 EKLARIYVKEIVRLHGVPANIV 1152
+A+ Y+ + +LHG P++IV
Sbjct: 587 MTVAQAYLDNVFKLHGCPSSIV 608
Score = 100 bits (248), Expect = 8e-21
Identities = 58/156 (37%), Positives = 90/156 (57%), Gaps = 5/156 (3%)
Query: 1232 MAPFEALYGRRCKTPLCWLSGEDKITLGPELLQEMTEKVRSIREKLRIAQDRQKSYYDKR 1291
M P+EA+YG+ L +L GE K+ + +QE + ++ L AQ R K D+
Sbjct: 625 MTPYEAVYGQPPPLHLPYLPGESKVAVVARSMQERESMILFLKFHLMRAQHRMKQLADQH 684
Query: 1292 HKPLEFQEGDHVFLRVTPITGVGRSIHS-KKLTPKYLGPYQILDRIGAVAYRIALPPSLS 1350
EF+ GD+VF+++ P + S +KL+PKY GPY+++DR G VAY++ LP + S
Sbjct: 685 ITEREFEVGDYVFVKLQPYRQQSVVMRSTQKLSPKYFGPYKVIDRCGEVAYKLQLPAN-S 743
Query: 1351 NLHDVFHISQLRKY---LPDSSHVIEPDNIELEENL 1383
+H VFH+SQLR + S+H++ + L ENL
Sbjct: 744 QVHPVFHVSQLRVLVGTVTTSTHLLRCYLMSLRENL 779
>At4g03650 putative reverse transcriptase
Length = 839
Score = 401 bits (1031), Expect = e-111
Identities = 183/294 (62%), Positives = 234/294 (79%)
Query: 613 DVQKTAFRTRYGHYEYLVMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDILIYSKSK 672
DV+KTAFRTRYGH+E++VMPFG+TNAPA FM MN +F +LD+FVI+FIDDIL+YSKS
Sbjct: 520 DVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSP 579
Query: 673 EEHVEHMQVVLKVLKDRKLYAKLSKCEFWLEQVQFLGHVVSEDGIAVDPAKVEAVNSWKV 732
EEH H++ V++ L+++KL+AKLSKC FW ++ FLGH+VS +G++VDP K+EA+ W
Sbjct: 580 EEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWPR 639
Query: 733 PETVTGVRSFLGLAGYYRRFIEGFSKIATPLTQLTKKDHPFVWTEKCE*SFQTLKERLTK 792
P T +RSFLGLAGYYRRFI+GF+ +A P+T+LT KD PFVW+ +CE F +LKE LT
Sbjct: 640 PTNATEIRSFLGLAGYYRRFIKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTS 699
Query: 793 APVLTLPDPSKDYDVYCDASKSGLGCVLMQERKVIAYASQQLRPHEQNYPTHDMELAAVV 852
PVL LP+ + Y VY DAS GLGC LMQ KVIAYAS+QLR HE NYPTHD+E+AAV+
Sbjct: 700 TPVLALPEHGEPYMVYTDASGVGLGCALMQRGKVIAYASRQLRKHEGNYPTHDLEMAAVI 759
Query: 853 FALKIWRHYLYGVKFTIYSDHQSLKYLFDQKTLNMRQRRWVEFLEDYDFKLQYH 906
FALKIWR YLYG K +++DH+SLKY+F Q LN+RQRRW+E + DYD ++ YH
Sbjct: 760 FALKIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQRRWMELVADYDLEIAYH 813
Score = 74.7 bits (182), Expect = 4e-13
Identities = 58/196 (29%), Positives = 87/196 (43%), Gaps = 8/196 (4%)
Query: 9 LANQAARAEQQNQPAED-DVYKGIDKFLKR-NPPLFDGGYDPEGANRWLRKIEQIYESLP 66
+A + +Q+ AE+ Y + K L+R F GG PE A+ W ++E+ + S
Sbjct: 121 VAPRVVEVQQRAAVAEEVPSYLRMMKQLQRIGTEYFSGGTSPEEADSWRSQVERNFGSSR 180
Query: 67 TSEDRMIAYASYLFHEEARNWWVHAKSRITPPDGVLTWSIFKEAFLEKYFPADVKGKKET 126
+ + + +A WW +R D ++W+ F F KYFP + + E
Sbjct: 181 CPAEYRVDLTVHFLEGDAHLWWRSVTARRRQAD--MSWADFMAEFNAKYFPREALDRMEA 238
Query: 127 EFLELKQGEMFVGQYAARFEELSQFHPYYGT-TADDASKCIRFECGLRPDIRAAIGHQQI 185
FLEL QG V +Y +F L Y G DD ++ RF GLRP++R Q
Sbjct: 239 RFLELTQGVRSVREYDRKFNRLL---VYAGRGMEDDQAQMRRFLRGLRPNLRVRCRVSQY 295
Query: 186 RTFTVLVEKCRIFEEN 201
T LVE EE+
Sbjct: 296 ATKAALVETAAEVEED 311
Score = 74.3 bits (181), Expect = 5e-13
Identities = 52/143 (36%), Positives = 78/143 (54%), Gaps = 13/143 (9%)
Query: 403 NLVCLPLEGLDIILGMNWLSINNVLLDC-RLRVPIFLQKYKEKHTASLPEKEPSAYLILF 461
+L+ +E D+ILGM+WL V LDC R RV E+ S + L
Sbjct: 387 DLIISHVELYDVILGMDWLDHYRVHLDCHRGRVSF------ERPEGSAGRENDREGLRGL 440
Query: 462 SSEGTK-----RPAMEDIPVVREFPEVFPEDMTELPPEREVEFAIDVIPGTTPISAAPYR 516
+ + A+ DI VV+EF VF + + LPP R F I++ P T P+S APYR
Sbjct: 441 PGDDIYAGVCGQVAVSDIRVVQEFQYVF-QSLQGLPPSRSDPFTIELEPKTAPLSKAPYR 499
Query: 517 ISPLELAELQKQVEELLSKGFIR 539
++P ++AEL+KQ+E+LL++ +R
Sbjct: 500 MAPAKMAELKKQLEDLLAEADVR 522
Score = 39.7 bits (91), Expect = 0.013
Identities = 15/23 (65%), Positives = 21/23 (91%)
Query: 1171 LGTRLSLSSAYHPQSDGQSERTI 1193
+GT++ +S+ YHPQ+DGQSERTI
Sbjct: 817 MGTKVQMSTTYHPQTDGQSERTI 839
>At4g03840 putative transposon protein
Length = 973
Score = 318 bits (815), Expect = 1e-86
Identities = 165/322 (51%), Positives = 208/322 (64%), Gaps = 9/322 (2%)
Query: 775 WTEKCE*SFQTLKERLTKAPVLTLPDPSKDYDVYCDASKSGLGCVLMQERKVIAYASQQL 834
W CE +F LK LT APVL LP+ + Y VY DAS GL CVLMQ+ VIAYAS+QL
Sbjct: 375 WQRSCEKNFLELKAMLTNAPVLVLPEEGEPYTVYTDASVVGLECVLMQKGSVIAYASRQL 434
Query: 835 RPHEQNYPTHDMELAAVVFALKIWRHYLYGVKFTIYSDHQSLKYLFDQKTLNMRQRRWVE 894
R HE+NYPTHD+E+AAVVFALKIWR YLYG K IY+DH+SLKY+F Q LN+RQRRW+E
Sbjct: 435 RKHEKNYPTHDLEMAAVVFALKIWRSYLYGAKVQIYTDHKSLKYIFTQPELNLRQRRWME 494
Query: 895 FLEDYDFKLQYHPGKANVVADALSRK--SLHAARLMIEETELIEKFRDMNLIMETLPQGT 952
+ DYD + YH GKAN VADALSR+ + A R ++ ++ L E P G
Sbjct: 495 LVADYDLDIAYHAGKANQVADALSRRRSEVEAERSQVDLVNMMGTLHVNALSKEVEPLG- 553
Query: 953 RLGTLTLTNEFIEEVKKEQARDENLQKEAHGRDSMSRPDFLKGPDGLWRYQGRLCVPEGG 1012
LG N + ++ Q RDE E G ++ ++ +G GR+CVP
Sbjct: 554 -LGAADQAN-LLSRIRLAQERDE----EIKGWALNNKTEYQTSNNGTIVVNGRVCVPNNR 607
Query: 1013 ELRQKILEEGHKSDFSIHPGTTKMYQDLKKMFWWPGMKKDIMKKVTSCLTCQKVKGEHQK 1072
L+++IL E H+S FSIHPG+ K+Y+DLK+ + W GMKKD+ + V C TCQ VK EHQ
Sbjct: 608 ALKEEILREAHQSKFSIHPGSNKIYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAEHQV 667
Query: 1073 PSGSLQPLSIPEWKWEGISMDF 1094
PSG LQ L IPEWKW+ I+MDF
Sbjct: 668 PSGLLQNLPIPEWKWDHITMDF 689
Score = 249 bits (636), Expect = 8e-66
Identities = 125/274 (45%), Positives = 176/274 (63%), Gaps = 20/274 (7%)
Query: 1163 FWGSLHEALGTRLSLSSAYHPQSDGQSERTIQTLEDMLRACVLDYKGSWEDFLPLAEFSY 1222
FW + +ALGTR++LS+AYHPQ+DGQSERTIQTLEDMLRAC LD+ G+WE +L LA
Sbjct: 690 FWKAFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACALDWGGNWEKYLRLA---- 745
Query: 1223 NNSYHSSLGMAPFEALYGRRCKTPLCWLSGEDKITLGPELLQEMTEKVRSIREKLRIAQD 1282
LYGR C+TPLCW ++ GP ++ E TE+++ ++ KL+ AQD
Sbjct: 746 ---------------LYGRACRTPLCWTPVGERRLFGPIIVDETTERMKFLKIKLKEAQD 790
Query: 1283 RQKSYYDKRHKPLEFQEGDHVFLRVTPITGVGRSIHSKKLTPKYLGPYQILDRIGAVAYR 1342
RQKSY +KR K LEFQ D V+L+ G GR KKL+P+Y+GPY++++R+GAVAY+
Sbjct: 791 RQKSYANKRRKELEFQVEDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYK 850
Query: 1343 IALPPSLSNLHDVFHISQLRKYLPDSSHVIEPDNIELEENLTYPTQPVKILERREKQLRK 1402
+ LPP L+ H+VFH+SQLRK L + +E L+EN+T PV+I++R K R
Sbjct: 851 LDLPPKLNAFHNVFHVSQLRKCLSNQEESVEDVPPGLKENMTVEAWPVQIMDRMTKGTRG 910
Query: 1403 RTVPLVKLAWS-DDNQDATWELEESARKRYPSLF 1435
++ L+K+ W+ + TWE E + + F
Sbjct: 911 KSRDLLKVLWNCGGREQYTWETENKMKANFSEWF 944
Score = 130 bits (326), Expect = 7e-30
Identities = 57/89 (64%), Positives = 74/89 (83%)
Query: 613 DVQKTAFRTRYGHYEYLVMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDILIYSKSK 672
DV+KTAF TRYGH+E++VMPFG+TNAP FM MN +F +LD+FVI+FIDDIL+YSKS
Sbjct: 287 DVRKTAFLTRYGHFEFVVMPFGLTNAPTAFMKMMNGVFRDFLDEFVIIFIDDILVYSKSW 346
Query: 673 EEHVEHMQVVLKVLKDRKLYAKLSKCEFW 701
E H EH++ VL+ L++ +L+AKLSKC FW
Sbjct: 347 EAHQEHLRAVLEQLREHELFAKLSKCSFW 375
Score = 60.5 bits (145), Expect = 7e-09
Identities = 32/110 (29%), Positives = 56/110 (50%), Gaps = 2/110 (1%)
Query: 42 FDGGYDPEGANRWLRKIEQIYESLPTSEDRMIAYASYLFHEEARNWWVHAKSRITPPDGV 101
F G DP A+ W ++++ ++S ED A + +A NWW+ + R D V
Sbjct: 147 FMGSTDPIVADEWRSRLKRNFKSTRCPEDYQRDIAVHFLERDAHNWWLTVEKR--RGDEV 204
Query: 102 LTWSIFKEAFLEKYFPADVKGKKETEFLELKQGEMFVGQYAARFEELSQF 151
+++ F++ F +KYFP + + E +L+L QG V +Y F L ++
Sbjct: 205 RSFADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTVREYDEEFNRLRRY 254
>At2g06470 putative retroelement pol polyprotein
Length = 899
Score = 318 bits (814), Expect = 2e-86
Identities = 166/322 (51%), Positives = 208/322 (64%), Gaps = 9/322 (2%)
Query: 775 WTEKCE*SFQTLKERLTKAPVLTLPDPSKDYDVYCDASKSGLGCVLMQERKVIAYASQQL 834
W CE SF LK LT APVL LP+ + Y VY DAS GLGCVLMQ+ VIAYAS+QL
Sbjct: 375 WQRSCEKSFLELKAMLTNAPVLVLPEEGEPYTVYTDASIVGLGCVLMQKGSVIAYASRQL 434
Query: 835 RPHEQNYPTHDMELAAVVFALKIWRHYLYGVKFTIYSDHQSLKYLFDQKTLNMRQRRWVE 894
HE+NYPTHD+E+AAVVFALKIWR YLYG K IY+DH+SLKY+F Q LN+RQRRW+E
Sbjct: 435 WKHEKNYPTHDLEMAAVVFALKIWRSYLYGAKVQIYTDHKSLKYIFIQPELNLRQRRWME 494
Query: 895 FLEDYDFKLQYHPGKANVVADALSRK--SLHAARLMIEETELIEKFRDMNLIMETLPQGT 952
+ DYD + YHPGKAN VADALSR+ + A R ++ ++ L E G
Sbjct: 495 LVADYDLDIAYHPGKANQVADALSRRRSEVEAERSQVDLVNMMGTLHVNALSKEVESLG- 553
Query: 953 RLGTLTLTNEFIEEVKKEQARDENLQKEAHGRDSMSRPDFLKGPDGLWRYQGRLCVPEGG 1012
LG + + ++ Q RDE E G ++ ++ +G GR+CVP
Sbjct: 554 -LGAADQA-DLLSRIRLAQERDE----EIKGWALNNKTEYQTSNNGTIVVNGRVCVPNDR 607
Query: 1013 ELRQKILEEGHKSDFSIHPGTTKMYQDLKKMFWWPGMKKDIMKKVTSCLTCQKVKGEHQK 1072
L+++IL E H+S FSIHPG+ KMY+DLK+ + W GMKKD+ + V C TCQ VK EHQ
Sbjct: 608 ALKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAEHQV 667
Query: 1073 PSGSLQPLSIPEWKWEGISMDF 1094
PSG LQ L IPEWKW+ I+MDF
Sbjct: 668 PSGLLQNLPIPEWKWDHITMDF 689
Score = 232 bits (591), Expect = 1e-60
Identities = 114/229 (49%), Positives = 156/229 (67%), Gaps = 19/229 (8%)
Query: 1166 SLHEALGTRLSLSSAYHPQSDGQSERTIQTLEDMLRACVLDYKGSWEDFLPLAEFSYNNS 1225
+ +ALGTR++LS+AYHPQ+DGQSERTIQTLEDMLRACVLD+ G+WE +L LA
Sbjct: 690 AFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLTLA------- 742
Query: 1226 YHSSLGMAPFEALYGRRCKTPLCWLSGEDKITLGPELLQEMTEKVRSIREKLRIAQDRQK 1285
LYGR C+TPLCW ++ GP ++ E TE+++ ++ KL+ A DRQK
Sbjct: 743 ------------LYGRACRTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKEAHDRQK 790
Query: 1286 SYYDKRHKPLEFQEGDHVFLRVTPITGVGRSIHSKKLTPKYLGPYQILDRIGAVAYRIAL 1345
SY +KR K LEFQ GD V+L+ G GR KKL+P+Y+GPY++++R+GAVAY++ L
Sbjct: 791 SYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDL 850
Query: 1346 PPSLSNLHDVFHISQLRKYLPDSSHVIEPDNIELEENLTYPTQPVKILE 1394
PP L+ H+VFH+SQLRK L D +E L+EN+T PV+I++
Sbjct: 851 PPKLNAFHNVFHVSQLRKCLSDQEESVEDVPPGLKENMTVEAWPVRIMD 899
Score = 134 bits (336), Expect = 5e-31
Identities = 59/89 (66%), Positives = 76/89 (85%)
Query: 613 DVQKTAFRTRYGHYEYLVMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDILIYSKSK 672
DV+KTAFRTRYGH+E++VMPFG+TNAPA FM MN +F +LD+FVI+FIDDIL+YSKS
Sbjct: 287 DVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVYSKSW 346
Query: 673 EEHVEHMQVVLKVLKDRKLYAKLSKCEFW 701
E H EH++ VL+ L++ +L+AKLSKC FW
Sbjct: 347 EAHQEHLRAVLERLREHELFAKLSKCSFW 375
Score = 59.7 bits (143), Expect = 1e-08
Identities = 32/110 (29%), Positives = 56/110 (50%), Gaps = 2/110 (1%)
Query: 42 FDGGYDPEGANRWLRKIEQIYESLPTSEDRMIAYASYLFHEEARNWWVHAKSRITPPDGV 101
F G DP A+ W ++++ ++S ED A + +A NWW+ + R D V
Sbjct: 147 FMGSTDPIVADEWRSRLKRNFKSTRCPEDYQRDIAVHFLEGDAHNWWLTVEKR--RGDEV 204
Query: 102 LTWSIFKEAFLEKYFPADVKGKKETEFLELKQGEMFVGQYAARFEELSQF 151
+++ F++ F +KYFP + + E +L+L QG V +Y F L ++
Sbjct: 205 RSFADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTVREYDEEFNRLRRY 254
>At4g08100 putative polyprotein
Length = 1054
Score = 312 bits (799), Expect = 1e-84
Identities = 159/348 (45%), Positives = 222/348 (63%), Gaps = 30/348 (8%)
Query: 564 DYRQLNKVTIKNRYPLPRIDDLMDQLKGARVFSKIDLRSGYHQIRVKSDDVQKTAFRTRY 623
D R +N +T+K R+P+PR+DD +D+L G+ +FSKIDL+SGYHQ R+K D KTA +T+
Sbjct: 527 DCRAINNITVKYRHPIPRLDDTLDKLHGSSIFSKIDLKSGYHQTRMKEGDEWKTAIKTKQ 586
Query: 624 GHYEYLVMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDILIYSKSKEEHVEHMQVVL 683
YE+LVMPFG+TNAP FM MN + ++ FVIV+ DDIL+YSK+ E HV H+++VL
Sbjct: 587 RLYEWLVMPFGLTNAPNTFMRLMNHVLRKHIGVFVIVYFDDILVYSKNLEYHVMHLKLVL 646
Query: 684 KVLKDRKLYAKLSKCEFWLEQVQFLGHVVSEDGIAVDPAKVEAVNSWKVPETVTGVRSFL 743
+L+ KLYA L KC F + + FLG VVS DGI VD KV+A+ W P+ V+
Sbjct: 647 DLLRKEKLYANLKKCTFCTDNLVFLGFVVSADGIKVDEEKVKAIREWPNPKNVS------ 700
Query: 744 GLAGYYRRFIEGFSKIATPLTQLTKKDHPFVWTEKCE*SFQTLKERLTKAPVLTLPDPSK 803
+KD F W + E +FQ LKE+LT + V LP+ K
Sbjct: 701 ------------------------EKDIGFKWEDAQENAFQALKEKLTNSSVPILPNFMK 736
Query: 804 DYDVYCDASKSGLGCVLMQERKVIAYASQQLRPHEQNYPTHDMELAAVVFALKIWRHYLY 863
+++ CDAS G+G VLMQ+ K IAY S++L NYPT+D EL A+V AL+ W+HYL+
Sbjct: 737 SFEIECDASGLGIGAVLMQDHKPIAYFSEKLGGATLNYPTYDKELYALVRALQTWQHYLW 796
Query: 864 GVKFTIYSDHQSLKYLFDQKTLNMRQRRWVEFLEDYDFKLQYHPGKAN 911
KF I++DH+SLK+L Q+ LN R RWVEF+E + + ++Y +A+
Sbjct: 797 PKKFVIHTDHESLKHLKGQQKLNKRHARWVEFIETFPYVIKYKKREAH 844
Score = 54.7 bits (130), Expect = 4e-07
Identities = 41/170 (24%), Positives = 84/170 (49%), Gaps = 12/170 (7%)
Query: 1227 HSSLGMAPFEALYGRRCKTPLCW--LSGEDKITLGPELLQEMTEKVRSIREKLRIAQDRQ 1284
HS+ +PFE +YG + +PL L ++ +L + ++ ++V ++ + + +
Sbjct: 851 HSATKFSPFEIVYGFKPTSPLDLIPLPLSERSSLDGKKKDDLVQQVLNLEARTK----QY 906
Query: 1285 KSYYDKRHKPLEFQEGDHVFLRVTPITGVGRSIHSKKLTPKYLGPYQILDRIGAVAYRIA 1344
K Y +K K + F EGD V++ + + S KL P+ GP+++L RI AY++
Sbjct: 907 KKYANKGRKEVIFNEGDQVWVHLRKKRFP--EVRSSKLMPRIDGPFKVLKRINNNAYKLD 964
Query: 1345 LPPSLSNLHDVFHISQLRKYLPDSSHVIEP----DNIELEENLTYPTQPV 1390
L + + F + + + + +++ +EP + + EE L P P+
Sbjct: 965 LQDTSDLRTNPFQVGEDDENMTNATKPLEPQEDEEQLVPEEALIVPAGPL 1014
Score = 50.4 bits (119), Expect = 7e-06
Identities = 48/221 (21%), Positives = 88/221 (39%), Gaps = 13/221 (5%)
Query: 10 ANQAARAEQQNQPAEDDVYKGIDKFLKRNPPLFDGGYDPEGANRWLRKIEQIYESLPTSE 69
++ + R ++ +P D G LK P F G +P+ W +KIE ++ +
Sbjct: 71 SHSSQRRHRRERPPPRDPLGG----LKLKIPAFHGTDNPDTYLEWEQKIELVFLCQECLQ 126
Query: 70 DRMIAYASYLFHEEARNWW--VHAKSRITPPDGVLTWSIFKEAFLEKYFPADVKGKKETE 127
+ A+ F+ A +WW + R T + TW+ K +++ P+ +
Sbjct: 127 SNKVKIAATKFYNYALSWWDQLVTSRRRTRDYPIKTWNQLKFVMRKRFVPSYYHRELHQR 186
Query: 128 FLELKQGEMFVGQYAARFEELSQFHPYYGTTADDASKCIRFECGLRPDIRAAIGHQQIRT 187
L QG V +Y F E+ D + RF GL +I+ + Q
Sbjct: 187 LRNLVQGSKTVEEY---FLEMETLMLRADLQEDGEAVMSRFMGGLNREIQDRLETQHYVE 243
Query: 188 FTVLVEKCRIFEENDRARREYYKSSKFN----KTSKRREER 224
++ K +FE+ + + +K N K S ++EE+
Sbjct: 244 LEEMLHKAVMFEQQIKRKNARSSHTKTNYSSGKPSYQKEEK 284
>At3g29490 hypothetical protein
Length = 438
Score = 308 bits (789), Expect = 1e-83
Identities = 161/388 (41%), Positives = 224/388 (57%), Gaps = 54/388 (13%)
Query: 1049 MKKDIMKKVTSCLTCQKVKGEHQKPSGSLQPLSIPEWKWEGISMDFVSGLPRTTTGHDAI 1108
MK+D+ V C CQ VK EHQ P G LQ L IPEW
Sbjct: 1 MKRDVANWVAECDVCQLVKAEHQVPGGMLQSLPIPEWNT--------------------- 39
Query: 1109 WVIVDRLTKSAHFIAVNMTFPSEKLARIYVKEIVRLHGVPANIVSDRDPRFVSKFWGSLH 1168
F+A+ T + LA+ YV EIV+LHGVPA
Sbjct: 40 ------------FLAIRKTDGAAVLAKKYVSEIVKLHGVPAE------------------ 69
Query: 1169 EALGTRLSLSSAYHPQSDGQSERTIQTLEDMLRACVLDYKGSWEDFLPLAEFSYNNSYHS 1228
+GT++ +S+ YHPQ+DGQ ERTIQTLEDMLR CVLD+ G W D L L EF+YNNSY +
Sbjct: 70 --MGTKVQMSTPYHPQTDGQFERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYQA 127
Query: 1229 SLGMAPFEALYGRRCKTPLCWLSGEDKITLGPELLQEMTEKVRSIREKLRIAQDRQKSYY 1288
+GMAPFEALYGR C+TPLCW ++ G + +QE TE++R ++ ++ AQDRQ SY
Sbjct: 128 GIGMAPFEALYGRPCRTPLCWTQVGERSIYGADYVQETTERIRVLKLNMKEAQDRQWSYA 187
Query: 1289 DKRHKPLEFQEGDHVFLRVTPITGVGRSIHSKKLTPKYLGPYQILDRIGAVAYRIALPPS 1348
DKR + LEF+ GD V+L++ + G RSI KL+ +Y+GP++I++R+G VAY + LP
Sbjct: 188 DKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLSLRYMGPFRIVERVGPVAYMLELPDV 247
Query: 1349 LSNLHDVFHISQLRKYLPDSSHVIEPDNIELEENLTYPTQPVKILERREKQLRKRTVPLV 1408
+ H VFH+S LRK L V+ +L+ N+T + V++LERR K+L+++ + L+
Sbjct: 248 MRAFHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARQVRVLERRIKELQRKKISLI 307
Query: 1409 KLAWSDDN-QDATWELEESARKRYPSLF 1435
K+ W D + TW+ E + R+ F
Sbjct: 308 KVLWDCDGVTEETWQPEARMKARFKKWF 335
>At3g29480 hypothetical protein
Length = 718
Score = 280 bits (717), Expect = 3e-75
Identities = 161/368 (43%), Positives = 218/368 (58%), Gaps = 32/368 (8%)
Query: 681 VVLKVLKDRKLYAKLSKCEFWLEQVQFLGH---VVSEDGIAVDPAKVEAVNSWKVPETVT 737
+V+ ++ K+ K CE +L + +E G++VD K+EA+ W P T
Sbjct: 379 LVISAVQAEKMIKK--DCEAYLVTISMKSDGEATGAEAGVSVDLEKIEAIKDWPRPTNAT 436
Query: 738 GVRSFLGLAGYYRRFIEGFSKIATPLTQLTKKDHPFVWTEKCE*SFQTLKERLTKAPVLT 797
+RSFLGLAGYYRRF++GF+ +A PLT+LT K+ PFVW+ +CE F +LKE LT PVL
Sbjct: 437 EIRSFLGLAGYYRRFVKGFASMAPPLTKLTGKNVPFVWSPECEEGFVSLKEILTSTPVLA 496
Query: 798 LPDPSKDYDVYCDASKSGLGCVLMQERKVIAYASQQLRPHEQNYPTHDMELAAVVFALKI 857
LP+ + Y VY DAS+ GLGCVLMQ KVIAYAS+QLR HE NYPTHD+E++AV
Sbjct: 497 LPEHGESYMVYTDASRVGLGCVLMQRGKVIAYASRQLRKHEGNYPTHDLEMSAV------ 550
Query: 858 WRHYLYGVKFTIYSDHQSLKYLFDQKTLNMRQRRWVEFLEDYDFKLQYHPGKANVVADAL 917
++DH+SLKY+ Q LN+RQRRW+E + DYD ++ YH GKA+VVADAL
Sbjct: 551 ------------FTDHKSLKYIITQPELNLRQRRWMELVADYDLEIAYHHGKASVVADAL 598
Query: 918 SRKSLHAARLMIEETELIEKFRDMNL-IMETLPQGTRLGTLTLTNEFIEEVKKEQARDEN 976
SRK + A E L+ + R + L + P G + + V+ Q +DE
Sbjct: 599 SRKRVGVAPGQSVEA-LVSEIRALRLCAVAREPLGLEAVDRV---DLLSRVRLAQEKDEG 654
Query: 977 LQKEAHGRDSMSRPDFLKGPDGLWRYQGRLCVPEGGELRQKILEEGHKSDFSIHPGTTKM 1036
L + S + +G GR+CV ELR++IL E H S FSIHPG TKM
Sbjct: 655 LIAASKAEGS----KYQFAANGTILVHGRVCVLNDEELRREILSEAHASMFSIHPGATKM 710
Query: 1037 YQDLKKMF 1044
Y+DLK+ +
Sbjct: 711 YRDLKEYY 718
Score = 61.6 bits (148), Expect = 3e-09
Identities = 38/142 (26%), Positives = 65/142 (45%), Gaps = 4/142 (2%)
Query: 9 LANQAARAEQQNQPAED--DVYKGIDKFLKRNPPLFDGGYDPEGANRWLRKIEQIYESLP 66
+A + A +Q+ AE+ + +++ + F GG PE A+ W ++E+ + S
Sbjct: 16 VAPRVAEVQQRAAVAEEVPSYLRMMEQLQRIGIGYFSGGTSPEEADSWRSRVERNFGSSR 75
Query: 67 TSEDRMIAYASYLFHEEARNWWVHAKSRITPPDGVLTWSIFKEAFLEKYFPADVKGKKET 126
+ + A + +A WW +R + ++W+ F F KYFP + + E
Sbjct: 76 CPVEYRVDLAVHFLEGDAHLWWKSVTTRRRQAN--MSWADFVAEFNAKYFPQEALDRMEA 133
Query: 127 EFLELKQGEMFVGQYAARFEEL 148
FLEL QGE V +Y F L
Sbjct: 134 HFLELTQGERSVREYDREFNRL 155
Score = 33.9 bits (76), Expect = 0.70
Identities = 35/115 (30%), Positives = 50/115 (43%), Gaps = 28/115 (24%)
Query: 403 NLVCLPLEGLDIILGMNWLSINNVLLDC-RLRVPIFLQKYKEKHTASLP----------- 450
NL P E D+ILGM+WL V LDC R RV K + + A P
Sbjct: 326 NLFLSPEELYDVILGMDWLDYYRVHLDCHRGRVSFERPKGRLIYQAVRPTLGSLVISAVQ 385
Query: 451 -----EKEPSAYLILFS--SEGTKRPA-------MEDIPVVREFPEVFPEDMTEL 491
+K+ AYL+ S S+G A +E I ++++P P + TE+
Sbjct: 386 AEKMIKKDCEAYLVTISMKSDGEATGAEAGVSVDLEKIEAIKDWPR--PTNATEI 438
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,573,582
Number of Sequences: 26719
Number of extensions: 1512715
Number of successful extensions: 4514
Number of sequences better than 10.0: 113
Number of HSP's better than 10.0 without gapping: 78
Number of HSP's successfully gapped in prelim test: 35
Number of HSP's that attempted gapping in prelim test: 4155
Number of HSP's gapped (non-prelim): 251
length of query: 1435
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1323
effective length of database: 8,326,068
effective search space: 11015387964
effective search space used: 11015387964
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0039.4


