

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0039.1
(821 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
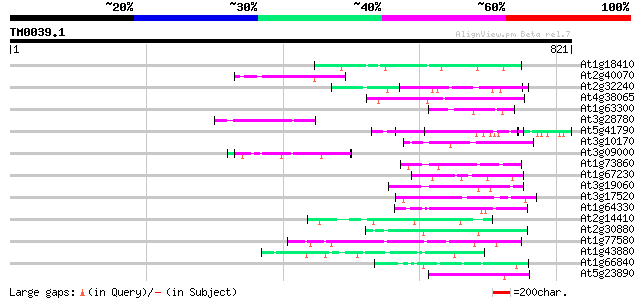
Score E
Sequences producing significant alignments: (bits) Value
At1g18410 kinesin-related protein, putative 51 2e-06
At2g40070 En/Spm-like transposon protein 50 5e-06
At2g32240 putative myosin heavy chain 49 9e-06
At4g38065 hypothetical protein 49 1e-05
At1g63300 hypothetical protein 47 3e-05
At3g28780 histone-H4-like protein 47 4e-05
At5g41790 myosin heavy chain-like protein 46 7e-05
At3g10170 hypothetical protein 46 1e-04
At3g09000 unknown protein 45 1e-04
At1g73860 kinesin-related protein 45 1e-04
At1g67230 unknown protein 45 1e-04
At3g19060 hypothetical protein 45 2e-04
At3g17520 unknown protein 45 2e-04
At1g64330 unknown protein 45 2e-04
At2g14410 hypothetical protein 44 3e-04
At2g30880 unknown protein 44 5e-04
At1g77580 unknown protein 44 5e-04
At1g43880 hypothetical protein 44 5e-04
At1g66840 hypothetical protein 43 6e-04
At5g23890 unknown protein 43 8e-04
>At1g18410 kinesin-related protein, putative
Length = 1081
Score = 51.2 bits (121), Expect = 2e-06
Identities = 87/381 (22%), Positives = 142/381 (36%), Gaps = 82/381 (21%)
Query: 447 SPPRQVTPPSPPPTNDERSLSPPPRQEERPSPGAATTS-------------EAAQIEQAP 493
S PR PS P + L+ Q P+ A S EA Q++Q
Sbjct: 169 STPRSPFSPSSPRERHNKGLADSRFQRPLPNSSALDPSSPGSMLHGGHKSHEAFQMKQGR 228
Query: 494 GNRAGSSSHFNMLPNAIE--PSEFLLTGLNRDGIEKEVLSRGINETKEETLACLLRA--- 548
+ + M N ++ P++ LL+ +N GI E + R E + +ACLLR
Sbjct: 229 FDLQAAKISELMKSNNLDNAPTQSLLSIVN--GILDETIERKNGELPQR-VACLLRKVVQ 285
Query: 549 ----------------GCIFAHAFDKFNSAVTEAERLKAESAQHQEAATAW--EKRFDKL 590
+F +K+ S + E L + +++ E + EK+ DK
Sbjct: 286 EIERRISTQSEHLRTQNSVFKAREEKYQSRIKVLETLASGTSEENETEKSKLEEKKKDKE 345
Query: 591 VAQAGKDKVHADKLIGAAGNRIGELEDHLKLMKEEADDLD-------ASLQACKKEKDQV 643
G +K + + + R ELE K +++ ++ A ++ KE +Q+
Sbjct: 346 EDMVGIEKENGHYNLEISTLR-RELETTKKAYEQQCLQMESKTKGATAGIEDRVKELEQM 404
Query: 644 EKDLAAKGKALAAKELEFETLGAELELAKKALAEQKKKSA-------------------- 683
KD + KAL + E E +G E + K L E+ K+
Sbjct: 405 RKDASVARKALEERVRELEKMGKEADAVKMNLEEKVKELQKYKDETITVTTSIEGKNREL 464
Query: 684 ----ESLALVKADLEAMRATSEEIKKVNEAHGATLVTKDAEI-----------TSLLARI 728
+ V LEA E+ K +L K+ E+ TSL A+
Sbjct: 465 EQFKQETMTVTTSLEAQNRELEQAIKETMTVNTSLEAKNRELEQSKKETMTVNTSLKAKN 524
Query: 729 KELEGELSVEKAKATEAREQA 749
+ELE L K+KA E E++
Sbjct: 525 RELEQNLVHWKSKAKEMEEKS 545
>At2g40070 En/Spm-like transposon protein
Length = 510
Score = 50.1 bits (118), Expect = 5e-06
Identities = 45/170 (26%), Positives = 76/170 (44%), Gaps = 16/170 (9%)
Query: 329 ASSSNPGAQPTAAAVAKGKKAASTAAIASSKSSTDGTSQLPTAEASAAAATELVTVTTLD 388
+SS PG++P A G+ + TA SS+ ST PT+ A+ ++AT +
Sbjct: 150 SSSGGPGSRP---ATPTGRSSTLTANSKSSRPST------PTSRATVSSATRPSLTNSRS 200
Query: 389 SAAVGTATGAAAKFTTIGAT--TTTADQPSTAGISAAAATTCSGAAAGDKERETEKEN-- 444
+ + T ++ T++ ++ T TA +P+T+ +A + T S + K +
Sbjct: 201 TVSATTKPTPMSRSTSLSSSRLTPTASKPTTSTARSAGSVTRSTPSTTTKSAGPSRSTTP 260
Query: 445 -PKSPPRQVTPPSPPPTNDERSLSPPPRQEERP--SPGAATTSEAAQIEQ 491
+S R TP S P +++S RP S AATT+ I Q
Sbjct: 261 LSRSTARSSTPTSRPTLPPSKTISRSSTPTRRPIASASAATTTANPTISQ 310
Score = 37.0 bits (84), Expect = 0.045
Identities = 43/175 (24%), Positives = 70/175 (39%), Gaps = 8/175 (4%)
Query: 308 AENQAEGAQAPKRRRLVKGSSASSSNPGAQPTAAAVAKGKKAASTAAIASSKSSTDGTSQ 367
A +++ P R V S+ S ++ T +A K + + +++SS+ T S+
Sbjct: 171 ANSKSSRPSTPTSRATVS-SATRPSLTNSRSTVSATTKPTPMSRSTSLSSSRL-TPTASK 228
Query: 368 LPTAEA-SAAAATELVTVTTLDSAAVGTATGAAAKFTTIGATTTTADQ-PSTAGISAAAA 425
T+ A SA + T TT SA +T ++ T +T T+ P + IS ++
Sbjct: 229 PTTSTARSAGSVTRSTPSTTTKSAGPSRSTTPLSRSTARSSTPTSRPTLPPSKTISRSST 288
Query: 426 TT----CSGAAAGDKERETEKENPKSPPRQVTPPSPPPTNDERSLSPPPRQEERP 476
T S +AA T + S P P P N S + P RP
Sbjct: 289 PTRRPIASASAATTTANPTISQIKPSSPAPAKPMPTPSKNPALSRAASPTVRSRP 343
Score = 29.3 bits (64), Expect = 9.3
Identities = 32/135 (23%), Positives = 55/135 (40%), Gaps = 21/135 (15%)
Query: 386 TLDSAAVGTATGAAAKFTTIGATTTTADQPSTAGISAAAATTCSGAAAGDKERETEKENP 445
TL S ++T +AA+ T+ Q S+ G+S+++ G R + P
Sbjct: 110 TLTSRLANSSTESAAR-----NHLTSRQQTSSPGLSSSS---------GASRRPSSSGGP 155
Query: 446 KSPPRQVTPPSPPPTNDERSLSPPPRQEE-------RPSPGAATTSEAAQIEQAPGNRAG 498
S P T S T + +S P RPS + ++ +A + P +R+
Sbjct: 156 GSRPATPTGRSSTLTANSKSSRPSTPTSRATVSSATRPSLTNSRSTVSATTKPTPMSRST 215
Query: 499 SSSHFNMLPNAIEPS 513
S S + P A +P+
Sbjct: 216 SLSSSRLTPTASKPT 230
>At2g32240 putative myosin heavy chain
Length = 1333
Score = 49.3 bits (116), Expect = 9e-06
Identities = 69/320 (21%), Positives = 124/320 (38%), Gaps = 44/320 (13%)
Query: 472 QEERPSPGAATTSEAAQIEQAPGNRAGSSSHFNMLPNAIEPSEFLLTGLNRDGIEKEVLS 531
+E+ ++S ++EQ G A + S L + ++ + E E+L+
Sbjct: 840 EEQLAEASGKSSSLKEKLEQTLGRLAAAESVNEKLKQEFDQAQEKSL---QSSSESELLA 896
Query: 532 RGINETKE--ETLACLLRAGCI--------FAHAFDKFNSAVTEAERLKAESAQHQEAAT 581
N+ K + L L+ +G + A ++FN TE+ L + H+
Sbjct: 897 ETNNQLKIKIQELEGLIGSGSVEKETALKRLEEAIERFNQKETESSDLVEKLKTHENQIE 956
Query: 582 AWEKRFDKLVAQAGKDKVHADKLIGAAGNRIGELED------------------HLKLMK 623
++K + A KV + + N +E+ +LKL
Sbjct: 957 EYKKLAHEASGVADTRKVELEDALSKLKNLESTIEELGAKCQGLEKESGDLAEVNLKLNL 1016
Query: 624 E------EADDLDASLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKALAE 677
E EA++L L A + EK+Q +L A + + + G +L+ + E
Sbjct: 1017 ELANHGSEANELQTKLSALEAEKEQTANELEASKTTIEDLTKQLTSEGEKLQSQISSHTE 1076
Query: 678 QKKKSAESLALVKADLEAMRATSEEIKKVNEAHGATLVTK-------DAEITSLLARIKE 730
+ + K +L+++ A EE V + TLV++ AE + L + +E
Sbjct: 1077 ENNQVNAMFQSTKEELQSVIAKLEEQLTVESSKADTLVSEIEKLRAVAAEKSVLESHFEE 1136
Query: 731 LEGELSVEKAKATEAREQAA 750
LE LS KA+ E E AA
Sbjct: 1137 LEKTLSEVKAQLKENVENAA 1156
Score = 44.3 bits (103), Expect = 3e-04
Identities = 44/194 (22%), Positives = 87/194 (44%), Gaps = 16/194 (8%)
Query: 571 AESAQHQEAATAWEKRFDKLVAQAGKDKVHADKLIGAAGNRIGELEDHLKLMKEEADDLD 630
+E+ + Q +A E ++ + K + L + +L+ + EE + ++
Sbjct: 1023 SEANELQTKLSALEAEKEQTANELEASKTTIEDLTKQLTSEGEKLQSQISSHTEENNQVN 1082
Query: 631 ASLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKALAEQKKKSAESLALVK 690
A Q+ K+E V +A + L + + +TL +E+E + AE+ ++++
Sbjct: 1083 AMFQSTKEELQSV---IAKLEEQLTVESSKADTLVSEIEKLRAVAAEK--------SVLE 1131
Query: 691 ADLEAMRATSEEIK---KVNEAHGATLVTKDAEITSLLARIKELEGELSV--EKAKATEA 745
+ E + T E+K K N + AT K AE+TS L + + GE V E+ +
Sbjct: 1132 SHFEELEKTLSEVKAQLKENVENAATASVKVAELTSKLQEHEHIAGERDVLNEQVLQLQK 1191
Query: 746 REQAADIAYDSRER 759
QAA + D +++
Sbjct: 1192 ELQAAQSSIDEQKQ 1205
Score = 40.0 bits (92), Expect = 0.005
Identities = 42/202 (20%), Positives = 87/202 (42%), Gaps = 22/202 (10%)
Query: 567 ERLKAESAQHQEAATAWEKRFDKLVAQAGKDKVHADKLIG-------AAGNRIGELEDHL 619
E L +E+ AAT + +V + + A I AA + ELE L
Sbjct: 458 EELHSEAGSAAAAATQKNLELEDVVRSSSQAAEEAKSQIKELETKFTAAEQKNAELEQQL 517
Query: 620 KLMKEEADDLDASLQACKKEKDQVEK--DLAAKGKALAAKEL-EFETLGAELELAKKALA 676
L++ ++ D + L+ ++ +++ ++A + K A ++ E++ +ELEL+ +
Sbjct: 518 NLLQLKSSDAERELKELSEKSSELQTAIEVAEEEKKQATTQMQEYKQKASELELSLTQSS 577
Query: 677 EQKKKSAESLALVKADLEAMRATSEEIKKVNEAHGATLV------TKDAEITSLLARIKE 730
+ + E L + A++ +E + N H ++ + ++ R+K+
Sbjct: 578 ARNSELEEDLRI------ALQKGAEHEDRANTTHQRSIELEGLCQSSQSKHEDAEGRLKD 631
Query: 731 LEGELSVEKAKATEAREQAADI 752
LE L EK + E EQ + +
Sbjct: 632 LELLLQTEKYRIQELEEQVSSL 653
Score = 39.3 bits (90), Expect = 0.009
Identities = 41/194 (21%), Positives = 90/194 (46%), Gaps = 25/194 (12%)
Query: 565 EAERLKAESAQHQ-EAATAWEKRFDKLVAQAGKDKVHADKLIGAAGNRIGELEDHLKLMK 623
EA ++K+ S + AT EK + + +K + + +I E E+ L+ ++
Sbjct: 679 EAFQVKSSSLEAALNIATENEKELTENLNAVTSEKKKLEATVDEYSVKISESENLLESIR 738
Query: 624 EEADDLDASLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKALAEQKKKSA 683
E + L++ +E DL A G L+ + +L+ A+++L EQK +
Sbjct: 739 NELNVTQGKLES-------IENDLKAAG-------LQESEVMEKLKSAEESL-EQKGREI 783
Query: 684 ESLALVKADLEAMRAT-----SEEIKKVNEAHGATLVTKDAEITSLLARIKELEGELSVE 738
+ + +LEA+ + ++K E ++D+E +SL ++++LEG++
Sbjct: 784 DEATTKRMELEALHQSLSIDSEHRLQKAMEE----FTSRDSEASSLTEKLRDLEGKIKSY 839
Query: 739 KAKATEAREQAADI 752
+ + EA +++ +
Sbjct: 840 EEQLAEASGKSSSL 853
Score = 38.5 bits (88), Expect = 0.015
Identities = 39/174 (22%), Positives = 70/174 (39%), Gaps = 18/174 (10%)
Query: 585 KRFDKLVAQAGKDKVHADKLIGAAGNRIGELEDHLKLMKEEADDLDASLQACKKEKDQVE 644
KR++ + + A + + + G+LE K +E+ + + + K +
Sbjct: 110 KRYESENTHLKDELLSAKEKLEETEKKHGDLEVVQKKQQEKIVEGEERHSSQLKSLEDAL 169
Query: 645 KDLAAKGKALAAKELEFETLGAELELAKKALAEQKKKSAESLALVKADLEAMRATSEEIK 704
+ AK K L + F+ LG ELE ++K L E + E ++ ++EE +
Sbjct: 170 QSHDAKDKELTEVKEAFDALGIELESSRKKLIELE--------------EGLKRSAEEAQ 215
Query: 705 KVNEAHGATLVTKDAEITSLLARIKELEGELSVEKAKATEAREQAADIAYDSRE 758
K E H + D+E L E L K A E E+ A + + +E
Sbjct: 216 KFEELHKQSASHADSESQKAL----EFSELLKSTKESAKEMEEKMASLQQEIKE 265
Score = 38.1 bits (87), Expect = 0.020
Identities = 50/210 (23%), Positives = 93/210 (43%), Gaps = 50/210 (23%)
Query: 567 ERLKAESAQHQEAATAWEKRFDKLVAQAGKDK-VHADKLIGAAGNRIGELEDHLKLMKEE 625
E L++ A+ +E T + D LV++ K + V A+K + LE H + +++
Sbjct: 1090 EELQSVIAKLEEQLTVESSKADTLVSEIEKLRAVAAEKSV---------LESHFEELEKT 1140
Query: 626 ADDLDASLQACKKEKDQVEKDLAAKGKA--LAAKELEFETLGAE--------------LE 669
++ A L K+ VE A K L +K E E + E L+
Sbjct: 1141 LSEVKAQL------KENVENAATASVKVAELTSKLQEHEHIAGERDVLNEQVLQLQKELQ 1194
Query: 670 LAKKALAEQKKKSAESLALVKADLE-AMRATSEEIKKVNEAHGATLVTKDAEITSLLARI 728
A+ ++ EQK+ ++ +++LE A++ + EEI+ K +T + +
Sbjct: 1195 AAQSSIDEQKQAHSQK----QSELESALKKSQEEIE-----------AKKKAVTEFESMV 1239
Query: 729 KELEGELSVEKAKATEAREQAADIAYDSRE 758
K+LE ++ + AK E +A D+ SR+
Sbjct: 1240 KDLEQKVQLADAKTKET--EAMDVGVKSRD 1267
Score = 37.0 bits (84), Expect = 0.045
Identities = 53/242 (21%), Positives = 95/242 (38%), Gaps = 48/242 (19%)
Query: 557 DKFNSAVTEAERLKAESAQHQEAATAWEKRFDKLVAQAGKDKVHADKLIGAAGNRIGELE 616
+K S E + L + +++++ A + +L A + + + +R+ E E
Sbjct: 254 EKMASLQQEIKELNEKMSENEKVEAALKSSAGELAA--------VQEELALSKSRLLETE 305
Query: 617 DHLKLMKEEADDLDASLQACK------KEKDQVEKDLAAKGKALAAKELEFETLGAEL-- 668
+ + D+L L+ K KE+ V +DL A+ K L AK E E + ++L
Sbjct: 306 QKVSSTEALIDELTQELEQKKASESRFKEELSVLQDLDAQTKGLQAKLSEQEGINSKLAE 365
Query: 669 ELAKKALAEQKKKSAES--------LALVKADLEAMR----------------------- 697
EL +K L E K E LA V + EA+
Sbjct: 366 ELKEKELLESLSKDQEEKLRTANEKLAEVLKEKEALEANVAEVTSNVATVTEVCNELEEK 425
Query: 698 -ATSEEIKKVNEAHGATLVTKDAEITSLLARIKELEGELSVEKAKATEAREQAADIAYDS 756
TS+E +A + ++ ++E+ L ++EL E A AT+ + D+ S
Sbjct: 426 LKTSDENFSKTDALLSQALSNNSELEQKLKSLEELHSEAGSAAAAATQKNLELEDVVRSS 485
Query: 757 RE 758
+
Sbjct: 486 SQ 487
Score = 31.6 bits (70), Expect = 1.9
Identities = 33/140 (23%), Positives = 58/140 (40%), Gaps = 11/140 (7%)
Query: 615 LEDHLKLMKEEADDLDASLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKA 674
+E+ ++++ + L +++ ++E +L L E E L EL AK+
Sbjct: 70 VEEQKEVIERSSSGSQRELHESQEKAKELELELERVAGELKRYESENTHLKDELLSAKEK 129
Query: 675 LAEQKKKSAESLALVKAD----LEAMRATSEEIKKVNEA---HGATLVTKDAEITSLLAR 727
L E +KK + + K +E S ++K + +A H A KD E+T +
Sbjct: 130 LEETEKKHGDLEVVQKKQQEKIVEGEERHSSQLKSLEDALQSHDA----KDKELTEVKEA 185
Query: 728 IKELEGELSVEKAKATEARE 747
L EL + K E E
Sbjct: 186 FDALGIELESSRKKLIELEE 205
>At4g38065 hypothetical protein
Length = 1050
Score = 48.9 bits (115), Expect = 1e-05
Identities = 56/243 (23%), Positives = 103/243 (42%), Gaps = 13/243 (5%)
Query: 522 RDGIEKEVLSRGINETK---EETLACLLRAGCIFAHAFDKFNSAVTEAE-RLKAESAQHQ 577
+ G E E SR I E K EE CL + D + E + + + +
Sbjct: 52 KHGFEIEEKSREIAELKRANEELQRCLREKDSVVKRVNDVNDKLRANGEDKYREFEEEKR 111
Query: 578 EAATAWEKRFDKLVAQAGKDKVHADKLIGAAG-------NRIGELEDHLKLMKEEADDLD 630
+ ++ +K + K+ V+ ++ G G RI E E +K MKE D
Sbjct: 112 NMMSGLDEASEKNIDLEQKNNVYRAEIEGLKGLLAVAETKRI-EAEKTVKGMKEMRGRDD 170
Query: 631 ASLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKALAEQKKKSAESLALVK 690
++ ++EK QVE+ L K + E +E L + +KK E+K K + + ++
Sbjct: 171 VVVKM-EEEKSQVEEKLKWKKEQFKHLEEAYEKLKNLFKDSKKEWEEEKSKLLDEIYSLQ 229
Query: 691 ADLEAMRATSEEIKKVNEAHGATLVTKDAEITSLLARIKELEGELSVEKAKATEAREQAA 750
L+++ SE+++K + L ++ L ++ E + + A+ +AR Q
Sbjct: 230 TKLDSVTRISEDLQKKLQMCNGALTQEETRRKHLEIQVSEFKAKYEDAFAECQDARTQLD 289
Query: 751 DIA 753
D+A
Sbjct: 290 DLA 292
Score = 42.4 bits (98), Expect = 0.001
Identities = 47/192 (24%), Positives = 81/192 (41%), Gaps = 7/192 (3%)
Query: 566 AERLKAESAQHQ------EAATAWEKRFDKLVAQAGKDKVHADKLIGAAGNRIGELEDHL 619
A+RLKAE Q+Q EA+ + + + Q + VH K+I A + E ++ L
Sbjct: 733 AKRLKAELEQNQNLRKRVEASLLEQVGVGEAIKQEKNELVHKLKVISHARSSDSEKKESL 792
Query: 620 KLMKEEA-DDLDASLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKALAEQ 678
K+E + L ++ +++ + E + + +EL+ E L+ + L E
Sbjct: 793 MRDKDEMLESLQREVELLEQDSLRRELEDVVLAHMIGERELQNEREICALQQKDQDLCEV 852
Query: 679 KKKSAESLALVKADLEAMRATSEEIKKVNEAHGATLVTKDAEITSLLARIKELEGELSVE 738
K + SL V L+ + ++K E A + E S I ELEGE+S
Sbjct: 853 KHELEGSLKSVSLLLQQKQNEVNMLRKTWEKLTARQILTAVETESKKMMIIELEGEISSL 912
Query: 739 KAKATEAREQAA 750
K + E +
Sbjct: 913 SQKLETSNESVS 924
>At1g63300 hypothetical protein
Length = 1029
Score = 47.4 bits (111), Expect = 3e-05
Identities = 40/148 (27%), Positives = 74/148 (49%), Gaps = 31/148 (20%)
Query: 614 ELEDHLKLMKEEADDLDASLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKK 673
ELE + LM++E++ L A LQ K KD+ K A++ + E ET+ ++ + K
Sbjct: 784 ELESKISLMRKESESLAAELQVIKLAKDE-------KETAISLLQTELETVRSQCDDLKH 836
Query: 674 ALAE---QKKKSAESLALVKADLEAMRATSEEI-KKVNEAHGATLVTKDA---------- 719
+L+E + +K + +A VK++L+ T + KK+ E+ T +TK A
Sbjct: 837 SLSENDLEMEKHKKQVAHVKSELKKKEETMANLEKKLKESR--TAITKTAQRNNINKGSP 894
Query: 720 --------EITSLLARIKELEGELSVEK 739
E+ + +IK LEG++ +++
Sbjct: 895 VGAHGGSKEVAVMKDKIKLLEGQIKLKE 922
Score = 39.3 bits (90), Expect = 0.009
Identities = 45/219 (20%), Positives = 101/219 (45%), Gaps = 25/219 (11%)
Query: 562 AVTEAERLKAESAQHQEAATAWEKRFDKLVAQAGKDKVHADKLIGAAGNRIGELEDHLKL 621
A+TEA L+ + Q +E + D+L A + + +L + ++E L+
Sbjct: 648 AMTEANELRMQKRQLEEMI---KDANDELRANQAEYEAKLHELSEKLSFKTSQMERMLEN 704
Query: 622 MKEEADDLD----------ASL-QACKKEKDQVEKDLAAKGKALAAKELEFETLGAELEL 670
+ E+++++D A+L Q K K+++E +L +L + + E L +LE
Sbjct: 705 LDEKSNEIDNQKRHEEDVTANLNQEIKILKEEIE-NLKKNQDSLMLQAEQAENLRVDLEK 763
Query: 671 AKKALAE---------QKKKSAES-LALVKADLEAMRATSEEIKKVNEAHGATLVTKDAE 720
KK++ E KK ES ++L++ + E++ A + IK + + E
Sbjct: 764 TKKSVMEAEASLQRENMKKIELESKISLMRKESESLAAELQVIKLAKDEKETAISLLQTE 823
Query: 721 ITSLLARIKELEGELSVEKAKATEAREQAADIAYDSRER 759
+ ++ ++ +L+ LS + + ++Q A + + +++
Sbjct: 824 LETVRSQCDDLKHSLSENDLEMEKHKKQVAHVKSELKKK 862
Score = 34.7 bits (78), Expect = 0.22
Identities = 34/141 (24%), Positives = 69/141 (48%), Gaps = 13/141 (9%)
Query: 621 LMKEEADDLDASLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKALAEQKK 680
+++++ DL ++ K++KD++E + + AL + L+ + +L + L EQ K
Sbjct: 466 ILEQKITDLYNEIEIYKRDKDELE--IQMEQLALDYEILKQQNHDISYKLEQSQLQEQLK 523
Query: 681 KSAESLALVKADLEAMRATSEEIKKVNEAHGATLVTKDAEITSLLARIKELEGEL-SVEK 739
++ + + E++ E+ A L + E + L RIKELE ++ ++E+
Sbjct: 524 --------IQYECSSSLVDVTELENQVESLEAELKKQSEEFSESLCRIKELESQMETLEE 575
Query: 740 AKATEAREQAADIAYDSRERG 760
+A+ ADI D+ RG
Sbjct: 576 EMEKQAQVFEADI--DAVTRG 594
>At3g28780 histone-H4-like protein
Length = 614
Score = 47.0 bits (110), Expect = 4e-05
Identities = 37/148 (25%), Positives = 64/148 (43%), Gaps = 8/148 (5%)
Query: 300 GDGKRRGGAENQAEGAQAPKRRRLVKGSSASSSNPGAQPTAAAVAKGKKAASTAAIASSK 359
G+ G AE E A A +S S+ G + + A G ++AS A +
Sbjct: 305 GEAASGGSAETGGESASAG-------AASGGSAETGGESGSGGAASGGESASGGATSGGS 357
Query: 360 SSTDGTSQLPTAEASAAAATELVTVTTLDSAAVGTATGAAAKFTTIGATTTTADQPSTAG 419
T G+++ AS AA+ + + +++ +G + T G + T+D+ S +G
Sbjct: 358 PETGGSAETGGESASGGAASGGESASGGAASSGSVESGGESTGATSGGSAETSDE-SASG 416
Query: 420 ISAAAATTCSGAAAGDKERETEKENPKS 447
+A+ + SG AA ET E+ S
Sbjct: 417 GAASGGESASGGAASGGSAETGGESTSS 444
Score = 42.4 bits (98), Expect = 0.001
Identities = 46/206 (22%), Positives = 73/206 (35%), Gaps = 6/206 (2%)
Query: 300 GDGKRRGGAENQAEGAQAPKRRRLVKGSSASSSNPGAQPTAAAVAKGKKAASTAAIASSK 359
G GGA + + G SA +S+ A AA+ G ++AS A +
Sbjct: 378 GGESASGGAASSGSVESGGESTGATSGGSAETSDESASGGAAS---GGESASGGAASGGS 434
Query: 360 SSTDGTSQLPTAEASAAAATELVTVTTLDSAAVGTATGAAAKFTTIGATTTTADQPSTAG 419
+ T G S + + +E + + GAAA +T + T+ + S G
Sbjct: 435 AETGGESTSSGVASGGSTGSESASAGAASGGSTEANGGAAAGGSTEAGSGTSTETSSMGG 494
Query: 420 ISAAAATTCSGAAAGDKERETEKENPKSPPRQVTPPSPPPTNDERSLSPPPRQEERPSPG 479
SAAA ++ G E+ S D SP G
Sbjct: 495 GSAAAGGVSESSSGGSTAAGGTSESASGGSATAGGASGGTYTDSTGGSPTGSPSAGGPSG 554
Query: 480 AATTSE---AAQIEQAPGNRAGSSSH 502
+A+ S Q+ G +AGS+S+
Sbjct: 555 SASESSMEGGTFGGQSMGGQAGSASY 580
Score = 40.0 bits (92), Expect = 0.005
Identities = 49/217 (22%), Positives = 74/217 (33%), Gaps = 18/217 (8%)
Query: 302 GKRRGGAENQAEGAQAPKRRRLVKGSSASSSNPGAQPTAAAVAKGKKAASTAAIASSKSS 361
G GGA + A+ A G++A+ ++ G+ T A G A+ A + + +
Sbjct: 241 GADSGGAAS-ADSGGAAAGETASGGAAAADTSGGSAETGGESASGGAASGAGAASGASAK 299
Query: 362 TDGTSQLPTAEASAAAATELVTVTTLDSAAVGT-----ATGAAAKFTTIGATTTTADQPS 416
T G S + SA E + + T + GAA+ + T+ P
Sbjct: 300 TGGESGEAASGGSAETGGESASAGAASGGSAETGGESGSGGAASGGESASGGATSGGSPE 359
Query: 417 TAGISAAAATTCSGAAAGDKE------RETEKENPKSPPRQVTPPSPPPTNDERSLSPPP 470
T G + + SG AA E + T T+DE +
Sbjct: 360 TGGSAETGGESASGGAASGGESASGGAASSGSVESGGESTGATSGGSAETSDESASGGAA 419
Query: 471 RQEERPSPGAA------TTSEAAQIEQAPGNRAGSSS 501
E S GAA T E+ A G GS S
Sbjct: 420 SGGESASGGAASGGSAETGGESTSSGVASGGSTGSES 456
Score = 39.7 bits (91), Expect = 0.007
Identities = 50/204 (24%), Positives = 73/204 (35%), Gaps = 23/204 (11%)
Query: 300 GDGKRRGGAENQAEGAQAPKRRRLVKGSSASSSNPGAQPTAAAVAKGKKAASTAAIASSK 359
GD G A A A G +A + + GA A A + G +A + A+ +
Sbjct: 209 GDSSSAGAAGESGSAATADS------GDAAGADSGGA---AGADSGGAASADSGGAAAGE 259
Query: 360 SSTDGTSQLPTAEASAAAATELVTVTTLDSAAVGTATGAAAKFTTIGATTTTADQPSTAG 419
+++ G + T+ SA E + A G A+GA+AK + T G
Sbjct: 260 TASGGAAAADTSGGSAETGGESASGGAASGA--GAASGASAKTGGESGEAASGGSAETGG 317
Query: 420 ISAAAATTCSGAA--AGDKERETEKENPKSPPRQVTPPSPPPTNDERSLSPPPRQEERPS 477
SA+A G+A G+ +S T P T E S
Sbjct: 318 ESASAGAASGGSAETGGESGSGGAASGGESASGGATSGGSPETGGSAETG-----GESAS 372
Query: 478 PGAATTSEAAQIEQAPGNRAGSSS 501
GAA+ E+A G A S S
Sbjct: 373 GGAASGGESAS-----GGAASSGS 391
Score = 38.1 bits (87), Expect = 0.020
Identities = 34/141 (24%), Positives = 62/141 (43%), Gaps = 19/141 (13%)
Query: 300 GDGKRRGGAENQAEGAQAPKRRRLVKGSSASSSNPGAQPTAAAVAKGKKAASTAAIASSK 359
G G G E+ + GA + + S+ G + + A G ++AS A +S
Sbjct: 337 GSGGAASGGESASGGATSGG-----SPETGGSAETGGESASGGAASGGESASGGAASSGS 391
Query: 360 SSTDGTSQLPTAEASAAAATELVTVTTLDSAAVGTATG--AAAKFTTIGATTTTADQPST 417
+ G S T+ SA + E SA+ G A+G +A+ G + T + ++
Sbjct: 392 VESGGESTGATSGGSAETSDE--------SASGGAASGGESASGGAASGGSAETGGESTS 443
Query: 418 AGISAAAAT----TCSGAAAG 434
+G+++ +T +GAA+G
Sbjct: 444 SGVASGGSTGSESASAGAASG 464
Score = 35.0 bits (79), Expect = 0.17
Identities = 31/134 (23%), Positives = 54/134 (40%), Gaps = 6/134 (4%)
Query: 297 NTEGDGKRRGGAENQAEGAQAPKRRRLVKGSSA----SSSNPGAQPTAAAVAKGKKAAS- 351
+TE +G G +A + + + GS+A S S+ G A ++ S
Sbjct: 466 STEANGGAAAGGSTEAGSGTSTETSSMGGGSAAAGGVSESSSGGSTAAGGTSESASGGSA 525
Query: 352 TAAIASSKSSTDGTSQLPTAEASAAAATELVTVTTLDSAAV-GTATGAAAKFTTIGATTT 410
TA AS + TD T PT SA + + ++++ G + G A + +T
Sbjct: 526 TAGGASGGTYTDSTGGSPTGSPSAGGPSGSASESSMEGGTFGGQSMGGQAGSASYQSTNY 585
Query: 411 TADQPSTAGISAAA 424
+AG S+ +
Sbjct: 586 QKTHSKSAGKSSVS 599
Score = 32.7 bits (73), Expect = 0.85
Identities = 31/122 (25%), Positives = 54/122 (43%), Gaps = 4/122 (3%)
Query: 307 GAENQAE-GAQAPKRRRLVKGSSASSSNPGAQPTAA-AVAKGKKAASTAAIASSKSSTDG 364
G +A GA A G+S +S+ G AA V++ STAA +S+S++ G
Sbjct: 464 GGSTEANGGAAAGGSTEAGSGTSTETSSMGGGSAAAGGVSESSSGGSTAAGGTSESASGG 523
Query: 365 TSQLPTAEASAAAATELVTVTTLDSAAVGTATGAAAKFTTIGATTTTADQPSTAGISAAA 424
++ AS T+ + S + G +G+A++ + G T AG ++
Sbjct: 524 SA--TAGGASGGTYTDSTGGSPTGSPSAGGPSGSASESSMEGGTFGGQSMGGQAGSASYQ 581
Query: 425 AT 426
+T
Sbjct: 582 ST 583
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 46.2 bits (108), Expect = 7e-05
Identities = 48/227 (21%), Positives = 97/227 (42%), Gaps = 19/227 (8%)
Query: 530 LSRGINETKEETLACLLRAGCIFAHAFDKFNSAVTEAERLKAESAQHQEAATAWEKRFDK 589
LS + +EE+ + + D+ + L A+S++ +E E +
Sbjct: 680 LSESLKAAEEESRTMSTK----ISETSDELERTQIMVQELTADSSKLKEQLAEKESKLFL 735
Query: 590 LVAQAGKDKVHADKLIGAAGNRIGELEDHLKLMKEEADDLDASLQACKKEKDQVEKDLAA 649
L + K +V +L + LE L+ ++ DL+ + + K V + L A
Sbjct: 736 LTEKDSKSQVQIKELEAT----VATLELELESVRARIIDLETEIAS----KTTVVEQLEA 787
Query: 650 KGKALAAK--ELE--FETLGAELELAKKALAEQKKKSAESLALVKADLEAMRATSEEIKK 705
+ + + A+ ELE E G EL + L + K+S+ S+ + A+++ +RA + +
Sbjct: 788 QNREMVARISELEKTMEERGTELSALTQKLEDNDKQSSSSIETLTAEIDGLRAELDSMSV 847
Query: 706 VNEAHGATLVTKDAEITSLLARIKELEGELSVEKAKATEAREQAADI 752
E +V K E + +IK L+ E++ + + Q A++
Sbjct: 848 QKEEVEKQMVCKSEEAS---VKIKRLDDEVNGLRQQVASLDSQRAEL 891
Score = 45.1 bits (105), Expect = 2e-04
Identities = 74/302 (24%), Positives = 118/302 (38%), Gaps = 56/302 (18%)
Query: 565 EAERLKAESAQHQEAATAWEKRFDKLVAQAGKDKVH-ADKLIGAAGNRIGELEDHLKLMK 623
+ L+ E+ Q + E R K DK++ A I A I L++ L ++
Sbjct: 938 KGRELELETLGKQRSELDEELRTKKEENVQMHDKINVASSEIMALTELINNLKNELDSLQ 997
Query: 624 EEADDLDASLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKALAEQKKKSA 683
+ + +A L+ K+EK ++ + KAL +E + TL E K + E K++
Sbjct: 998 VQKSETEAELEREKQEKSELSNQITDVQKALVEQEAAYNTLEEE----HKQINELFKETE 1053
Query: 684 ESLALVKADL-EAMRATSEEIKKVNE------AHGAT-------LVTKDAEITSLLARIK 729
+L V D EA R E K+V H T L K EI +L+ +I
Sbjct: 1054 ATLNKVTVDYKEAQRLLEERGKEVTSRDSTIGVHEETMESLRNELEMKGDEIETLMEKIS 1113
Query: 730 ELEGELSVEKAKATEAREQAADIAYDSRERGFYLAKDQAQH------LFPNLDF---SAM 780
+E +L + K + + +E F K++A+H L NL +
Sbjct: 1114 NIEVKLRLSNQKL-----RVTEQVLTEKEEAF--RKEEAKHLEEQALLEKNLTMTHETYR 1166
Query: 781 GVMKE------ITAEGLVGPGDPPLIDQN------------LWTATE---EEEEEEEERN 819
G++KE IT +G + Q LWTAT E E+E+ N
Sbjct: 1167 GMIKEIADKVNITVDGFQSMSEKLTEKQGRYEKTVMEASKILWTATNWVIERNHEKEKMN 1226
Query: 820 NE 821
E
Sbjct: 1227 KE 1228
Score = 43.5 bits (101), Expect = 5e-04
Identities = 35/147 (23%), Positives = 74/147 (49%), Gaps = 17/147 (11%)
Query: 608 AGNRIGELEDHLKLMKEEADDLDASLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAE 667
+ R+ ELE LKL+++ DL ASL A ++EK + + L + + + L E
Sbjct: 483 SSTRLSELETQLKLLEQRVVDLSASLNAAEEEKKSLSSMILEITDELKQAQSKVQELVTE 542
Query: 668 LELAKKALAEQKKKSAESLALVKA----------DLEA-MRATSEEIKKVNEAHGATLVT 716
L +K L +++ + + + + +A +LEA + + E++K++N+ L +
Sbjct: 543 LAESKDTLTQKENELSSFVEVHEAHKRDSSSQVKELEARVESAEEQVKELNQ----NLNS 598
Query: 717 KDAEITSLLARIKELEGELSVEKAKAT 743
+ E L +I E+ + +++A++T
Sbjct: 599 SEEEKKILSQQISEM--SIKIKRAEST 623
Score = 42.7 bits (99), Expect = 8e-04
Identities = 49/232 (21%), Positives = 96/232 (41%), Gaps = 28/232 (12%)
Query: 530 LSRGINETKEETLACLLRAGCIFAHAFDKFNSAVTEAERLKAESAQHQEAATAWEKRFDK 589
LS +N +EE + + D+ A ++ + L E A+ ++ T E
Sbjct: 504 LSASLNAAEEEKKSL----SSMILEITDELKQAQSKVQELVTELAESKDTLTQKENELSS 559
Query: 590 LVAQAGKDKVHADKLIGAAGNRIGELEDHLKLMKEEADDLDASLQACKKEKDQVEKDLAA 649
V +VH ++ +++ ELE ++ +E+ +L+ +L + ++EK + + ++
Sbjct: 560 FV------EVHEAHKRDSS-SQVKELEARVESAEEQVKELNQNLNSSEEEKKILSQQISE 612
Query: 650 KGKALAAKELEFETLGAELELAKKALAEQKKK----------SAESLALVKADLEAMRAT 699
+ E + L +E E K + AE+ + L+ LEA +
Sbjct: 613 MSIKIKRAESTIQELSSESERLKGSHAEKDNELFSLRDIHETHQRELSTQLRGLEAQLES 672
Query: 700 SE-------EIKKVNEAHGATLVTKDAEITSLLARIKELEGELSVEKAKATE 744
SE E K E T+ TK +E + L R + + EL+ + +K E
Sbjct: 673 SEHRVLELSESLKAAEEESRTMSTKISETSDELERTQIMVQELTADSSKLKE 724
Score = 40.0 bits (92), Expect = 0.005
Identities = 41/203 (20%), Positives = 84/203 (41%), Gaps = 27/203 (13%)
Query: 558 KFNSAVTEAERLKAESAQHQEAATAWEKRFDKLVAQAGKDKVHA-DKLIGAAGNRIGELE 616
+ +S + E ++ + E A+ KR D D+V+ + + + ++ ELE
Sbjct: 841 ELDSMSVQKEEVEKQMVCKSEEASVKIKRLD--------DEVNGLRQQVASLDSQRAELE 892
Query: 617 DHLKLMKEEADDLDASLQACKKE-------KDQVEKDLAAKGKALAAKELEFETLGAELE 669
L+ EE + + + K+E + + +++ + + +ELE ETLG +
Sbjct: 893 IQLEKKSEEISEYLSQITNLKEEIINKVKVHESILEEINGLSEKIKGRELELETLGKQRS 952
Query: 670 LAKKALAEQKKKSAESLALVKADLEAMRATSEEIKKVNEAHGATLVTKDAEITSLLARIK 729
+ L +K+++ + + + A +E I + E+ SL +
Sbjct: 953 ELDEELRTKKEENVQMHDKINVASSEIMALTELINNLKN-----------ELDSLQVQKS 1001
Query: 730 ELEGELSVEKAKATEAREQAADI 752
E E EL EK + +E Q D+
Sbjct: 1002 ETEAELEREKQEKSELSNQITDV 1024
Score = 38.9 bits (89), Expect = 0.012
Identities = 85/391 (21%), Positives = 145/391 (36%), Gaps = 103/391 (26%)
Query: 499 SSSHFNMLPNAIEPSEFLLT----GLNRDGIEKEVLSRGINETKEETLACLLRAGCIFAH 554
SSS L IE SE L+ LN EK++LS+ I E E
Sbjct: 64 SSSQVKELEAHIESSEKLVADFTQSLNNAEEEKKLLSQKIAELSNE-------------- 109
Query: 555 AFDKFNSAVTEAERLKAESAQHQEAATAWEKRFDKLVAQAGKDKVHADKLIGAAGNRIGE 614
A + L +ES Q +E+ + E+ +L + ++H + R E
Sbjct: 110 ----IQEAQNTMQELMSESGQLKESHSVKER---ELFSLRDIHEIHQRD----SSTRASE 158
Query: 615 LEDHLKLMKEEADDLDASLQACKKE------------------KDQVEKDLAAKGK---- 652
LE L+ K++ DL ASL+A ++E ++ +++ +A GK
Sbjct: 159 LEAQLESSKQQVSDLSASLKAAEEENKAISSKNVETMNKLEQTQNTIQELMAELGKLKDS 218
Query: 653 ----------------------ALAAKELEFET-----LGAELELAKKALAEQKKKSAES 685
++ KELE + L AEL E+KK ++
Sbjct: 219 HREKESELSSLVEVHETHQRDSSIHVKELEEQVESSKKLVAELNQTLNNAEEEKKVLSQK 278
Query: 686 LALVKADLEAMRATSEEIKKVNEAHGATLVTKDAEITSL-----------LARIKELEGE 734
+A + +++ + T +E+ + + KD ++ SL R+ ELE +
Sbjct: 279 IAELSNEIKEAQNTIQELVSESGQLKESHSVKDRDLFSLRDIHETHQRESSTRVSELEAQ 338
Query: 735 LSVEKAKATEAREQAADI-----AYDSRERGFYLAKDQAQHLFPNLDFSAMGVM------ 783
L + + ++ D A S+ +QAQ+ L +G +
Sbjct: 339 LESSEQRISDLTVDLKDAEEENKAISSKNLEIMDKLEQAQNTIKEL-MDELGELKDRHKE 397
Query: 784 KEITAEGLVGPGDPPLID--QNLWTATEEEE 812
KE LV D + D Q+L A EE++
Sbjct: 398 KESELSSLVKSADQQVADMKQSLDNAEEEKK 428
Score = 35.0 bits (79), Expect = 0.17
Identities = 42/195 (21%), Positives = 81/195 (41%), Gaps = 3/195 (1%)
Query: 558 KFNSAVTEAERLKAESAQHQEAATAWEKRFDKLVAQAGKDKVHADKLIGAAGNRIGELED 617
+ +S V E + +S+ H + + KLVA+ + +A++ +I EL +
Sbjct: 225 ELSSLVEVHETHQRDSSIHVKELEEQVESSKKLVAELNQTLNNAEEEKKVLSQKIAELSN 284
Query: 618 HLKLMKEEADDLDASLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKALAE 677
+K + +L + K+ ++DL + + E T +ELE ++ +
Sbjct: 285 EIKEAQNTIQELVSESGQLKESHSVKDRDLFSLRDIHETHQRESSTRVSELEAQLESSEQ 344
Query: 678 QKKKSAESLALVKADLEAMRATSEEIKKVNEAHGATLVTKDAEITSLLARIKELEGELSV 737
+ L + + +A+ + + EI E T+ E+ L R KE E ELS
Sbjct: 345 RISDLTVDLKDAEEENKAISSKNLEIMDKLEQAQNTIKELMDELGELKDRHKEKESELS- 403
Query: 738 EKAKATEAREQAADI 752
+ A +Q AD+
Sbjct: 404 --SLVKSADQQVADM 416
Score = 32.3 bits (72), Expect = 1.1
Identities = 32/191 (16%), Positives = 86/191 (44%), Gaps = 8/191 (4%)
Query: 567 ERLKAESAQHQEAATAWEKRFDKLVAQAGKDKVH---ADKLIGAAGNRIGELEDHLKLMK 623
E+ K + ++H + + +Q + + H ++KL+ + E+ KL+
Sbjct: 41 EKYKEKESEHSSLVELHKTHERESSSQVKELEAHIESSEKLVADFTQSLNNAEEEKKLLS 100
Query: 624 EEADDLDASLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKALAEQKKKSA 683
++ +L +Q + ++ + ++ + KE E +L E+ ++ + +++
Sbjct: 101 QKIAELSNEIQEAQNTMQELMSESGQLKESHSVKERELFSLRDIHEIHQR---DSSTRAS 157
Query: 684 ESLALVKADLEAMRATSEEIKKVNEAHGATLVTKDAEITSLLARIKELEGELSVEKAKAT 743
E A +++ + + S +K E + A + +K+ E + L + + EL E K
Sbjct: 158 ELEAQLESSKQQVSDLSASLKAAEEENKA-ISSKNVETMNKLEQTQNTIQELMAELGKLK 216
Query: 744 EA-REQAADIA 753
++ RE+ ++++
Sbjct: 217 DSHREKESELS 227
Score = 32.3 bits (72), Expect = 1.1
Identities = 34/165 (20%), Positives = 69/165 (41%)
Query: 572 ESAQHQEAATAWEKRFDKLVAQAGKDKVHADKLIGAAGNRIGELEDHLKLMKEEADDLDA 631
ES+ + A + +KLVA + +A++ +I EL + ++ + +L +
Sbjct: 63 ESSSQVKELEAHIESSEKLVADFTQSLNNAEEEKKLLSQKIAELSNEIQEAQNTMQELMS 122
Query: 632 SLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKALAEQKKKSAESLALVKA 691
K+ E++L + + + T +ELE ++ +Q + SL +
Sbjct: 123 ESGQLKESHSVKERELFSLRDIHEIHQRDSSTRASELEAQLESSKQQVSDLSASLKAAEE 182
Query: 692 DLEAMRATSEEIKKVNEAHGATLVTKDAEITSLLARIKELEGELS 736
+ +A+ + + E E T+ AE+ L +E E ELS
Sbjct: 183 ENKAISSKNVETMNKLEQTQNTIQELMAELGKLKDSHREKESELS 227
>At3g10170 hypothetical protein
Length = 634
Score = 45.8 bits (107), Expect = 1e-04
Identities = 45/193 (23%), Positives = 82/193 (42%), Gaps = 19/193 (9%)
Query: 577 QEAATAWEKRFDKLVAQAGKDKVHADKLIGAAGNRIGELEDHLKLMKEEADDLDASLQAC 636
+E W + +K + +A ++K+ K I ++E K M EE +L++ C
Sbjct: 406 EEEKAIWSSK-EKALTEAVEEKIRLYKNI--------QIESLSKEMSEEKKELESCRLEC 456
Query: 637 KKEKDQV---EKDLAAKGKALAAKELEFETLGAELELAKKALAEQKKKSAESLALVKADL 693
D++ E++ ++ K LE + LG EL A S +S ++K+D+
Sbjct: 457 VTLADRLRCSEENAKQDKESSLEKSLEIDRLGDELRSADAV-------SKQSQEVLKSDI 509
Query: 694 EAMRATSEEIKKVNEAHGATLVTKDAEITSLLARIKELEGELSVEKAKATEAREQAADIA 753
+ +++ + K+++ + +E LLARI+EL EL+ A E
Sbjct: 510 DILKSEVQHACKMSDTFQREMDYVTSERQGLLARIEELSKELASSNRWQDAAAENKEKAK 569
Query: 754 YDSRERGFYLAKD 766
R RG D
Sbjct: 570 LKMRLRGMQARLD 582
Score = 36.2 bits (82), Expect = 0.076
Identities = 64/277 (23%), Positives = 109/277 (39%), Gaps = 40/277 (14%)
Query: 500 SSHFNMLPNAIEPSEFLLTGLNRDGIEKEVLSRGINETKEET--LACLLRAGCIFAHAFD 557
+ +FN L N E L + + E +S +NE ++ + L+ +F
Sbjct: 64 NENFNSLVNVATEIEVLESEFQKYKASVETISSVMNEGLQDFAFFSPLIHDFTLFVRQSS 123
Query: 558 K-----FNSAVTEAERLKA-------ESAQHQEAATAWEKRFDKLVAQAGKDKVHADKLI 605
+ NS T LK E QE + + ++L +A K + L
Sbjct: 124 EQHDSLINSYQTVQSSLKKKVLDVENEKLLLQEQCAGLQSQIEELNQEAQKHETSLKMLS 183
Query: 606 GAAGNRIGELEDHLKLMKEEADDLDASLQACKKEKDQVEKDLAAKGKALAAKE------- 658
+ +L H++ ++++ L +S A KEK+ + KD L E
Sbjct: 184 EHHESERSDLLSHIECLEKDIGSLSSSSLA--KEKENLRKDFEKTKTKLKDTESKLKNSM 241
Query: 659 -----LEFETLGAELEL----AKKALAEQKKKSAESLALVKAD-LEAMRATSEEIKKVNE 708
LE E AE EL ++KAL E+ ES A + D L R+ ++ +++ E
Sbjct: 242 QDKTKLEAEKASAERELKRLHSQKALLERDISKQESFAGKRRDSLLVERSANQSLQE--E 299
Query: 709 AHGATLVTKDAEITSLLARIKELEGELSVEKAKATEA 745
++ + E T I LE EL+ E+ + EA
Sbjct: 300 FKQLEVLAFEMETT-----IASLEEELAAERGEKEEA 331
>At3g09000 unknown protein
Length = 541
Score = 45.4 bits (106), Expect = 1e-04
Identities = 50/198 (25%), Positives = 73/198 (36%), Gaps = 21/198 (10%)
Query: 320 RRRLVKGSSASSSNPGAQPT---AAAVAKGKKAASTAAIASSKSSTDGTSQLPTAEASAA 376
RR GSS S+S P A PT + +T A S S+ + L A A+ +
Sbjct: 145 RRPSSSGSSRSTSRP-ATPTRRSTTPTTSTSRPVTTRASNSRSSTPTSRATLTAARATTS 203
Query: 377 AATELVTVTTLDSAAVGTAT------GAAAKFTTIGATTTTADQPSTAGISAAAATTCSG 430
A T T+ SA T T +A+ + T +PST + + +
Sbjct: 204 TAAPRTTTTSSGSARSATPTRSNPRPSSASSKKPVSRPATPTRRPSTP--TGPSIVSSKA 261
Query: 431 AAAGDKERETEKENPKSPPRQVTP--------PSPPPTNDERSLSPPPRQEERPSPGAAT 482
+ G T K+P R +P P PP SL PP + +
Sbjct: 262 PSRGTSPSPTVNSLSKAPSRGTSPSPTLNSSRPWKPPEMPGFSLEAPPNLRTTLADRPVS 321
Query: 483 TSEAAQ-IEQAPGNRAGS 499
S + APG+R+GS
Sbjct: 322 ASRGRPGVASAPGSRSGS 339
Score = 44.3 bits (103), Expect = 3e-04
Identities = 42/171 (24%), Positives = 73/171 (42%), Gaps = 6/171 (3%)
Query: 330 SSSNPGAQPTAAAVAKGKKAASTAAIASSKSSTDGTSQLPTAEASAAAATELVTVTTLDS 389
S +N Q ++++VA ++ +S+ SS+S++ + PT ++ + VTT S
Sbjct: 128 SGNNNKPQTSSSSVAGLRRPSSSG---SSRSTSRPAT--PTRRSTTPTTSTSRPVTTRAS 182
Query: 390 AAVGTATGAAAKFTTIGATTTTADQPSTAGISAAAATTCSGAAAGDKERETEKENPKSPP 449
+ + + A T ATT+TA P T S+ +A + + + + + P S P
Sbjct: 183 NSRSSTPTSRATLTAARATTSTA-APRTTTTSSGSARSATPTRSNPRPSSASSKKPVSRP 241
Query: 450 RQVTPPSPPPTNDERSLSPPPRQEERPSPGAATTSEAAQIEQAPGNRAGSS 500
T PT S P + PSP + S+A +P SS
Sbjct: 242 ATPTRRPSTPTGPSIVSSKAPSRGTSPSPTVNSLSKAPSRGTSPSPTLNSS 292
>At1g73860 kinesin-related protein
Length = 1050
Score = 45.4 bits (106), Expect = 1e-04
Identities = 49/190 (25%), Positives = 82/190 (42%), Gaps = 28/190 (14%)
Query: 573 SAQHQEAATA---WEKRFDKLVAQAGKDKVHADKLIGAAGNRIGELEDHLKLMKEE---- 625
SA QE T +E+++ ++ +Q K K K E ED KL+KE
Sbjct: 268 SALKQELETTKRKYEQQYSQIESQTKKSKWEEQKK--------NEEEDMDKLLKENDQFN 319
Query: 626 ------ADDLDASLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKALAEQK 679
+L+ + +A +++ Q+E L ++ E E G + AK AL E+
Sbjct: 320 LQISALRQELETTRKAYEQQCSQMESQTMVATTGLESRLKELEQEGKVVNTAKNALEERV 379
Query: 680 KKSAESLALVKADLEAMRATSEEIKKVNEAHGATLVTKDAEITSLLARIKELEGELSVEK 739
K+ + + K A A E+IK++ + T TSL +I+ELE L + K
Sbjct: 380 KELEQ---MGKEAHSAKNALEEKIKQLQQMEKETKTAN----TSLEGKIQELEQNLVMWK 432
Query: 740 AKATEAREQA 749
K E +++
Sbjct: 433 TKVREMEKKS 442
Score = 37.4 bits (85), Expect = 0.034
Identities = 65/279 (23%), Positives = 107/279 (38%), Gaps = 56/279 (20%)
Query: 512 PSEFLLTGLNRDGIEKEVLSRGINETKEETLACLLRAGCIFAHAFDKFNSAV-------- 563
P++ LL+ LN GI E + R E + LL G F ++++ A
Sbjct: 134 PTQSLLSVLN--GILDESIERKNGEIPQIPSFVLLLLGKCFDKSYNEQRVACLLRKVVQE 191
Query: 564 ------TEAERLKAESAQHQEAATAWEKRFDKLVAQA-GKDKVHADKLIGAAGNRIGELE 616
T+AE L+ ++ + ++ R + L A A G H + + E E
Sbjct: 192 IERRISTQAEHLRTQNNIFKTREEKYQSRINVLEALASGTGVEHETEKSMWEEKKKHEEE 251
Query: 617 DHLKLMKE------EADDLDASLQACKKEKDQVEKDLAAKGKALAAKE------------ 658
D +KLMK+ E L L+ K++ +Q + ++ K +E
Sbjct: 252 DMVKLMKQNDQHNLEISALKQELETTKRKYEQQYSQIESQTKKSKWEEQKKNEEEDMDKL 311
Query: 659 --------LEFETLGAELELAKKALAEQKKKSAESLALVKADLEA-MRATSEEIKKVNEA 709
L+ L ELE +KA +Q + + LE+ ++ +E K VN A
Sbjct: 312 LKENDQFNLQISALRQELETTRKAYEQQCSQMESQTMVATTGLESRLKELEQEGKVVNTA 371
Query: 710 HGATLVTKDAEITSLLARIKELEGELSVEKAKATEAREQ 748
A L R+KELE ++ E A A E+
Sbjct: 372 KNA-----------LEERVKELE-QMGKEAHSAKNALEE 398
Score = 30.0 bits (66), Expect = 5.5
Identities = 30/105 (28%), Positives = 49/105 (46%), Gaps = 5/105 (4%)
Query: 309 ENQAEGAQAPKRRRLVKGSSASSSNPGAQPTAAAVAKGKKAASTAAIASSKSSTDGTSQL 368
E+ A A+A RL + +S ++S G Q + + + + A ++KSST T L
Sbjct: 943 ESLASSAEAEYDERLSEITSDAASM-GTQGSIDVTKRPPRISDRAKSVTAKSSTSVTRPL 1001
Query: 369 PTAEASAAAATELVT-VTTLDSAAVGTATGAAAKFTTIGATTTTA 412
A T V VT L S++ G A+ + K G+T++ A
Sbjct: 1002 DKLRKVATRTTSTVAKVTGLTSSSKGLASSSIKK---TGSTSSLA 1043
>At1g67230 unknown protein
Length = 1132
Score = 45.4 bits (106), Expect = 1e-04
Identities = 52/186 (27%), Positives = 84/186 (44%), Gaps = 27/186 (14%)
Query: 588 DKLVAQAGKDKVHADKLIGAAGNRIGELED-------HLKLMKEEADDLDASLQACKKEK 640
DK++ Q GK+ A K I AA + +LED L L ++E D L S++ +E
Sbjct: 264 DKIIKQKGKELEEAQKKIDAANLAVKKLEDDVSSRIKDLALREQETDVLKKSIETKAREL 323
Query: 641 DQVEKDLAAKGKALAAKEL------EFETLGAELELAKKALAEQKKKSA-ESLALVKADL 693
+++ L A+ K +A ++L + ++ E EL EQK+KS +SL A++
Sbjct: 324 QALQEKLEAREK-MAVQQLVDEHQAKLDSTQREFELE----MEQKRKSIDDSLKSKVAEV 378
Query: 694 EAMRA----TSEEIKKVNEAHGATLVTKDAEITSLLARIKELEGE----LSVEKAKATEA 745
E A E++ K +A L + R+K + G S EKA TE
Sbjct: 379 EKREAEWKHMEEKVAKREQALDRKLEKHKEKENDFDLRLKGISGREKALKSEEKALETEK 438
Query: 746 REQAAD 751
++ D
Sbjct: 439 KKLLED 444
Score = 37.7 bits (86), Expect = 0.026
Identities = 47/206 (22%), Positives = 88/206 (41%), Gaps = 19/206 (9%)
Query: 567 ERLKAESAQHQEAATAWEKRFDKLVAQAGKDKVHADKLIGAAGNRIGELEDHLKLMKEEA 626
E L+ ++ A EK +L A+ + K AD + A + +E+ ++ +
Sbjct: 117 EGLRKALGIEKQCALDLEKALKELRAENAEIKFTADSKLTEANALVRSVEEKSLEVEAKL 176
Query: 627 DDLDASLQACKKEKDQVE---KDLAAKGKAL--------AAKELEFETLGAELELAK--- 672
+DA L ++ VE K++ A+ +L A +E + TL + E +
Sbjct: 177 RAVDAKLAEVSRKSSDVERKAKEVEARESSLQRERFSYIAEREADEATLSKQREDLREWE 236
Query: 673 KALAEQKKKSAESLALVKADLEAMRATSEEIKKVNEAHGATLVTKDAEITSLLARIKELE 732
+ L E +++ A+S +VK + E K+ + G L +I + +K+LE
Sbjct: 237 RKLQEGEERVAKSQMIVK----QREDRANESDKIIKQKGKELEEAQKKIDAANLAVKKLE 292
Query: 733 GELSVEKAKATEAREQAADIAYDSRE 758
++S + K REQ D+ S E
Sbjct: 293 DDVS-SRIKDLALREQETDVLKKSIE 317
Score = 37.4 bits (85), Expect = 0.034
Identities = 35/184 (19%), Positives = 86/184 (46%), Gaps = 13/184 (7%)
Query: 611 RIGELEDHLKLMKEEADDLDASLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAELEL 670
+I + +L+++EA+DL A ++ +KE +++++ A G L + E L + L
Sbjct: 496 QIEKCRSQQELLQKEAEDLKAQRESFEKEWEELDERKAKIGNELKNITDQKEKLERHIHL 555
Query: 671 AKKALAEQKKKSAESLALVKADLEAMRATSEEIKKVNEAHGATLVTKDAE------ITSL 724
++ L ++K+ + E++ LE +A+ E + + ++++K AE + +
Sbjct: 556 EEERLKKEKQAANENMERELETLEVAKASFAETME----YERSMLSKKAESERSQLLHDI 611
Query: 725 LARIKELEGELSV---EKAKATEAREQAADIAYDSRERGFYLAKDQAQHLFPNLDFSAMG 781
R ++LE ++ EK + +A+++ + + +D A+ ++
Sbjct: 612 EMRKRKLESDMQTILEEKERELQAKKKLFEEEREKELSNINYLRDVARREMMDMQNERQR 671
Query: 782 VMKE 785
+ KE
Sbjct: 672 IEKE 675
Score = 32.3 bits (72), Expect = 1.1
Identities = 35/179 (19%), Positives = 82/179 (45%), Gaps = 9/179 (5%)
Query: 575 QHQEAATAWEKRFDKLVAQAGKDKVHADKLIGAAGNRIGELEDHLKLMKEEADDLDASLQ 634
+HQ + ++ F+ + Q K D + + + + E K M+E+ + +L
Sbjct: 344 EHQAKLDSTQREFELEMEQKRKS---IDDSLKSKVAEVEKREAEWKHMEEKVAKREQALD 400
Query: 635 ACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKALAEQKKKSAESLALVKADLE 694
++ + E D + K ++ +E ++ LE KK L E K E + +KA +E
Sbjct: 401 RKLEKHKEKENDFDLRLKGISGREKALKSEEKALETEKKKLLEDK----EIILNLKALVE 456
Query: 695 AMRATSE-EIKKVNEAHGATLVTKDAEITSLLARIKELEGELSVEKAKATEAREQAADI 752
+ ++ ++ ++N+ VT++ E + L EL+ ++ +++ +++A D+
Sbjct: 457 KVSGENQAQLSEINKEKDELRVTEE-ERSEYLRLQTELKEQIEKCRSQQELLQKEAEDL 514
>At3g19060 hypothetical protein
Length = 1647
Score = 44.7 bits (104), Expect = 2e-04
Identities = 48/204 (23%), Positives = 91/204 (44%), Gaps = 20/204 (9%)
Query: 555 AFDKFNSAVTEAERLKAESAQHQEAATAWEKRFDKLVAQAGKDKVHADKLIGAAGNRIGE 614
A D+ ++ E LK E + + A E R+ + A K +AD E
Sbjct: 1378 AIDERDNLQDEVLNLKEEFGKMKSEAKEMEARYIEAQQIAESRKTYAD-----------E 1426
Query: 615 LEDHLKLMKEEADDLDASLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKA 674
E+ +KL++ ++L+ ++ + + + V+ + + E+E T+ ++E A+ A
Sbjct: 1427 REEEVKLLEGSVEELEYTINVLENKVNVVKDEAERQRLQREELEMELHTIRQQMESARNA 1486
Query: 675 LAEQKKKSAE---SLALVKADLEAM-RATSE---EIKKVNEAHGATLVTKDAEITSLLAR 727
E K+ E LA K +EA+ R T++ EI +++E + +A+ + + +
Sbjct: 1487 DEEMKRILDEKHMDLAQAKKHIEALERNTADQKTEITQLSEHISELNLHAEAQASEYMHK 1546
Query: 728 IKELEGELSVEKAKATEAREQAAD 751
KELE E+ K QA D
Sbjct: 1547 FKELEA--MAEQVKPEIHVSQAID 1568
>At3g17520 unknown protein
Length = 298
Score = 44.7 bits (104), Expect = 2e-04
Identities = 60/218 (27%), Positives = 93/218 (42%), Gaps = 28/218 (12%)
Query: 565 EAERLKAESA---QHQEAATAWEKRFDKLVAQAGKDKVHADKLIGAAGNRIGELEDHLKL 621
EA + KAE A ++A +A+E K+ K A + +AG ++ K
Sbjct: 81 EAAKRKAEEAVGAAKEKAGSAYETAKSKVEEGLASVKDKASQSYDSAGQVKDDVSHKSKQ 140
Query: 622 MKEE--ADDLDASLQACKKEKDQVE-KDLAAK--GKALAAKELEFETLGAELELAKKALA 676
+K+ D+ D S KEK ++ +D+ + G+ ++ K E + E AK+
Sbjct: 141 VKDSLSGDENDESWTGWAKEKIGIKNEDINSPNLGETVSEKAKEAK------EAAKRKAG 194
Query: 677 EQKKKSAESLALVKADLEAMR-ATSEEIKKVNEAHGATLVTKDAEITSLLARIKELEGEL 735
+ K+K AE++ K M A E+ +K+ E + E S +IKE
Sbjct: 195 DAKEKLAETVETAKEKASDMTSAAKEKAEKLKEE-------AERESKSAKEKIKE---SY 244
Query: 736 SVEKAKATEAREQAADIA---YDSRERGFYLAKDQAQH 770
K+KA E E A D A YDS R AKD H
Sbjct: 245 ETAKSKADETLESAKDKASQSYDSAARKSEEAKDTVSH 282
>At1g64330 unknown protein
Length = 555
Score = 44.7 bits (104), Expect = 2e-04
Identities = 55/204 (26%), Positives = 88/204 (42%), Gaps = 21/204 (10%)
Query: 564 TEAERLKAESAQHQEAATAWEKRFDKLVAQAGKDKVHADKLIGAAGNRIGELEDHLKLMK 623
T E +A ++HQE K+ + G +V +KL + N+ EL + L++
Sbjct: 143 TTDEHKEAVESEHQEIL----KKLKESDEICGNLRVETEKL--TSENK--ELNEKLEVAG 194
Query: 624 EEADDLDASLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKALAEQKKKSA 683
E DL+ L+ KKE+D +E +LA+K K + E L + + L +K++
Sbjct: 195 ETESDLNQKLEDVKKERDGLEAELASKAKDHESTLEEVNRLQGQKNETEAELEREKQEKP 254
Query: 684 ESLALV----KADLE---AMRATSEEIKKVNEAHGATLVTKDAEITSLLARIKELEGELS 736
L + KA LE A S+E K++N ++A I L K+ L
Sbjct: 255 ALLNQINDVQKALLEQEAAYNTLSQEHKQIN----GLFEEREATIKKLTDDYKQAREMLE 310
Query: 737 --VEKAKATEAREQAADIAYDSRE 758
+ K + TE R Q SRE
Sbjct: 311 EYMSKMEETERRMQETGKDVASRE 334
Score = 38.9 bits (89), Expect = 0.012
Identities = 26/141 (18%), Positives = 66/141 (46%), Gaps = 3/141 (2%)
Query: 611 RIGELEDHLKLMKEEADDLDASLQACKKE---KDQVEKDLAAKGKALAAKELEFETLGAE 667
R G E+ ++L+K++ +D + + K + D+ ++ + ++ + + K E + +
Sbjct: 112 RNGRGENEIELLKKQMEDANLEIADLKMKLATTDEHKEAVESEHQEILKKLKESDEICGN 171
Query: 668 LELAKKALAEQKKKSAESLALVKADLEAMRATSEEIKKVNEAHGATLVTKDAEITSLLAR 727
L + + L + K+ E L + + E++KK + A L +K + S L
Sbjct: 172 LRVETEKLTSENKELNEKLEVAGETESDLNQKLEDVKKERDGLEAELASKAKDHESTLEE 231
Query: 728 IKELEGELSVEKAKATEAREQ 748
+ L+G+ + +A+ +++
Sbjct: 232 VNRLQGQKNETEAELEREKQE 252
Score = 34.7 bits (78), Expect = 0.22
Identities = 53/229 (23%), Positives = 93/229 (40%), Gaps = 33/229 (14%)
Query: 522 RDGIEKEVLSRGINETKEETLACLLRAGCIFAHAFDKFNSAVTEAERLKAESA------- 574
RDG+E E+ S+ + E TL + R + N E ER K E
Sbjct: 211 RDGLEAELASKA--KDHESTLEEVNRLQ-------GQKNETEAELEREKQEKPALLNQIN 261
Query: 575 QHQEAATAWEKRFDKLVAQAGKDKVHADKLIGAAGNRIGELEDHLKLMKEEADDLDASLQ 634
Q+A E ++ L ++ + L I +L D K +E ++ + ++
Sbjct: 262 DVQKALLEQEAAYNTL----SQEHKQINGLFEEREATIKKLTDDYKQAREMLEEYMSKME 317
Query: 635 ACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKALAEQKKKSAESLALVKADLE 694
++ + KD+A++ A+ E E+L E+ E+K ESL +++E
Sbjct: 318 ETERRMQETGKDVASRESAIVDLEETVESLRNEV--------ERKGDEIESLMEKMSNIE 369
Query: 695 AMRATSEEIKKVNEAHGATLVTKDAEITSLLARIKELEGELSVEKAKAT 743
S + +V E L K+ E+ + A K LE + +E+ AT
Sbjct: 370 VKLRLSNQKLRVTE---QVLTEKEGELKRIEA--KHLEEQALLEEKIAT 413
>At2g14410 hypothetical protein
Length = 679
Score = 44.3 bits (103), Expect = 3e-04
Identities = 67/291 (23%), Positives = 112/291 (38%), Gaps = 55/291 (18%)
Query: 436 KERETEKENPKSPPRQV----TPPSPPPTNDERSLSPPPRQEERPSPGAATTSEAAQIEQ 491
K+R+ N SP R T SP P++ RS+ PPP PS G+
Sbjct: 297 KKRDRPSRNSSSPKRDKARARTDRSPRPSSPPRSMGPPPPVTVSPSSGS----------- 345
Query: 492 APGNRAGSSSHFNMLPNAIEPSEFLLTGLNRDGIEKEVL-----SRGINETKEETLACLL 546
G + + +P E RD K VL S ET+ CL
Sbjct: 346 -----GGEKAERSAIPRRDETEV-------RDAGTKAVLVPQTKSLTAMETERNVFRCLR 393
Query: 547 RAGCIFAHAFDKFNSAVTEAERLKA-ESAQHQEAATAWEKRFDKLV------AQAGKDKV 599
FD++ A+ E L A S++ + +++L+ A KDK
Sbjct: 394 TRKDTILPTFDRWRPAIRERFLLPAHHSSRANSELNDMIEYYERLLLDREKDVMAWKDKS 453
Query: 600 HA-DKLIGAAGNRIGELEDHLKLMKEEADDLDASLQACKKEKDQVEKDLAAKGKALAAKE 658
+ + + ++ + +LED L + E + LQ + D+++++L+ L+ E
Sbjct: 454 SSLESDLHSSKDARLKLEDQLDNLSTELMKSNGELQEQYQRYDKIQEELSTSRDRLSECE 513
Query: 659 ---LEFETLGAELELAKKALAEQKKKSAESLALVKADLEAMRATSEEIKKV 706
E +EL+L KA+A + D E ++TS+ K+V
Sbjct: 514 SSAYELSNQLSELQLKYKAVANYR------------DAELAKSTSKARKEV 552
>At2g30880 unknown protein
Length = 504
Score = 43.5 bits (101), Expect = 5e-04
Identities = 59/257 (22%), Positives = 104/257 (39%), Gaps = 26/257 (10%)
Query: 521 NRDGIEKEVLSRGINETKEETLACLLRAGCIFAHAFDKFNSAV-TEAERLKAESAQHQEA 579
+RD + KE+ + ++ET E +A A H D+ V E ERL +S + QEA
Sbjct: 226 SRDSMIKEIADK-LSETAEAAVAAASAA-----HTMDEQRKIVCVEFERLTTDSQRQQEA 279
Query: 580 ATAWEKRFDKLVAQAGKDKVHADK--------------LIGAAGNRIGELEDHLKLMKEE 625
K ++ K+K K +G A R+ LE + +E+
Sbjct: 280 TKLKLKELEEKTFTLSKEKDQLVKERDAALQEAHMWRSELGKARERVVILEGAVVRAEEK 339
Query: 626 ADDLDASLQACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKALAEQKKKSAE- 684
+AS +A KE Q E + + L A +T +L K + E+K +S
Sbjct: 340 VRVAEASGEAKSKEASQREATAWTEKQELLAYVNMLQTQLQRQQLETKQVCEEKTESTNG 399
Query: 685 --SLALVKADLEAMRATSEEIKKVNEAHGATLV-TKDAEITSLLARIKELE-GELSVEKA 740
SL + K + + I + G +V + ++ + + E E ++ +A
Sbjct: 400 EASLPMTKETEKNVDKACLSISRTASIPGENVVHMSEEQVVNAQPPVGENEWNDIQATEA 459
Query: 741 KATEAREQAADIAYDSR 757
+ ++ RE +A+ D R
Sbjct: 460 RVSDVREISAETERDRR 476
>At1g77580 unknown protein
Length = 779
Score = 43.5 bits (101), Expect = 5e-04
Identities = 82/361 (22%), Positives = 150/361 (40%), Gaps = 32/361 (8%)
Query: 407 ATTTTADQPSTAGISAAAATTCSGAAAGDKER-----ETEKENPKSPPRQVTPPSPPPTN 461
A++T ++ S G S++ G + ER ET N KS P VT P+
Sbjct: 280 ASSTLIEKRSLQGTSSSIELDLMGDFL-EMERLVALPETPDGNGKSGPESVTEEVVVPS- 337
Query: 462 DERSLSPP-----PRQEERPSPGAATTSEAAQIE-QAPGNRAGSSSHFNMLPNAIEPSEF 515
E SL+ R +E +E ++E + NR + H ++
Sbjct: 338 -ENSLASEIEVLTSRIKELEEKLEKLEAEKHELENEVKCNREEAVVHIENSEVLTSRTKE 396
Query: 516 LLTGLNRDGIEKEVLSRGINETKEETLACLLRAGCIFAHAFDKFNSAVTEAE-RLKAESA 574
L L + EKE L + +E+ + + + A + S E E +L+ A
Sbjct: 397 LEEKLEKLEAEKEELKSEVKCNREKAVVHVENS---LAAEIEVLTSRTKELEEQLEKLEA 453
Query: 575 QHQEAATAWEKRFDKLVAQAGKDKVHADKLIGAAGNRIGELEDHLKLMKEEADDLDASLQ 634
+ E + + ++ VAQ +++ RI +LE+ L+ ++ E D+L + ++
Sbjct: 454 EKVELESEVKCNREEAVAQVENSLATEIEVLTC---RIKQLEEKLEKLEVEKDELKSEVK 510
Query: 635 ACKKEKDQVEKDLAAKGKALAAKELEFETLGAELELAKKALAEQ----KKKSAESLALVK 690
C +E VE L + +A+A +++E E +LE+ K L K K ES ++
Sbjct: 511 -CNRE---VESTLRFELEAIACEKMELENKLEKLEVEKAELQISFDIIKDKYEESQVCLQ 566
Query: 691 ADLEAMRATSEEIKKVNEAHG---ATLVTKDAEITSLLARIKELEGELSVEKAKATEARE 747
+ E+K VNE + + +A+ + A+I+ LE ++ E+ E R
Sbjct: 567 EIETKLGEIQTEMKLVNELKAEVESQTIAMEADAKTKSAKIESLEEDMRKERFAFDELRR 626
Query: 748 Q 748
+
Sbjct: 627 K 627
>At1g43880 hypothetical protein
Length = 409
Score = 43.5 bits (101), Expect = 5e-04
Identities = 91/352 (25%), Positives = 141/352 (39%), Gaps = 49/352 (13%)
Query: 369 PTAEASAAAATELVTVTTLDSAAVGTATGAAAKFTTIGATTTTADQPSTAGISAAAATTC 428
P+ A A L L SAAV A A + +T+ + G SAAA +
Sbjct: 4 PSLRVQRANAGALARAGDL-SAAVTAAQNILAVSSASASTSHPSLPEVNTGTSAAAGQSV 62
Query: 429 SGAAA---GDKERETEKENPKSPPRQVTPPSPPPT-----NDERSLSPPPRQEERPSPGA 480
+ A G P SP R T P P +D+ SP +EE G
Sbjct: 63 NILVAQVFGPSAVVRSDGAPSSPKRARTQPEVNPAAELLVSDQGVASP---REEIVGAGR 119
Query: 481 ATTSEAAQIEQAPG--NRAGSSSHFNMLP---NAIEPSEFLLTGLNRDGIEKEVLSRGIN 535
Q PG N A ++++ +M+ +AI + L+ L + + + +
Sbjct: 120 NC--------QVPGVNNMAEAAAYADMMAKEGHAIAATNKLVA-LYEQRLSQVPSANELE 170
Query: 536 ETKE--ETLACLLRAGCIFAHAFDKFNSAVTEAERLKAESAQHQEAATAWEKRFDKLVAQ 593
E K L ++AG D+ S E ERLK E + ++ ++++ + +
Sbjct: 171 EGKTLIRELTSSVKAGQ------DREVSFQVEIERLKMELSTSKDLEKGYDEKIGFMEME 224
Query: 594 AGKDKVHADKLIGAAGNRIGELEDHLKLMKEEADDLDASLQACKKEKDQVEKDLAA---- 649
G + ADK A N+I LE + + ++ DL ++ Q KK V+ +LAA
Sbjct: 225 IGG--LQADKQ--TARNQIHRLEQRREELSKKVMDLTSTAQGAKKAVHDVKVELAAAYSK 280
Query: 650 -----KGKALAAKELE-FETLGAELELAKKALAEQKKKSAESLALVKADLEA 695
K K +A KE E AE+E AL +Q K A L + K L+A
Sbjct: 281 LLAGVKEKWVAKKEYTVLEVQAAEVE-TNLALIDQITKVAIDLTVEKPRLQA 331
>At1g66840 hypothetical protein
Length = 607
Score = 43.1 bits (100), Expect = 6e-04
Identities = 65/245 (26%), Positives = 98/245 (39%), Gaps = 37/245 (15%)
Query: 534 INETKEETLACLLRAGCIFAHAFDKFNSAVTEAERLKAESAQHQEAATAWEKRFDKLVAQ 593
+ E E T A I A F N+ T + H + TAW DK++
Sbjct: 274 LKEVTEATEAKKAELASINAELFCLVNTMDTLRKEF-----DHAKKETAW---LDKMIQ- 324
Query: 594 AGKDKVHADKL-------------IGAAGNRIGELEDHLKLMKEEADDLDASLQACKKEK 640
KD V ++L + A RI L D+L E+ L + +A KKE+
Sbjct: 325 --KDDVMLERLNTKLLIAKDQLEAVSKAEERISYLADNLTTSFEK---LKSDREAAKKEE 379
Query: 641 DQVEKDLAAKGKALAAKELEFETLGAELELAKKALAEQKKKSAESLALVKADLEAMRATS 700
++ ++ + E F+ G E EL K +K K AESLAL K LE M +
Sbjct: 380 LKLREEARIINNEIQKTETGFD--GKEKELLSKLDELEKAKHAESLALEK--LETMVEKT 435
Query: 701 EEIKKVNEAHGATLVTKDAEITSLLARIKELE--GELSVEKA----KATEAREQAADIAY 754
E +++ +T+ E L + E E VE A +A +A +A I
Sbjct: 436 METREMESRRNSTITISRFEYEYLSGKACHAEETAEKKVEAAMAWVEALKASTKAIMIKT 495
Query: 755 DSRER 759
+S +R
Sbjct: 496 ESLKR 500
Score = 32.3 bits (72), Expect = 1.1
Identities = 42/221 (19%), Positives = 90/221 (40%), Gaps = 45/221 (20%)
Query: 591 VAQAGKDKVHADKLIGAAGNRIGELEDHLKLMKEEADDLDAS--------------LQAC 636
V ++KV A+K + +R +E++LKL++ ++D + L+ C
Sbjct: 128 VVYVSREKVVAEKEVMELESR---MEENLKLLESLKLEVDVANEEHVLVEVAKIEALKEC 184
Query: 637 KKEKDQVEKDLAAKGKALAAKELEFETLGAELELAK--------------------KALA 676
K+ ++Q EK+ ++L ++ + E+E +K K +
Sbjct: 185 KEVEEQREKERKEVSESLHKRKKRIREMIREIERSKNFENELAETLLDIEMLETQLKLVK 244
Query: 677 EQKKKSAESLALVKADLEAMR------ATSEEIKKVNEAHGATLVTKDAEITSLLARIKE 730
E ++K + ++ ++ A + +E+ + EA A L + +AE+ L+ +
Sbjct: 245 EMERKVQRNESMSRSKNRAFERGKDNLSVLKEVTEATEAKKAELASINAELFCLVNTMDT 304
Query: 731 LEGELSVEKAKAT--EAREQAADIAYDSRERGFYLAKDQAQ 769
L E K + + Q D+ + +AKDQ +
Sbjct: 305 LRKEFDHAKKETAWLDKMIQKDDVMLERLNTKLLIAKDQLE 345
>At5g23890 unknown protein
Length = 946
Score = 42.7 bits (99), Expect = 8e-04
Identities = 49/169 (28%), Positives = 82/169 (47%), Gaps = 21/169 (12%)
Query: 613 GELEDHL--KLMKEEADDL-DASLQACKKEKDQVEKDLAAK-GKALAAKELEFETLGAEL 668
GE D + +L + EA+ + + ++ A +VEKD+ A K L+ + + E +
Sbjct: 630 GEASDIVSEELARIEAESMAEKAVSAHNALVAEVEKDVNASFEKELSMEREKIEAVEKMA 689
Query: 669 ELAKKALAE-QKKKSAESLALVK--ADLEAMRATSEEIKKVNEAHGATLVTKDAEIT--- 722
ELAK L + ++K+ E+LALVK A +E+ +++ E L++ AEIT
Sbjct: 690 ELAKVELEQLREKREEENLALVKERAAVESEMEVLSRLRRDAEEKLEDLMSNKAEITFEK 749
Query: 723 -----------SLLARIKELEGELSVEKAKATEAREQAADIAYDSRERG 760
RI +L+ EL VE+ + AR A + A +RE+G
Sbjct: 750 ERVFNLRKEAEEESQRISKLQYELEVERKALSMARSWAEEEAKKAREQG 798
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.128 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,627,207
Number of Sequences: 26719
Number of extensions: 869542
Number of successful extensions: 8008
Number of sequences better than 10.0: 418
Number of HSP's better than 10.0 without gapping: 116
Number of HSP's successfully gapped in prelim test: 308
Number of HSP's that attempted gapping in prelim test: 5750
Number of HSP's gapped (non-prelim): 1626
length of query: 821
length of database: 11,318,596
effective HSP length: 108
effective length of query: 713
effective length of database: 8,432,944
effective search space: 6012689072
effective search space used: 6012689072
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0039.1


