

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0039.13
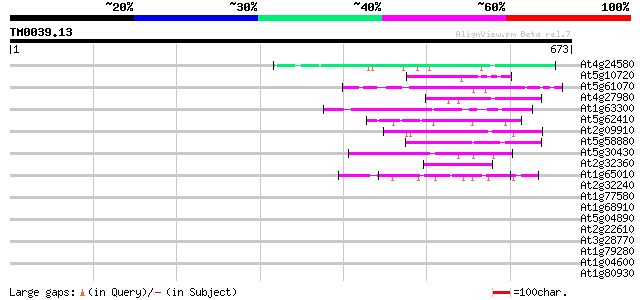
(673 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g24580 putative protein 47 5e-05
At5g10720 histidine kinase - like protein 44 3e-04
At5g61070 HDA18 43 5e-04
At4g27980 putative protein 43 5e-04
At1g63300 hypothetical protein 43 5e-04
At5g62410 chromosomal protein - like 43 7e-04
At2g09910 pseudogene 43 7e-04
At5g58880 putative protein 42 9e-04
At5g30430 putative protein 42 9e-04
At2g32360 hypothetical protein 42 9e-04
At1g65010 hypothetical protein 42 9e-04
At2g32240 putative myosin heavy chain 42 0.001
At1g77580 unknown protein 42 0.001
At1g68910 unknown protein 42 0.001
At5g04890 unknown protein 41 0.002
At2g22610 putative kinesin heavy chain 41 0.002
At3g28770 hypothetical protein 41 0.002
At1g79280 hypothetical protein 41 0.002
At1g04600 putative myosin heavy chain 41 0.002
At1g80930 unknown protein 40 0.004
>At4g24580 putative protein
Length = 790
Score = 46.6 bits (109), Expect = 5e-05
Identities = 88/389 (22%), Positives = 141/389 (35%), Gaps = 62/389 (15%)
Query: 317 EDGQSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAEVGGTSGSAKNVSLEADHGSDP 376
E+G S DEE D + D +QG E + + + ED G S SA + AD DP
Sbjct: 367 EEG-SDDEEYDDDDDGSQGSEDYTDEE-----EDLENESNGSYSESAASEDKYAD-SIDP 419
Query: 377 VQGKGRSVSEEKDTRGP-ASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPS------- 428
K RS +K G S R +D + D V+G + V V S
Sbjct: 420 DDHKARSKEPKKLLSGSRRSSLPRHDDGKKDEDIVVKGVNNTEVKAVVEVSTSEDKNSST 479
Query: 429 ---------PERLS--------IWGPRAGNCREHLAQIDDLVLTENDQKYLGDREPAGL- 470
P +LS WG G + ID V + D + E L
Sbjct: 480 SDVASDTQKPSKLSDAPGGSKRHWGRTPGKKNLSMESIDFSVEVDEDNADIERLESTKLE 539
Query: 471 ----------LNYVLRASLKTASAVRY-----LQGSTIPELEELREK-------TEKDDQ 508
N VL+ASL+ Y L+ +L+E+ + K D
Sbjct: 540 LQSRITEEVKSNAVLQASLERRKKALYGRRQALEQDLKKDLQEVAQAEADIAKLEHKVDD 599
Query: 509 LISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEG---K 565
L + + + + SK + N ++K+K +D +A + R + G +
Sbjct: 600 LENRLGHHDGKASGSTHSASKESRKLPEHNAKMKEKQKDTEAASTHISERSTSKTGNILQ 659
Query: 566 AETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAATVSKINKASFNNAVS 625
GAAR E E K+++ S ++ +R SS K + T + ++ N +
Sbjct: 660 DGQGAARENETE----KQQDSRSKSSQQETSRGSSKLVGLSKRSGTKGEERRSQIANELQ 715
Query: 626 QLAVVNPGLETAPVGYRKVVSGGTDRDCG 654
+ + +P + VS T++ G
Sbjct: 716 NMDKGKTLGQPSPTSGQNRVSEETEKGSG 744
>At5g10720 histidine kinase - like protein
Length = 950
Score = 43.9 bits (102), Expect = 3e-04
Identities = 37/129 (28%), Positives = 63/129 (48%), Gaps = 11/129 (8%)
Query: 477 ASLKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKL 536
A L+ +AVR S + + + E+T + Q++++MS E+ S + +++ L KL
Sbjct: 334 AKLREDNAVRKAMESELNKTIHITEETMRAKQMLATMSHEIRSPLSGVVGMAEILSTTKL 393
Query: 537 ENEQ---LKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKE 593
+ EQ L + GD V + + L + K E+G RL EA + RE VK
Sbjct: 394 DKEQRQLLNVMISSGDLVLQLINDILDLS--KVESGVMRL---EATKFRPRE---VVKHV 445
Query: 594 LENRVSSLE 602
L+ +SL+
Sbjct: 446 LQTAAASLK 454
>At5g61070 HDA18
Length = 682
Score = 43.1 bits (100), Expect = 5e-04
Identities = 60/272 (22%), Positives = 111/272 (40%), Gaps = 34/272 (12%)
Query: 400 VEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSIWGPRAGNCREHLAQIDDLVLTENDQ 459
+ED Q H E E R V+ ERL + P + EN +
Sbjct: 381 LEDKQIHGSSETYPLESTRR----VIQAVRERLCTYWPSLDA---------SMASNENLK 427
Query: 460 KYLGDREPAGLLNYVLRASLKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEE 519
+R A L L+ ++ L + ELE R++ + ++ + + KELE
Sbjct: 428 NPSAERNSADAL-------LREVEELKSLMAARDGELEARRKELKAKNKELEANEKELEA 480
Query: 520 GCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDL-----EGRLKAAEGKAETG---AA 571
G I + + + E L+++ + A E + E R ++ E K +T +
Sbjct: 481 GLMLIRAREDVICGLHAKIESLQQERDEAVAKAERIDKELQEDRARSQEFKEDTEFCLST 540
Query: 572 RLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAATVSKINKASFNNAVSQLAVVN 631
+E E A + + + A +KELE R+ + A + K+ A + ++ + + AV++ ++
Sbjct: 541 LRREKELAIMAKNKDLEAKEKELEARLMLVHAREDKIHAKIERLQQER-DEAVAKAERID 599
Query: 632 PGLETAPVGYRKVVSGGTDRDCGFPREIYPDL 663
L+ R V G+ F +E Y D+
Sbjct: 600 KELQED--RSRSRVGNGS---FAFSQEFYEDM 626
>At4g27980 putative protein
Length = 565
Score = 43.1 bits (100), Expect = 5e-04
Identities = 40/151 (26%), Positives = 67/151 (43%), Gaps = 14/151 (9%)
Query: 499 LREKTEKDDQLISSMSKELEEGCSAIE---ELSKALDNMKLE--------NEQLKKKVQD 547
LR T+K ++L+S EE C E EL KA L+ NE+ + + +D
Sbjct: 123 LRSTTKKLEELVSEFDGRKEEACRVSEKLCELEKAEKEFHLKQRAETERRNEESEAREKD 182
Query: 548 GDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKK 607
A+ E ++ + + K ET ++KE K RE++ ++K LE + +LE K+
Sbjct: 183 LRALEEAVKEKTAELKRKEETLELKMKEEAE---KLREETELMRKGLEIKEKTLEKRLKE 239
Query: 608 LAATVSKINKASFNNAVSQLAVVNPGLETAP 638
L ++ + S V + LE P
Sbjct: 240 LELKQMELEETSRPQLVEAESRKRSNLEIEP 270
>At1g63300 hypothetical protein
Length = 1029
Score = 43.1 bits (100), Expect = 5e-04
Identities = 59/255 (23%), Positives = 113/255 (44%), Gaps = 34/255 (13%)
Query: 377 VQGKGRSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSIWG 436
V+ + R++ E+ R K V G++Q +E + + S E++++
Sbjct: 596 VEQEQRAIQAEETLRKTRWKNASVA-------GKLQDEFKRLSEQMDSMFTSNEKMAMKA 648
Query: 437 -PRAGNCREHLAQIDDLVLTENDQKYLGDREPAGLLNYVL-RASLKTASAVRYLQGSTIP 494
A R Q+++++ ND+ E L+ + + S KT+ R L+
Sbjct: 649 MTEANELRMQKRQLEEMIKDANDELRANQAEYEAKLHELSEKLSFKTSQMERMLENLDEK 708
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRED 554
E +K ++D + +++++E++ IE L K D++ L+ EQ + +R D
Sbjct: 709 SNEIDNQKRHEED-VTANLNQEIKILKEEIENLKKNQDSLMLQAEQAEN-------LRVD 760
Query: 555 LEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAA--TV 612
LE K+ + EAEA+ +E + K ELE+++S + + + LAA V
Sbjct: 761 LEKTKKS-----------VMEAEASLQRENMK----KIELESKISLMRKESESLAAELQV 805
Query: 613 SKINKASFNNAVSQL 627
K+ K A+S L
Sbjct: 806 IKLAKDEKETAISLL 820
Score = 32.3 bits (72), Expect = 0.88
Identities = 28/109 (25%), Positives = 49/109 (44%), Gaps = 7/109 (6%)
Query: 483 SAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLK 542
S +R S EL+ ++ ++ + IS + ELE S ++L +L LE E+ K
Sbjct: 790 SLMRKESESLAAELQVIKLAKDEKETAISLLQTELETVRSQCDDLKHSLSENDLEMEKHK 849
Query: 543 KKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVK 591
K+V ++ LK E +LKE+ A K ++++ K
Sbjct: 850 KQV-------AHVKSELKKKEETMANLEKKLKESRTAITKTAQRNNINK 891
Score = 30.4 bits (67), Expect = 3.4
Identities = 64/316 (20%), Positives = 128/316 (40%), Gaps = 43/316 (13%)
Query: 321 STDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAEVGGTSGSA-----------KNVSLE 369
STD+ + D+ + A N D ++ E+ G + A K + E
Sbjct: 264 STDDSTNSSNDIVARDT--AINSSDEDEVEKLKNELVGLTRQADLSELELQSLRKQIVKE 321
Query: 370 ADHGSDPVQGKGRSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSP 429
D ++ + S+ +E+D+ ++++V D Q + E +P ++ +
Sbjct: 322 TKRSQDLLR-EVNSLKQERDSLKEDCERQKVSDKQKGETKTRNRLQFEGRDPWVLLEETR 380
Query: 430 ERLSIWGPRAGNCREHLAQIDD----LVLTENDQKYLGD---REPAGLLNYVLRASLKTA 482
E L R N R L + + L+L D + + + +E A + +R S ++
Sbjct: 381 EELDYEKDRNFNLRLQLEKTQESNSELILAVQDLEEMLEEKSKEGADNIEESMRRSCRSE 440
Query: 483 SAVRYLQGSTIPELEELR---EKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENE 539
+ + +L + + T +Q I+ + E+E +EL ++ + L+ E
Sbjct: 441 TDEDDHDQKALEDLVKKHVDAKDTHILEQKITDLYNEIEIYKRDKDELEIQMEQLALDYE 500
Query: 540 QLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVK-KELENRV 598
LK++ D+ +L+ ++ + E ++ SS V ELEN+V
Sbjct: 501 ILKQQ-------NHDISYKLEQSQLQ-----------EQLKIQYECSSSLVDVTELENQV 542
Query: 599 SSLEADKKKLAATVSK 614
SLEA+ KK + S+
Sbjct: 543 ESLEAELKKQSEEFSE 558
>At5g62410 chromosomal protein - like
Length = 1175
Score = 42.7 bits (99), Expect = 7e-04
Identities = 51/202 (25%), Positives = 88/202 (43%), Gaps = 20/202 (9%)
Query: 429 PERLSIWGPRAGNCREHLAQIDDLVLTEND----QKYLGDREPAGLLNYVLRASLKTASA 484
P L G R G + L ++ DL E++ QK L D E + + +K
Sbjct: 657 PSGLLTGGSRKGG-GDRLRKLHDLAEAESELQGHQKRLADVESQ--IKELQPLQMKFTDV 713
Query: 485 VRYLQGSTIPELEELREKTEKDD-----QLISSMSKELEEGCSAIEELSKALDNMKLENE 539
L+ T +L ++ E+++ + + + +ELEE S I+E A N
Sbjct: 714 YAQLELKTY-DLSLFLKRAEQNEHHKLGEAVKKLEEELEEAKSQIKEKELAYKNCFDAVS 772
Query: 540 QLKKKVQDGDAVRE----DLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKE-- 593
+L+ ++D D RE DLE +K + + + + LK E K + A+K+E
Sbjct: 773 KLENSIKDHDKNREGRLKDLEKNIKTIKAQMQAASKDLKSHENEKEKLVMEEEAMKQEQS 832
Query: 594 -LENRVSSLEADKKKLAATVSK 614
LE+ ++SLE L + V +
Sbjct: 833 SLESHLTSLETQISTLTSEVDE 854
Score = 34.7 bits (78), Expect = 0.18
Identities = 29/144 (20%), Positives = 56/144 (38%), Gaps = 6/144 (4%)
Query: 475 LRASLKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNM 534
L ++KT A + E +EK +++ + LE S + L + +
Sbjct: 792 LEKNIKTIKAQMQAASKDLKSHENEKEKLVMEEEAMKQEQSSLE---SHLTSLETQISTL 848
Query: 535 KLENEQLKKKVQDGDAVREDLEGRLKAAEGK---AETGAARLKEAEAAWLKEREQSSAVK 591
E ++ + KV + ++ LK K +T + + L++ +
Sbjct: 849 TSEVDEQRAKVDALQKIHDESLAELKLIHAKMKECDTQISGFVTDQEKCLQKLSDMKLER 908
Query: 592 KELENRVSSLEADKKKLAATVSKI 615
K+LEN V +E D K + V K+
Sbjct: 909 KKLENEVVRMETDHKDCSVKVDKL 932
>At2g09910 pseudogene
Length = 985
Score = 42.7 bits (99), Expect = 7e-04
Identities = 50/217 (23%), Positives = 92/217 (42%), Gaps = 30/217 (13%)
Query: 449 IDDLVLTENDQKYL-GDREPAGLLNYV-------LRASL----------KTASAVRYLQG 490
+DDLVL E QK+ E A L N + L+ L K A + +
Sbjct: 513 LDDLVLKEGYQKFARSSLETAALANDLIATYDRKLKLKLADREAFDNLKKCADQAKAIYA 572
Query: 491 STIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQ-DGD 549
+ E+ LR+ E +SS++ E++ +L K + ++++ +K+ + + +
Sbjct: 573 KDMKEMAALRDAAEIHKAEMSSLNDEVKRLNGREADLQKEISDLQVALVAVKEHGEKECN 632
Query: 550 AVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLE------- 602
+R DL ++ KA+ ++K A+LKE+E K ++EN+ E
Sbjct: 633 RLRNDLVAKVARTTKKAQARLDKVK----AYLKEQEDLVGPKVDVENQAKGAEEIELTKR 688
Query: 603 ADKKKLAATVSKINKASFNNAVSQLAVVNPGLETAPV 639
A + A V ++ N + QL + + APV
Sbjct: 689 ATDEVNALNVIELGDDDLNMSPDQLGFLRQASQIAPV 725
>At5g58880 putative protein
Length = 1088
Score = 42.4 bits (98), Expect = 9e-04
Identities = 40/163 (24%), Positives = 71/163 (43%), Gaps = 5/163 (3%)
Query: 476 RASLKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMK 535
R LK +VR + E + + EKD +++S+ +L EE KAL+
Sbjct: 92 RCHLKQQRSVRRNARMKVEEWDSQTSEEEKDKVILTSLYNDLLGRTPQFEESPKALETNV 151
Query: 536 LENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELE 595
+E E+ K+K G+ V DL G L E + E++ ++E S+A E
Sbjct: 152 VEEEEDKEKEFLGEGVSRDL-GHLNVEEPMVCNCEIKYGESDGKVEMKQEMSNA----NE 206
Query: 596 NRVSSLEADKKKLAATVSKINKASFNNAVSQLAVVNPGLETAP 638
+ +S +E +K+ + + + F A+ Q + T+P
Sbjct: 207 HGISEIERNKRLESLIARRRARRRFRLALDQKNKLQAEETTSP 249
>At5g30430 putative protein
Length = 623
Score = 42.4 bits (98), Expect = 9e-04
Identities = 47/211 (22%), Positives = 94/211 (44%), Gaps = 18/211 (8%)
Query: 407 ADGEVQGSEPERAEPVTVVPPSPERLSIWGPRA-GNCREHLAQIDDLVLTENDQKYLGDR 465
AD + +EP+ A P + P+ S+ GN + D +L D+ + R
Sbjct: 298 ADPRQEEAEPQDANPKSKAVLVPQTKSLAAMETEGNVFQCFKTRKDTILPTFDRWHPAIR 357
Query: 466 EPAGL-LNYVLRASLKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAI 524
E L ++ RA+++ ++Y + + +++ +K SS+ +L +
Sbjct: 358 ERFLLHAHHSSRANIELNDMIKYYERLLLDREQDVMAWKDK----FSSLESDLRSSTEVM 413
Query: 525 EELSKALDNMKLE----NEQLKKKVQDGDAVREDL---EGRLKAAEGKAETGAARLKEAE 577
++L LDN+ + N +L+ + Q D ++E+L GRL +E A + +L E +
Sbjct: 414 QKLEDQLDNLSFKIMKSNGELQDQYQRYDKIQEELSNARGRLSESESSAYDLSNQLSELQ 473
Query: 578 A-----AWLKEREQSSAVKKELENRVSSLEA 603
A A L++ E + + K + S++A
Sbjct: 474 AKYKAIAKLRDAELARSASKARKEISKSMKA 504
>At2g32360 hypothetical protein
Length = 175
Score = 42.4 bits (98), Expect = 9e-04
Identities = 28/84 (33%), Positives = 44/84 (52%), Gaps = 1/84 (1%)
Query: 497 EELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDA-VREDL 555
+E R K E DD + +S E E G EE+ KA + + E++ +K + +D D V ED
Sbjct: 86 KEERSKGEDDDDPMEEVSSEAESGRGNEEEVEKAKIDGEEEDQAMKDEEEDRDVKVEEDE 145
Query: 556 EGRLKAAEGKAETGAARLKEAEAA 579
E + K +G+A+ +E E A
Sbjct: 146 EEKEKEKDGEAKYVKEEAREVEEA 169
Score = 31.2 bits (69), Expect = 2.0
Identities = 16/55 (29%), Positives = 30/55 (54%)
Query: 553 EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKK 607
ED + ++ +AE+G +E E A + E+ A+K E E+R +E D+++
Sbjct: 93 EDDDDPMEEVSSEAESGRGNEEEVEKAKIDGEEEDQAMKDEEEDRDVKVEEDEEE 147
>At1g65010 hypothetical protein
Length = 1318
Score = 42.4 bits (98), Expect = 9e-04
Identities = 62/270 (22%), Positives = 113/270 (40%), Gaps = 47/270 (17%)
Query: 395 SKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSIWGPRAGNCREHLAQIDDLVL 454
S K V+ Q+ + G E + + V S E S + +E ++++ +L+
Sbjct: 491 SLKSTVDSIQNEFENSKAGWEQKELHLMGCVKKSEEENS-------SSQEEVSRLVNLLK 543
Query: 455 TENDQKYLGDREPAGLLNYVLRASLKTASA-VRYLQ---GSTIPELEELREKT-EKDDQL 509
+ E A L N +LK A V+YLQ G E +L+E +K++ L
Sbjct: 544 ESEEDACARKEEEASLKN-----NLKVAEGEVKYLQETLGEAKAESMKLKESLLDKEEDL 598
Query: 510 ------ISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAE 563
ISS+ + IEELSK +++ + +L+ Q+ + ++ +K E
Sbjct: 599 KNVTAEISSLREWEGSVLEKIEELSKVKESLVDKETKLQSITQEAEELKGREAAHMKQIE 658
Query: 564 GKAETGAAR----------------LKEAEAAWLKEREQSSAVKKELENRVSSLEA---D 604
+ A+ LKE EA +LK+ E+ S + L + V+ L++ +
Sbjct: 659 ELSTANASLVDEATKLQSIVQESEDLKEKEAGYLKKIEELSVANESLADNVTDLQSIVQE 718
Query: 605 KKKLAATVSKINKASFNNAVSQLAVVNPGL 634
K L K + ++ + +L+V N L
Sbjct: 719 SKDL-----KEREVAYLKKIEELSVANESL 743
Score = 42.4 bits (98), Expect = 9e-04
Identities = 48/174 (27%), Positives = 88/174 (49%), Gaps = 24/174 (13%)
Query: 443 REHLAQIDDLVLTEN---DQKYLGDREPAGL--LNYVLRASLKTASAVRYLQGSTIPELE 497
+E+ +D++ +N + K L +RE A L ++ + A+ A V LQ + E
Sbjct: 776 KENENLVDNVANMQNIAEESKDLREREVAYLKKIDELSTANGTLADNVTNLQNIS----E 831
Query: 498 ELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLK-------KKVQDGDA 550
E +E E++ L+ + E S +++ SK L + ENE+L+ KK+++
Sbjct: 832 ENKELRERETTLLKKAEELSELNESLVDKASK-LQTVVQENEELRERETAYLKKIEELSK 890
Query: 551 VRE---DLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSL 601
+ E D E +L+ + + E LKE E A+LK+ E+ S V+++L N+ + L
Sbjct: 891 LHEILSDQETKLQISNHEKE----ELKERETAYLKKIEELSKVQEDLLNKENEL 940
Score = 41.6 bits (96), Expect = 0.001
Identities = 44/173 (25%), Positives = 80/173 (45%), Gaps = 33/173 (19%)
Query: 483 SAVRYLQGSTIPELEEL---REKTEKDDQLISSMSKELEE-------GCSAIEELSKALD 532
S++R +GS + ++EEL +E + + S+++E EE IEELS A
Sbjct: 606 SSLREWEGSVLEKIEELSKVKESLVDKETKLQSITQEAEELKGREAAHMKQIEELSTANA 665
Query: 533 NMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAAR----------------LKEA 576
++ E +L+ VQ+ + ++E G LK E + + LKE
Sbjct: 666 SLVDEATKLQSIVQESEDLKEKEAGYLKKIEELSVANESLADNVTDLQSIVQESKDLKER 725
Query: 577 EAAWLKEREQSSAVKKEL---ENRVSSLEADKKKL----AATVSKINKASFNN 622
E A+LK+ E+ S + L E ++ ++ + ++L A+ + KI + S N
Sbjct: 726 EVAYLKKIEELSVANESLVDKETKLQHIDQEAEELRGREASHLKKIEELSKEN 778
Score = 40.0 bits (92), Expect = 0.004
Identities = 40/179 (22%), Positives = 79/179 (43%), Gaps = 9/179 (5%)
Query: 442 CREHLAQIDDLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQGSTIPELEELRE 501
C+E L + V D L +E +L + +++ S E E +
Sbjct: 453 CQEELKNCESQV----DSLKLASKETNEKYEKMLEDARNEIDSLKSTVDSIQNEFENSKA 508
Query: 502 KTE-KDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLK 560
E K+ L+ + K EE S+ EE+S+ ++ +K E + ++ L+ LK
Sbjct: 509 GWEQKELHLMGCVKKSEEENSSSQEEVSRLVNLLKESEEDACARKEE----EASLKNNLK 564
Query: 561 AAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAATVSKINKAS 619
AEG+ + L EA+A +K +E +++L+N + + + ++ + + KI + S
Sbjct: 565 VAEGEVKYLQETLGEAKAESMKLKESLLDKEEDLKNVTAEISSLREWEGSVLEKIEELS 623
Score = 34.7 bits (78), Expect = 0.18
Identities = 43/172 (25%), Positives = 78/172 (45%), Gaps = 35/172 (20%)
Query: 482 ASAVRYLQGSTIPELEELREKTEK------DDQLISSMSKELEEGCSA----IEELSKA- 530
A +R + S + ++EEL ++ E + Q I+ SK+L E A I+ELS A
Sbjct: 757 AEELRGREASHLKKIEELSKENENLVDNVANMQNIAEESKDLREREVAYLKKIDELSTAN 816
Query: 531 ---------LDNMKLENEQLK-------KKVQDGDAVREDLEGRLKAAEGKAETGAARLK 574
L N+ EN++L+ KK ++ + E L + + + L+
Sbjct: 817 GTLADNVTNLQNISEENKELRERETTLLKKAEELSELNESLVDKASKLQTVVQENE-ELR 875
Query: 575 EAEAAWLKEREQSSAVKKELENRVSSLEAD-------KKKLAATVSKINKAS 619
E E A+LK+ E+ S + + L ++ + L+ K++ A + KI + S
Sbjct: 876 ERETAYLKKIEELSKLHEILSDQETKLQISNHEKEELKERETAYLKKIEELS 927
Score = 32.0 bits (71), Expect = 1.2
Identities = 51/213 (23%), Positives = 90/213 (41%), Gaps = 43/213 (20%)
Query: 475 LRASLKTASAVRYLQGSTIPELEELREKTEK------DDQLISSMSKELEEGCSA----I 524
L++ ++ + ++ + + ++EEL E D Q I SK+L+E A I
Sbjct: 674 LQSIVQESEDLKEKEAGYLKKIEELSVANESLADNVTDLQSIVQESKDLKEREVAYLKKI 733
Query: 525 EELSKA----------LDNMKLENEQLK-------KKVQDGDAVREDLEGRLKAAEGKAE 567
EELS A L ++ E E+L+ KK+++ E+L + + AE
Sbjct: 734 EELSVANESLVDKETKLQHIDQEAEELRGREASHLKKIEELSKENENLVDNVANMQNIAE 793
Query: 568 TGAARLKEAEAAWLKEREQSSAVKKELENRVSSL-------------EADKKKLAATVSK 614
+ L+E E A+LK+ ++ S L + V++L E K A +S+
Sbjct: 794 E-SKDLREREVAYLKKIDELSTANGTLADNVTNLQNISEENKELRERETTLLKKAEELSE 852
Query: 615 INKASFNNAVSQLAVV--NPGLETAPVGYRKVV 645
+N++ + A VV N L Y K +
Sbjct: 853 LNESLVDKASKLQTVVQENEELRERETAYLKKI 885
>At2g32240 putative myosin heavy chain
Length = 1333
Score = 42.0 bits (97), Expect = 0.001
Identities = 67/351 (19%), Positives = 141/351 (40%), Gaps = 50/351 (14%)
Query: 313 ALRVEDGQSTDEEVDQETDVAQGEEGHAEN----------------QPQPDPEDRGGAEV 356
++++ + ++ E + E +V QG+ EN + + ++ G E+
Sbjct: 724 SVKISESENLLESIRNELNVTQGKLESIENDLKAAGLQESEVMEKLKSAEESLEQKGREI 783
Query: 357 GGTSGSAKNVSLEADHGSDPVQGKGRSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEP 416
+ K + LEA H S + + R ++ S+ + + +G+++ E
Sbjct: 784 D--EATTKRMELEALHQSLSIDSEHRLQKAMEEFTSRDSEASSLTEKLRDLEGKIKSYEE 841
Query: 417 ERAEPVTVVPPSPERLSIWGPR---AGNCREHLAQIDD--------------LVLTENDQ 459
+ AE E+L R A + E L Q D L+ N+Q
Sbjct: 842 QLAEASGKSSSLKEKLEQTLGRLAAAESVNEKLKQEFDQAQEKSLQSSSESELLAETNNQ 901
Query: 460 KYLGDREPAGLLNYVLRASLKTASAVRYLQGSTIPELEELREKTEKDDQLISSMS---KE 516
+ +E GL+ S++ +A++ L+ + +E +K + L+ + +
Sbjct: 902 LKIKIQELEGLIG---SGSVEKETALKRLEEA----IERFNQKETESSDLVEKLKTHENQ 954
Query: 517 LEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEA 576
+EE E S D K+E E K+++ ++ E+L + + E + + L E
Sbjct: 955 IEEYKKLAHEASGVADTRKVELEDALSKLKNLESTIEELGAKCQGLEKE----SGDLAEV 1010
Query: 577 EAAWLKEREQSSAVKKELENRVSSLEADKKKLAATVSKINKASFNNAVSQL 627
E + EL+ ++S+LEA+K++ A + + +K + + QL
Sbjct: 1011 NLKLNLELANHGSEANELQTKLSALEAEKEQTANEL-EASKTTIEDLTKQL 1060
Score = 38.9 bits (89), Expect = 0.009
Identities = 39/183 (21%), Positives = 85/183 (46%), Gaps = 30/183 (16%)
Query: 474 VLRASLKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSK------------------ 515
+L+++ ++A + S E++EL EK +++++ +++
Sbjct: 241 LLKSTKESAKEMEEKMASLQQEIKELNEKMSENEKVEAALKSSAGELAAVQEELALSKSR 300
Query: 516 --ELEEGCSA----IEELSKALDNMKLENEQLKKK---VQDGDAVREDLEGRLKAAEGKA 566
E E+ S+ I+EL++ L+ K + K++ +QD DA + L+ +L EG
Sbjct: 301 LLETEQKVSSTEALIDELTQELEQKKASESRFKEELSVLQDLDAQTKGLQAKLSEQEGIN 360
Query: 567 ETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAATVSKI--NKASFNNAV 624
A LKE E ++Q ++ E +++ + +K+ L A V+++ N A+
Sbjct: 361 SKLAEELKEKELLESLSKDQEEKLRTANE-KLAEVLKEKEALEANVAEVTSNVATVTEVC 419
Query: 625 SQL 627
++L
Sbjct: 420 NEL 422
Score = 37.4 bits (85), Expect = 0.027
Identities = 37/150 (24%), Positives = 71/150 (46%), Gaps = 18/150 (12%)
Query: 500 REKTEKDDQLISSMSKELEEGCSA-------IEELSKALDNMKLENEQLKKKVQDGDAVR 552
+EK + ++ SS K LE+ + + E+ +A D + +E E +KK+ + +
Sbjct: 148 QEKIVEGEERHSSQLKSLEDALQSHDAKDKELTEVKEAFDALGIELESSRKKLIE---LE 204
Query: 553 EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVK------KELENRVSSLEADKK 606
E L+ + A+ E A++ K E S +K KE+E +++SL+ + K
Sbjct: 205 EGLKRSAEEAQKFEELHKQSASHADSESQKALEFSELLKSTKESAKEMEEKMASLQQEIK 264
Query: 607 KLAATVSKINK--ASFNNAVSQLAVVNPGL 634
+L +S+ K A+ ++ +LA V L
Sbjct: 265 ELNEKMSENEKVEAALKSSAGELAAVQEEL 294
Score = 35.4 bits (80), Expect = 0.10
Identities = 29/106 (27%), Positives = 49/106 (45%), Gaps = 9/106 (8%)
Query: 477 ASLKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKL 536
AS+K A LQ E E + + + ++ + + KEL+ S+I+E +A +
Sbjct: 1158 ASVKVAELTSKLQ-----EHEHIAGERDVLNEQVLQLQKELQAAQSSIDEQKQAHSQKQS 1212
Query: 537 ENEQLKKKVQDGDAVRE----DLEGRLKAAEGKAETGAARLKEAEA 578
E E KK Q+ ++ + E +K E K + A+ KE EA
Sbjct: 1213 ELESALKKSQEEIEAKKKAVTEFESMVKDLEQKVQLADAKTKETEA 1258
Score = 35.0 bits (79), Expect = 0.14
Identities = 31/137 (22%), Positives = 67/137 (48%), Gaps = 10/137 (7%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRED 554
++EE ++ + + + ELE+ S ++ L ++ + + + L+K + GD +
Sbjct: 954 QIEEYKKLAHEASGVADTRKVELEDALSKLKNLESTIEELGAKCQGLEK--ESGDLAEVN 1011
Query: 555 LEGRLKAAEGKAETGAARLKEAEAAWLKEREQSS----AVKKELENRVSSLEADKKKLAA 610
L+ L+ A +E A L+ +A E+EQ++ A K +E+ L ++ +KL +
Sbjct: 1012 LKLNLELANHGSE--ANELQTKLSALEAEKEQTANELEASKTTIEDLTKQLTSEGEKLQS 1069
Query: 611 TVSKINKASFNNAVSQL 627
+S + NN V+ +
Sbjct: 1070 QIS--SHTEENNQVNAM 1084
Score = 33.9 bits (76), Expect = 0.30
Identities = 59/313 (18%), Positives = 132/313 (41%), Gaps = 26/313 (8%)
Query: 316 VEDGQSTDEEVDQETD--VAQGEEGHAENQPQPDPEDRGGAEVGGTSGSAKNVSLEAD-- 371
+E+ T +E +TD ++Q ++E + + + +E G + +A +LE +
Sbjct: 422 LEEKLKTSDENFSKTDALLSQALSNNSELEQKLKSLEELHSEAGSAAAAATQKNLELEDV 481
Query: 372 -HGSDPVQGKGRSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPER---------AEP 421
S + +S +E +T+ A++++ E Q +++ S+ ER +E
Sbjct: 482 VRSSSQAAEEAKSQIKELETKFTAAEQKNAELEQQLNLLQLKSSDAERELKELSEKSSEL 541
Query: 422 VTVVPPSPERLSIWGPRAGNCREHLAQIDDLVLTENDQKYLGDREPAGLLNYVLRASLKT 481
T + + E + ++ ++++ L LT++ + + E L L+ +
Sbjct: 542 QTAIEVAEEEKKQATTQMQEYKQKASELE-LSLTQSSAR---NSELEEDLRIALQKGAEH 597
Query: 482 ASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQL 541
+I ELE L + ++ + K+LE ++ + ++ + L
Sbjct: 598 EDRANTTHQRSI-ELEGLCQSSQSKHEDAEGRLKDLE---LLLQTEKYRIQELEEQVSSL 653
Query: 542 KKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSL 601
+KK + +A + G++ + E + EAA E +KEL ++++
Sbjct: 654 EKKHGETEADSKGYLGQVAELQSTLEAFQVKSSSLEAALNIATEN----EKELTENLNAV 709
Query: 602 EADKKKLAATVSK 614
++KKKL ATV +
Sbjct: 710 TSEKKKLEATVDE 722
Score = 33.9 bits (76), Expect = 0.30
Identities = 62/314 (19%), Positives = 128/314 (40%), Gaps = 22/314 (7%)
Query: 294 RRNAKLLRGMADIDRTLLDALR-VEDGQSTDEEVDQETDVAQGEEGHAENQPQPDPEDRG 352
++NA+L + + + DA R +++ E+ +VA+ E+ A Q Q +
Sbjct: 508 QKNAELEQQLNLLQLKSSDAERELKELSEKSSELQTAIEVAEEEKKQATTQMQEYKQKAS 567
Query: 353 GAEVGGTSGSAKNVSLEADHGSDPVQGKGRSVSEEKDTRGPASKKRRVEDHQDHADGEVQ 412
E+ T SA+N LE D +G E + R + +R +E +G Q
Sbjct: 568 ELELSLTQSSARNSELEEDLRIALQKGA------EHEDRANTTHQRSIE-----LEGLCQ 616
Query: 413 GSEPERAEPVTVVPPSPERLSIWGPRAGNCREHLAQIDDLV-LTENDQK-YLGDREPAGL 470
S+ + + + L R E ++ ++ TE D K YLG + A L
Sbjct: 617 SSQSKHEDAEGRLKDLELLLQTEKYRIQELEEQVSSLEKKHGETEADSKGYLG--QVAEL 674
Query: 471 LNYVLRASLKTASAVRYLQGSTIPE------LEELREKTEKDDQLISSMSKELEEGCSAI 524
+ + +K++S L +T E L + + +K + + S ++ E + +
Sbjct: 675 QSTLEAFQVKSSSLEAALNIATENEKELTENLNAVTSEKKKLEATVDEYSVKISESENLL 734
Query: 525 EELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKER 584
E + L+ + + E ++ ++ ++ +LK+AE E + EA ++
Sbjct: 735 ESIRNELNVTQGKLESIENDLKAAGLQESEVMEKLKSAEESLEQKGREIDEATTKRMELE 794
Query: 585 EQSSAVKKELENRV 598
++ + E+R+
Sbjct: 795 ALHQSLSIDSEHRL 808
Score = 33.1 bits (74), Expect = 0.52
Identities = 68/364 (18%), Positives = 145/364 (39%), Gaps = 44/364 (12%)
Query: 312 DALRVEDGQSTDEEVDQETDVAQG-------EEGHAENQPQPDPEDRGGAEVGGTSGSAK 364
DAL +E S + ++ E + + EE H ++ D E + E S K
Sbjct: 187 DALGIELESSRKKLIELEEGLKRSAEEAQKFEELHKQSASHADSESQKALEFSELLKSTK 246
Query: 365 NVSLEADHGSDPVQGKGRSVSE--------EKDTRGPASKKRRVEDHQDHADGEVQGSEP 416
+ E + +Q + + ++E E + A + V++ + + +E
Sbjct: 247 ESAKEMEEKMASLQQEIKELNEKMSENEKVEAALKSSAGELAAVQEELALSKSRLLETEQ 306
Query: 417 ERAEPVTVVPPSPERLSIWGPRAGNCREHLAQIDDL-VLTENDQKYLGDREPAGLLNYVL 475
+ + ++ + L +E L+ + DL T+ Q L ++E +N L
Sbjct: 307 KVSSTEALIDELTQELEQKKASESRFKEELSVLQDLDAQTKGLQAKLSEQEG---INSKL 363
Query: 476 RASLKTASAVRYLQGSTIPEL----EELREKTEKDDQL------ISSMSKELEEGCSAIE 525
LK + L +L E+L E ++ + L ++S + E C+ +E
Sbjct: 364 AEELKEKELLESLSKDQEEKLRTANEKLAEVLKEKEALEANVAEVTSNVATVTEVCNELE 423
Query: 526 ELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEG-KAETGAA---------RLKE 575
E K D + + L + ++ +LE +LK+ E +E G+A L++
Sbjct: 424 EKLKTSDENFSKTDALLSQALSNNS---ELEQKLKSLEELHSEAGSAAAAATQKNLELED 480
Query: 576 AEAAWLKEREQSSAVKKELENRVSSLEADKKKLAATVS--KINKASFNNAVSQLAVVNPG 633
+ + E++ + KELE + ++ E +L ++ ++ + + +L+ +
Sbjct: 481 VVRSSSQAAEEAKSQIKELETKFTAAEQKNAELEQQLNLLQLKSSDAERELKELSEKSSE 540
Query: 634 LETA 637
L+TA
Sbjct: 541 LQTA 544
Score = 32.3 bits (72), Expect = 0.88
Identities = 36/135 (26%), Positives = 58/135 (42%), Gaps = 16/135 (11%)
Query: 481 TASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQ 540
T Y Q ++ EL L + + ++ +L + L++G E+ + +E E
Sbjct: 556 TTQMQEYKQKASELELS-LTQSSARNSELEEDLRIALQKGAEH-EDRANTTHQRSIELEG 613
Query: 541 LKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSS 600
L Q + ED EGRLK E +T R++E E EQ S+ LE +
Sbjct: 614 L---CQSSQSKHEDAEGRLKDLELLLQTEKYRIQELE-------EQVSS----LEKKHGE 659
Query: 601 LEADKKKLAATVSKI 615
EAD K V+++
Sbjct: 660 TEADSKGYLGQVAEL 674
Score = 30.8 bits (68), Expect = 2.6
Identities = 28/135 (20%), Positives = 58/135 (42%), Gaps = 13/135 (9%)
Query: 493 IPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVR 552
+ LEEL + + + ELE+ + S+A + K + ++L+ K +
Sbjct: 454 LKSLEELHSEAGSAAAAATQKNLELED---VVRSSSQAAEEAKSQIKELETKFTAAEQKN 510
Query: 553 EDLEGRLKAAEGKAETGAARLKE----------AEAAWLKEREQSSAVKKELENRVSSLE 602
+LE +L + K+ LKE A +E++Q++ +E + + S LE
Sbjct: 511 AELEQQLNLLQLKSSDAERELKELSEKSSELQTAIEVAEEEKKQATTQMQEYKQKASELE 570
Query: 603 ADKKKLAATVSKINK 617
+ +A S++ +
Sbjct: 571 LSLTQSSARNSELEE 585
>At1g77580 unknown protein
Length = 779
Score = 41.6 bits (96), Expect = 0.001
Identities = 40/137 (29%), Positives = 64/137 (46%), Gaps = 13/137 (9%)
Query: 491 STIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDA 550
S I ELEE EK E + + + K E E S+ L + ++L++K++ +A
Sbjct: 350 SRIKELEEKLEKLEAEKHELENEVKCNREEAVVHIENSEVLTS---RTKELEEKLEKLEA 406
Query: 551 VREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAA 610
+E+L+ +K KA AE L R KELE ++ LEA+K +L +
Sbjct: 407 EKEELKSEVKCNREKAVVHVENSLAAEIEVLTSRT------KELEEQLEKLEAEKVELES 460
Query: 611 TVSKINKASFNNAVSQL 627
V K + AV+Q+
Sbjct: 461 EV----KCNREEAVAQV 473
Score = 35.8 bits (81), Expect = 0.080
Identities = 33/142 (23%), Positives = 65/142 (45%), Gaps = 26/142 (18%)
Query: 491 STIPELEELREKTEKDD----------------QLISSMSKELEEGCSAIEELSKALDNM 534
S ELEE EK E + Q+ +S++ E+E I++L + L+ +
Sbjct: 439 SRTKELEEQLEKLEAEKVELESEVKCNREEAVAQVENSLATEIEVLTCRIKQLEEKLEKL 498
Query: 535 KLENEQLKKKVQDGDAVREDLEGRLKA-------AEGKAETGAARLKEAEAAW--LKER- 584
++E ++LK +V+ V L L+A E K E E + ++ +K++
Sbjct: 499 EVEKDELKSEVKCNREVESTLRFELEAIACEKMELENKLEKLEVEKAELQISFDIIKDKY 558
Query: 585 EQSSAVKKELENRVSSLEADKK 606
E+S +E+E ++ ++ + K
Sbjct: 559 EESQVCLQEIETKLGEIQTEMK 580
Score = 34.3 bits (77), Expect = 0.23
Identities = 35/130 (26%), Positives = 63/130 (47%), Gaps = 10/130 (7%)
Query: 493 IPELEELREKTEKD-DQLISSMSKELE-EGCSAIEELSKALDNMKLENEQLKKKVQDGDA 550
I +LEE EK E + D+L S + E E E + A + M+LEN+ K +V+
Sbjct: 488 IKQLEEKLEKLEVEKDELKSEVKCNREVESTLRFELEAIACEKMELENKLEKLEVE---- 543
Query: 551 VREDLEGRLKAAEGKAETGAARLKEAEAAWLK---EREQSSAVKKELENRVSSLEADKKK 607
+ +L+ + K E L+E E + E + + +K E+E++ ++EAD K
Sbjct: 544 -KAELQISFDIIKDKYEESQVCLQEIETKLGEIQTEMKLVNELKAEVESQTIAMEADAKT 602
Query: 608 LAATVSKINK 617
+A + + +
Sbjct: 603 KSAKIESLEE 612
Score = 33.1 bits (74), Expect = 0.52
Identities = 66/305 (21%), Positives = 123/305 (39%), Gaps = 56/305 (18%)
Query: 320 QSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAEVGGTSGSAKNVSLEADHGSDPVQG 379
+S++ + V+ E +E QP+ E R T VSLE + + ++
Sbjct: 7 ESSERSFGESESVSSLSEKDSEIQPESTMESRDDEIQSPT------VSLEVETEKEELKD 60
Query: 380 KGRSVSEEKDTR--GPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSIWGP 437
++++E+ ++K V+ H A+ V G E E V +
Sbjct: 61 SMKTLAEKLSAALANVSAKDDLVKQHVKVAEEAVAGWEKAENEVVEL------------- 107
Query: 438 RAGNCREHLAQIDDLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQGSTIPELE 497
+E L DD + L DR L+ L+ ++ R Q I +
Sbjct: 108 -----KEKLEAADD------KNRVLEDR--VSHLDGALKECVRQLRQARDEQEQRIQDA- 153
Query: 498 ELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEG 557
+ E+T++ +S+ ++ E + EELS+ +++ EN L+ ++ A E+LE
Sbjct: 154 -VIERTQELQSSRTSLENQIFETATKSEELSQMAESVAKENVMLRHELL---ARCEELE- 208
Query: 558 RLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAATVSKINK 617
++ E T +AA ++Q ++KK V+ LEA+ +K +
Sbjct: 209 -IRTIERDLST--------QAAETASKQQLDSIKK-----VAKLEAECRKFRMLAK--SS 252
Query: 618 ASFNN 622
ASFN+
Sbjct: 253 ASFND 257
Score = 32.3 bits (72), Expect = 0.88
Identities = 23/102 (22%), Positives = 47/102 (45%), Gaps = 25/102 (24%)
Query: 511 SSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGA 570
+S++ E+E S I+EL + L+ ++ E +L+ +V+
Sbjct: 339 NSLASEIEVLTSRIKELEEKLEKLEAEKHELENEVKCN---------------------- 376
Query: 571 ARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAATV 612
+E ++ E ++ KELE ++ LEA+K++L + V
Sbjct: 377 ---REEAVVHIENSEVLTSRTKELEEKLEKLEAEKEELKSEV 415
>At1g68910 unknown protein
Length = 534
Score = 41.6 bits (96), Expect = 0.001
Identities = 35/137 (25%), Positives = 66/137 (47%), Gaps = 17/137 (12%)
Query: 493 IPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVR 552
+ E+ LRE + +Q + + EL+ ++ +E+ L M+ NE +K+ + +
Sbjct: 307 VSEVLTLREYVKSAEQKLKNTDLELKSVNASKQEILVHLAEMENANESVKENLFE----- 361
Query: 553 EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVK---KELENRVSSLEADKKKLA 609
AE +AE+G A++KE +AA L+ E+ + +K + +V+SLE ++L
Sbjct: 362 ---------AESRAESGEAKIKELDAANLELTEELNFLKDADDKKTKKVNSLEKQVRELE 412
Query: 610 ATVSKINKASFNNAVSQ 626
V +S N Q
Sbjct: 413 VQVQNSKVSSEANQEQQ 429
>At5g04890 unknown protein
Length = 366
Score = 41.2 bits (95), Expect = 0.002
Identities = 49/164 (29%), Positives = 72/164 (43%), Gaps = 12/164 (7%)
Query: 459 QKYLGDREPAGLLNYVLRASL--KTASAVRYLQGSTIPELEELRE--KTEKDDQLISSMS 514
Q L D+ N VL ++ +T + V YL ++ E L + K E+ L S
Sbjct: 81 QNCLVDKIHGSFKNNVLTITMPKETITKVAYLPETSRTEAAALEKAAKLEEKRLLEESRR 140
Query: 515 KELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLK 574
KE EE EE + + E E L +K+Q+ +E+ E R E KA+ AA K
Sbjct: 141 KEKEE-----EEAKQMKKQLLEEKEALIRKLQEEAKAKEEAEMRKLQEEAKAKEEAAAKK 195
Query: 575 EAEAAWLKER-EQSSAVKKELENRVSSLEADKKKLAATVSKINK 617
E KE+ E+ ++ LE R LE K A + KI +
Sbjct: 196 LQEEIEAKEKLEERKLEERRLEER--KLEDMKLAEEAKLKKIQE 237
Score = 30.8 bits (68), Expect = 2.6
Identities = 49/207 (23%), Positives = 89/207 (42%), Gaps = 18/207 (8%)
Query: 444 EHLAQIDDLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQGSTIPELEELREKT 503
E A++++ L E ++ + E A + L K A + + + E E+R+
Sbjct: 124 EKAAKLEEKRLLEESRRKEKEEEEAKQMKKQLLEE-KEALIRKLQEEAKAKEEAEMRKLQ 182
Query: 504 EKDDQLISSMSKELEEGCSAIEELS-----------KALDNMKLENEQLKKKVQDGDAVR 552
E+ + +K+L+E A E+L + L++MKL E KK+Q+ +V
Sbjct: 183 EEAKAKEEAAAKKLQEEIEAKEKLEERKLEERRLEERKLEDMKLAEEAKLKKIQERKSVD 242
Query: 553 E--DLEGRLKAAEGKAETG-AARLKEAEAAWLKEREQS-SAVKKELENRVSSL-EADKKK 607
E + E LK ++G A K + LK V K E ++ +L E +KK
Sbjct: 243 ESGEKEKILKPEVVYTKSGHVATPKPESGSGLKSGFGGVGEVVKSAEEKLGNLVEKEKKM 302
Query: 608 LAATVSKINKASFNNAVSQLAVVNPGL 634
+ KI + + +L ++N G+
Sbjct: 303 GKGIMEKIRRKEITSEEKKL-MMNVGV 328
>At2g22610 putative kinesin heavy chain
Length = 1068
Score = 41.2 bits (95), Expect = 0.002
Identities = 32/126 (25%), Positives = 62/126 (48%), Gaps = 12/126 (9%)
Query: 498 ELREKTEKDDQLISSMS---KELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVR-- 552
+L+E+ + D++ S++ KELE + A +N K+++ + K +G ++
Sbjct: 807 QLQERLKSRDEICSNLQQKVKELECKLRERHQSDSAANNQKVKDLENNLKESEGSSLVWQ 866
Query: 553 ---EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVK----KELENRVSSLEADK 605
+D E +LK +EG + ++KE E E+ Q + + KELE R+ E
Sbjct: 867 QKVKDYENKLKESEGNSLVWQQKIKELEIKHKDEQSQEAVLLRQKIKELEMRLKEQEKHI 926
Query: 606 KKLAAT 611
+++A T
Sbjct: 927 QEMATT 932
Score = 39.3 bits (90), Expect = 0.007
Identities = 42/179 (23%), Positives = 79/179 (43%), Gaps = 19/179 (10%)
Query: 466 EPAGLLNYVLRA-SLKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAI 524
E LN+ R ++ A + + I +L+ + EK ++ + K++EE +
Sbjct: 710 ETLSSLNFATRVRGVELGPARKQVDTGEIQKLKAMVEKARQESRSKDESIKKMEENIQNL 769
Query: 525 EELSKALDNMKLENEQLKKKVQDG-DAVRE-------DLEGRLKAAEGKAETGAARLKEA 576
E +K DN ++ K +Q+ D+V L+ RLK+ + ++KE
Sbjct: 770 EGKNKGRDNSYRSLQEKNKDLQNQLDSVHNQSEKQYAQLQERLKSRDEICSNLQQKVKEL 829
Query: 577 EAAWLKEREQSSAVK-----KELENRVSSLEAD----KKKLAATVSKINKASFNNAVSQ 626
E L+ER QS + K+LEN + E ++K+ +K+ ++ N+ V Q
Sbjct: 830 ECK-LRERHQSDSAANNQKVKDLENNLKESEGSSLVWQQKVKDYENKLKESEGNSLVWQ 887
>At3g28770 hypothetical protein
Length = 2081
Score = 40.8 bits (94), Expect = 0.002
Identities = 65/302 (21%), Positives = 123/302 (40%), Gaps = 35/302 (11%)
Query: 312 DALRVEDGQSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAEVG-GTSGSAKNVSLEA 370
D + G+ E+ ++ D Q E +N+ GG + G +K+ L+
Sbjct: 814 DEAKERSGEDNKEDKEESKDY-QSVEAKEKNE-------NGGVDTNVGNKEDSKD--LKD 863
Query: 371 DHGSDPVQGKGRSVSEEKDT--RGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPS 428
D + K S+ ++++ R S + V D ++ D +VQ E +V
Sbjct: 864 DRSVEVKANKEESMKKKREEVQRNDKSSTKEVRDFANNMDIDVQKGSGE-----SVKYKK 918
Query: 429 PERLSIWGPRAGNCREHLAQIDDLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYL 488
E+ + GN E+ D + T + QK D++ + K Y+
Sbjct: 919 DEK------KEGNKEEN----KDTINTSSKQKGK-DKKKKKKESKNSNMKKKEEDKKEYV 967
Query: 489 QGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDG 548
+ + +E T+ ++ + +K+ +E + + SK + K E E+ K K ++
Sbjct: 968 NNELKKQEDNKKETTKSENSKLKEENKDNKEKKESEDSASK--NREKKEYEEKKSKTKE- 1024
Query: 549 DAVREDLEGRLKAAEGKAETGAARLKEAEAAW---LKEREQSSAVKKELENRVSSLEADK 605
+A +E + + K E K KE E + K++E+ + KKE EN S + DK
Sbjct: 1025 EAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSKKKEDK 1084
Query: 606 KK 607
K+
Sbjct: 1085 KE 1086
Score = 38.1 bits (87), Expect = 0.016
Identities = 65/339 (19%), Positives = 138/339 (40%), Gaps = 45/339 (13%)
Query: 312 DALRVEDGQSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGA---EVGGTSGSAKNVSL 368
D+ ++D +S + + ++E + + E N E R A ++ GS ++V
Sbjct: 857 DSKDLKDDRSVEVKANKEESMKKKREEVQRNDKSSTKEVRDFANNMDIDVQKGSGESVKY 916
Query: 369 EADHGSD------------PVQGKGRSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEP 416
+ D + + KG+ ++K ++ K++ ED +++ + E++ E
Sbjct: 917 KKDEKKEGNKEENKDTINTSSKQKGKDKKKKKKESKNSNMKKKEEDKKEYVNNELKKQED 976
Query: 417 ERAEPVTVVPPSPERLSIWGPRAGNCREHLAQIDDLVLTENDQKYLGDREPAGLLNYVLR 476
+ E E + N +E D ++N +K + + + +
Sbjct: 977 NKKETT-----KSENSKLKEENKDN-KEKKESEDSA--SKNREKKEYEEKKSKTKEEAKK 1028
Query: 477 ASLKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKL 536
K+ R + S EE + K EK++ K+ EE + K +N K
Sbjct: 1029 EKKKSQDKKREEKDS-----EERKSKKEKEESRDLKAKKKEEE-----TKEKKESENHKS 1078
Query: 537 ENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKERE-QSSAVKKELE 595
+ ++ KK+ +D +++++ + K + K E +R KE + +++ E Q+S KKE +
Sbjct: 1079 KKKEDKKEHEDNKSMKKEED---KKEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDK 1135
Query: 596 N--------RVSSLEADKKKLAATVSKINKASFNNAVSQ 626
N ++ E+DKK+ K ++ SQ
Sbjct: 1136 NEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQ 1174
Score = 33.1 bits (74), Expect = 0.52
Identities = 55/301 (18%), Positives = 118/301 (38%), Gaps = 50/301 (16%)
Query: 319 GQSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAEVG-------GTSGSAKNVSLEAD 371
G+ST+ D+ + +E H + + E+ G + G G S K +LE
Sbjct: 398 GESTN---DKMVNATTNDEDHKKENKEETHENNGESVKGENLENKAGNEESMKGENLENK 454
Query: 372 HGSDPVQGKGRSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPER 431
G++ ++G + S E T +SK+ + E+ Q ++ E + E V + +
Sbjct: 455 VGNEELKG---NASVEAKTNNESSKEEKREESQ-RSNEVYMNKETTKGENVNI-----QG 505
Query: 432 LSIWGPRAGNCREHLAQIDDLV-LTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQG 490
SI N E+ + V E+D +R +N + K +
Sbjct: 506 ESIGDSTKDNSLENKEDVKPKVDANESDGNSTKERHQEAQVNNGVSTEDKNLDNIG---- 561
Query: 491 STIPELEELREKTEKDDQLISSMSKELEEGCSAIEELS----KALDNMKLENEQLKKKVQ 546
++ +K+D+ + + + + EE +++ N LEN++ KK+++
Sbjct: 562 ---------ADEQKKNDKSVEVTTNDGDHTKEKREETQGNNGESVKNENLENKEDKKELK 612
Query: 547 DGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKK 606
D ++V E + +T ++ +S K ++N+ + +++K+
Sbjct: 613 DDESVGAKTNNETSLEEKREQT-------------QKGHDNSINSKIVDNKGGNADSNKE 659
Query: 607 K 607
K
Sbjct: 660 K 660
Score = 32.3 bits (72), Expect = 0.88
Identities = 32/135 (23%), Positives = 67/135 (48%), Gaps = 11/135 (8%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKL-----ENEQLKKKVQDGD 549
E+++ +K+ KD Q KE++E S ++L K ++ K EN++ K+ ++ +
Sbjct: 1177 EVDKKEKKSSKDQQ--KKKEKEMKE--SEEKKLKKNEEDRKKQTSVEENKKQKETKKEKN 1232
Query: 550 AVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLA 609
++D + K + GK E+ + KEAE K + + A E +N + ++AD + +
Sbjct: 1233 KPKDDKKNTTKQSGGKKESMESESKEAENQ-QKSQATTQADSDESKNEI-LMQADSQADS 1290
Query: 610 ATVSKINKASFNNAV 624
+ S+ + N +
Sbjct: 1291 HSDSQADSDESKNEI 1305
Score = 31.2 bits (69), Expect = 2.0
Identities = 30/118 (25%), Positives = 50/118 (41%), Gaps = 7/118 (5%)
Query: 497 EELREKTEKDDQLISSMS--KELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRED 554
EE +EK E ++ KE E+ S +E K E++ KK+ D + +
Sbjct: 1065 EETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKLE 1124
Query: 555 LEGRLKAAEGKAETGAAR-----LKEAEAAWLKEREQSSAVKKELENRVSSLEADKKK 607
+ K E K E ++ KE++ KE E+ S K+ ++ E DKK+
Sbjct: 1125 DQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKKE 1182
Score = 30.4 bits (67), Expect = 3.4
Identities = 30/118 (25%), Positives = 61/118 (51%), Gaps = 7/118 (5%)
Query: 495 ELEELREKTE-KDDQLISSMSKELE-EGCSAIEELSKALDNMKLENEQLKKKVQDGDAVR 552
+LE +T+ KDD+ + +E + G + ++ S K E+++ KK + + VR
Sbjct: 710 KLENKESQTDSKDDKSVDDKQEEAQIYGGESKDDKSVEAKGKKKESKENKKTKTNENRVR 769
Query: 553 ---EDLEGRLKAAEGKAETGAAR-LKEAEAAWLKEREQSSAVKKELENRVSSLEADKK 606
E+++G K +E K E G + K+A++ K+ ++ S+ + E + S E +K+
Sbjct: 770 NKEENVQGNKKESE-KVEKGEKKESKDAKSVETKDNKKLSSTENRDEAKERSGEDNKE 826
Score = 30.4 bits (67), Expect = 3.4
Identities = 29/112 (25%), Positives = 51/112 (44%), Gaps = 10/112 (8%)
Query: 501 EKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLK 560
+K EK + S +KE+E S E+ K K +Q KKK ++ ++E E +LK
Sbjct: 1152 DKKEKKENEEKSETKEIESSKSQKNEVDK--KEKKSSKDQQKKKEKE---MKESEEKKLK 1206
Query: 561 AAE--GKAETGA---ARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKK 607
E K +T + KE + K ++ K+ + S+E++ K+
Sbjct: 1207 KNEEDRKKQTSVEENKKQKETKKEKNKPKDDKKNTTKQSGGKKESMESESKE 1258
Score = 30.4 bits (67), Expect = 3.4
Identities = 46/297 (15%), Positives = 106/297 (35%), Gaps = 31/297 (10%)
Query: 317 EDGQSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAEVGGT-----SGSAKNVSL-EA 370
+DG+ + D+E + +G + N D +D E+ G +KN + E
Sbjct: 1715 KDGKINEIHGDKEATMEEGSKDGGTNSTGKDSKDSKSVEINGVKDDSLKDDSKNGDINEI 1774
Query: 371 DHGSD--------PVQGKGRSVSE--------EKDTRGPASKKRRVEDHQDHADGEVQGS 414
++G + +QG S++ +K S K + Q ++G+
Sbjct: 1775 NNGKEDSVKDNVTEIQGNDNSLTNSTSSEPNGDKLDTNKDSMKNNTMEAQGGSNGDSTNG 1834
Query: 415 EPERAEPVTVVPPSPERLSIWGPRAGNCREHLAQIDDLVLTENDQKYLGDREPAGLLNYV 474
E E + V + + + DD++ T D + +E ++ +
Sbjct: 1835 ETEETKESNVSMNNQNMQDVGSNENSMNNQTTGTGDDIISTTTDTESNTSKEVTSFISNL 1894
Query: 475 LRASLKTASAVRYLQG--------STIPELEELREKTEKDDQLISSMSKELEEGCSAIEE 526
S T + Q + E ++ ++I SM+ +L + + ++
Sbjct: 1895 EEKSPGTQEFQSFFQKLKDYMKYLCPVSSTFEAKDSRSYMSEMI-SMATKLSDAMAVLQA 1953
Query: 527 LSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKE 583
MK + +++V + + + G+ + ++G+ L ++ +KE
Sbjct: 1954 KKSGSGQMKTTLQGYQQEVMKTLTILQSVMGKAVTEQQSKDSGSLTLTLSQQQAIKE 2010
>At1g79280 hypothetical protein
Length = 2111
Score = 40.8 bits (94), Expect = 0.002
Identities = 38/234 (16%), Positives = 103/234 (43%), Gaps = 22/234 (9%)
Query: 399 RVEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSIWGPRAGNCREHLAQ----IDDLVL 454
R++D + +++ + + V+ ++S+ NC++ L++ +DD
Sbjct: 1379 RLKDEVRQLEEKLKAKDAHAEDCKKVLLEKQNKISLLEKELTNCKKDLSEREKRLDDAQQ 1438
Query: 455 TENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQGSTIPELEELREKTEKDDQLISSMS 514
+ + +++ + L+ + Y T + E+ +++ K +Q ++
Sbjct: 1439 AQATMQSEFNKQ---------KQELEKNKKIHYTLNMTKRKYEKEKDELSKQNQSLAKQL 1489
Query: 515 KELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLK 574
+E +E ++ E E+ +K++Q D L+ ++ K E LK
Sbjct: 1490 EEAKEEAGKRTTTDAVVEQSVKEREEKEKRIQILDKYVHQLKDEVRK---KTED----LK 1542
Query: 575 EAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAATVSKINKASFNNAVSQLA 628
+ + KER + +V+KE+ + ++ ++ +K K+ ++K+ + + A++ L+
Sbjct: 1543 KKDEELTKERSERKSVEKEVGDSLTKIKKEKTKVDEELAKLER--YQTALTHLS 1594
Score = 37.7 bits (86), Expect = 0.021
Identities = 72/331 (21%), Positives = 128/331 (37%), Gaps = 42/331 (12%)
Query: 316 VEDGQSTDEEVDQETDVAQGEEGHAENQPQP-DPEDRGGAEVGGTSGSAKNVSLEADHGS 374
V+ +S +VD G+EG QP +PE + V + ++ S
Sbjct: 1727 VKPEESPKVDVDMPEAEGTGDEG---KQPAAHEPESQVTTSVRPVQTLVRKRQADS-LVS 1782
Query: 375 DPVQGKGRSVSEEKDTRGPASKKRR-VEDHQDHADGEVQGSEPERAEPVTVVPPSPERLS 433
+P Q + PASKK + E H D ++GE EP E + +
Sbjct: 1783 EPQQDSLTQGETSSEIAPPASKKAKGSESHPDTSEGENLAKEPAIDELMDATTTTD---- 1838
Query: 434 IWGPRAGNCREHLAQIDDLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQGSTI 493
G+ E A+ + E ++Y+ ++ V + +T TI
Sbjct: 1839 ------GDNEETEAENAE----EKTEEYVEAQQDNEADEPVEESPTET---------ETI 1879
Query: 494 PELEELREKTEKDDQ-LISSMSKELEEG---CSAIEELSKALDNMKLENEQLKKKVQDGD 549
P EE R++TE+++Q ++ M + EEG +E+L + D + K++VQ
Sbjct: 1880 PTEEESRDQTEEENQEPLTDMESDKEEGELDLDTLEDLEEGTDVASMMRSPEKEEVQPET 1939
Query: 550 AVR-----EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEAD 604
+E ++ AE ET K E E+++ + ++ + E D
Sbjct: 1940 LATPTQSPSRMETAMEEAETTIETPVEDDKTDEGG--DAAEEAADIPNNANDQQEAPETD 1997
Query: 605 KK--KLAATVSKINKASFNNAVSQLAVVNPG 633
K AAT S ++ A ++ A+ + G
Sbjct: 1998 IKPETSAATTSPVSTAPTTSSTLASAITSSG 2028
Score = 33.9 bits (76), Expect = 0.30
Identities = 29/128 (22%), Positives = 53/128 (40%), Gaps = 7/128 (5%)
Query: 497 EELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLE 556
E LRE K L +S + + E+ + L + K+ ++ +LE
Sbjct: 245 ERLRELETKIGSLQEDLSSCKDAATTTEEQYTAELFTANKLVDLYKESSEEWSRKAGELE 304
Query: 557 GRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAATVSKIN 616
G +KA E ARL + E+++ + ++ + K+ LE L+ +K A + K
Sbjct: 305 GVIKALE-------ARLSQVESSYKERLDKEVSTKQLLEKENGDLKQKLEKCEAEIEKTR 357
Query: 617 KASFNNAV 624
K N +
Sbjct: 358 KTDELNLI 365
Score = 30.8 bits (68), Expect = 2.6
Identities = 23/104 (22%), Positives = 42/104 (40%), Gaps = 4/104 (3%)
Query: 480 KTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENE 539
K ++ LQ +EL+E+ S ++ L +EE+ K L N
Sbjct: 839 KQEEHIKQLQREWAEAKKELQEERSNARDFTSDRNQTLNNAVMQVEEMGKELANALKAVS 898
Query: 540 QLKKKVQDGDAVREDLEGRLKAAEGKA----ETGAARLKEAEAA 579
+ + +A DLE ++++++ K G L + EAA
Sbjct: 899 VAESRASVAEARLSDLEKKIRSSDPKTLDMDSGGIVSLSDKEAA 942
>At1g04600 putative myosin heavy chain
Length = 1730
Score = 40.8 bits (94), Expect = 0.002
Identities = 35/113 (30%), Positives = 57/113 (49%), Gaps = 15/113 (13%)
Query: 498 ELREKTEKDDQLISSMSKELEEGCSAIEELSKALD---NMKLENEQLKKKVQDGDAVRED 554
+LR+ E + IS + L++ IEELSK L+ ++ ENEQLK+ V
Sbjct: 960 QLRDTQETKSKEISDLQSALQDMQLEIEELSKGLEMTNDLAAENEQLKESV-------SS 1012
Query: 555 LEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENR-----VSSLE 602
L+ ++ +E K E + +E + +QS+ +K E EN+ VSS+E
Sbjct: 1013 LQNKIDESERKYEEISKISEERIKDEVPVIDQSAIIKLETENQKLKALVSSME 1065
>At1g80930 unknown protein
Length = 900
Score = 40.0 bits (92), Expect = 0.004
Identities = 80/342 (23%), Positives = 127/342 (36%), Gaps = 42/342 (12%)
Query: 319 GQSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAEVGGTSGSAKNVSLEADHGSDPVQ 378
G D E + V + E E + +DR A G K + +++D +
Sbjct: 61 GTVADGEGRRGDKVRRKETSDDEELARRSRKDRKEANSGSEDDRGKRIEVDSDGDGERRV 120
Query: 379 GKGR---------SVSEEKDTRGPASKKRRVEDHQDHADG-------EVQGSE-PERAEP 421
KGR S EE D +G + V+D E QG E R
Sbjct: 121 NKGRNTDRVRADTSSDEEDDLKGNKKEPMEVDDDYGRRGRRRSPKVMEKQGRERSHRGSR 180
Query: 422 VTVVPPSPERLSIWGPRAG------NCREHLAQIDD---LVLTENDQKYLGDREPAGLLN 472
V PS E R G R+H A DD + +E K DR GLL
Sbjct: 181 VIADKPSDEEDDRQRSRGGRRESQRKRRDHRASDDDEEGEIRSERRGKEKNDRGSEGLLK 240
Query: 473 YVLRASLKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEE--GCSAIEELSKA 530
R T + GS E E ++++ +DD + + + S ++L K
Sbjct: 241 RDRRERDLTDG---HENGSRRRE-SERKDRSRRDDGVRDEKERRHNDKYDDSQRDKLRKE 296
Query: 531 LDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAE-AAWLKEREQSSA 589
D+ K E +KK++ +L A +TG + + A +KE E S+
Sbjct: 297 -DSRKRE----EKKIEVPKPKLAELNPSENNAMALGKTGGVYIPPFKLARMMKEVEDKSS 351
Query: 590 VKKELENRVSSLEADKKKLAATVSKINKASFNNAVSQLAVVN 631
V E + + +A +K + V+K+N ++ N + +L N
Sbjct: 352 V----EYQRLTWDALRKSINGLVNKVNASNIKNIIPELFAEN 389
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,330,115
Number of Sequences: 26719
Number of extensions: 765718
Number of successful extensions: 3257
Number of sequences better than 10.0: 295
Number of HSP's better than 10.0 without gapping: 59
Number of HSP's successfully gapped in prelim test: 238
Number of HSP's that attempted gapping in prelim test: 2682
Number of HSP's gapped (non-prelim): 710
length of query: 673
length of database: 11,318,596
effective HSP length: 106
effective length of query: 567
effective length of database: 8,486,382
effective search space: 4811778594
effective search space used: 4811778594
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0039.13


