

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0037a.7
(1027 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
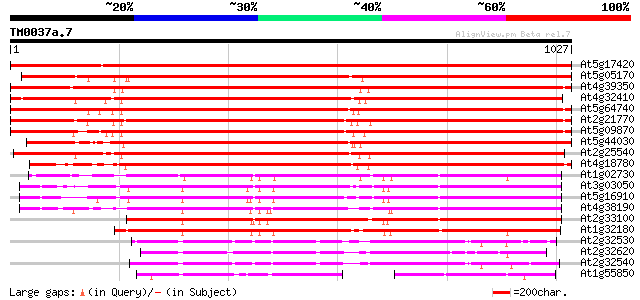
Score E
Sequences producing significant alignments: (bits) Value
At5g17420 cellulose synthase catalytic subunit (IRX3) 1890 0.0
At5g05170 cellulose synthase catalytic subunit (Ath-B) 1466 0.0
At4g39350 cellulose synthase catalytic subunit (Ath-A) 1466 0.0
At4g32410 cellulose synthase catalytic subunit (RSW1) 1463 0.0
At5g64740 cellulose synthase 1436 0.0
At2g21770 putative cellulose synthase catalytic subunit 1428 0.0
At5g09870 cellulose synthase catalytic subunit 1425 0.0
At5g44030 cellulose synthase catalytic subunit-like protein 1406 0.0
At2g25540 cellulose synthase catalytic subunit like protein 1388 0.0
At4g18780 cellulose synthase catalytic subunit (IRX1) 1306 0.0
At1g02730 cellulose synthase catalytic subunit like protein 865 0.0
At3g03050 putative cellulose synthase catalytic subunit 859 0.0
At5g16910 cellulose synthase catalytic subunit -like protein 855 0.0
At4g38190 unknown protein 843 0.0
At2g33100 putative cellulose synthase 839 0.0
At1g32180 cellulose synthase catalytic subunit, putative 811 0.0
At2g32530 cellulose synthase like protein 397 e-110
At2g32620 putative cellulose synthase 395 e-110
At2g32540 putative cellulose synthase 393 e-109
At1g55850 putative cellulose synthase catalytic subunit (At1g55850) 283 4e-76
>At5g17420 cellulose synthase catalytic subunit (IRX3)
Length = 1026
Score = 1890 bits (4896), Expect = 0.0
Identities = 886/1028 (86%), Positives = 960/1028 (93%), Gaps = 3/1028 (0%)
Query: 1 MEASAGLVAGSHNRNELVVIHGHEEHKPLKNLDGQVCEICGDGVGLTVDGDLFVACNECG 60
MEASAGLVAGSHNRNELVVIH HEE KPLKNLDGQ CEICGD +GLTV+GDLFVACNECG
Sbjct: 1 MEASAGLVAGSHNRNELVVIHNHEEPKPLKNLDGQFCEICGDQIGLTVEGDLFVACNECG 60
Query: 61 FPVCRPCYEYERREGSQNCPQCKTRYKRLKGSPRVEGDDDEEDVDDIEHEFNIDDQKNKQ 120
FP CRPCYEYERREG+QNCPQCKTRYKRL+GSPRVEGD+DEED+DDIE+EFNI+ +++K
Sbjct: 61 FPACRPCYEYERREGTQNCPQCKTRYKRLRGSPRVEGDEDEEDIDDIEYEFNIEHEQDKH 120
Query: 121 VNVVEALLHGKMSYGRGLEDDENSQFPPVISGGRSGEFPVGSHYGEQMLSSSLHKRIHPY 180
+ EA+L+GKMSYGRG EDDEN +FPPVI+GG SGEFPVG YG LHKR+HPY
Sbjct: 121 KHSAEAMLYGKMSYGRGPEDDENGRFPPVIAGGHSGEFPVGGGYGNG--EHGLHKRVHPY 178
Query: 181 PMSESGKEEGWKDRMDDWKLQQGNLGPEADEDTDASMLDEARQPLSRKVPIASSKINPYR 240
P SE+G E GW++RMDDWKLQ GNLGPE D+D + ++DEARQPLSRKVPIASSKINPYR
Sbjct: 179 PSSEAGSEGGWRERMDDWKLQHGNLGPEPDDDPEMGLIDEARQPLSRKVPIASSKINPYR 238
Query: 241 MVIVARLVILAFFLRYRILNPVHDALGLWLTSIICEIWFAFSWILDQFPKWFPIDRETYL 300
MVIVARLVILA FLRYR+LNPVHDALGLWLTS+ICEIWFA SWILDQFPKWFPI+RETYL
Sbjct: 239 MVIVARLVILAVFLRYRLLNPVHDALGLWLTSVICEIWFAVSWILDQFPKWFPIERETYL 298
Query: 301 DRLSIRYEREGEPNMLAPVDFFVSTVDPMKEPPLVTANTILSILAVDYPVDKVSCYISDD 360
DRLS+RYEREGEPNMLAPVD FVSTVDP+KEPPLVT+NT+LSILA+DYPV+K+SCY+SDD
Sbjct: 299 DRLSLRYEREGEPNMLAPVDVFVSTVDPLKEPPLVTSNTVLSILAMDYPVEKISCYVSDD 358
Query: 361 GASMCTFEALSETAEFARKWVPFCKKFSIEPRAPEMYFSEKIDYLKDKVQPTFVKERRAM 420
GASM TFE+LSETAEFARKWVPFCKKFSIEPRAPEMYF+ K+DYL+DKV PTFVKERRAM
Sbjct: 359 GASMLTFESLSETAEFARKWVPFCKKFSIEPRAPEMYFTLKVDYLQDKVHPTFVKERRAM 418
Query: 421 KREYEEFKVRINALVAKAQKVPPEGWIMQDGTPWPGNNTKDHPGMIQVFLGNSGGVDTEG 480
KREYEEFKVRINA VAKA KVP EGWIMQDGTPWPGNNTKDHPGMIQVFLG+SGG D EG
Sbjct: 419 KREYEEFKVRINAQVAKASKVPLEGWIMQDGTPWPGNNTKDHPGMIQVFLGHSGGFDVEG 478
Query: 481 NQLPRLVYVSREKRPGFQHHKKAGAMNALVRVSAVLTNAPFMLNLDCDHYINNSKAVREA 540
++LPRLVYVSREKRPGFQHHKKAGAMNALVRV+ VLTNAPFMLNLDCDHY+NNSKAVREA
Sbjct: 479 HELPRLVYVSREKRPGFQHHKKAGAMNALVRVAGVLTNAPFMLNLDCDHYVNNSKAVREA 538
Query: 541 MCFLMDPQTGKKVCYVQFPQRFDGIDTHDRYANRNTVFFDINMKGLDGIQGPVYVGTGCV 600
MCFLMDPQ GKKVCYVQFPQRFDGIDT+DRYANRNTVFFDINMKGLDGIQGPVYVGTGCV
Sbjct: 539 MCFLMDPQIGKKVCYVQFPQRFDGIDTNDRYANRNTVFFDINMKGLDGIQGPVYVGTGCV 598
Query: 601 FRRQALYGYNPPKGPKRPKMVSCDCCPCFGSRKK-LKHAKSDVNGEAASLKGMDDDKEVL 659
F+RQALYGY PPKGPKRPKM+SC CCPCFG R+K K +K+D+NG+ A+L G + DKE L
Sbjct: 599 FKRQALYGYEPPKGPKRPKMISCGCCPCFGRRRKNKKFSKNDMNGDVAALGGAEGDKEHL 658
Query: 660 MSQMNFEKKFGQSSIFVTSTLMEEGGVPPSSSPAGMLKEAIHVISCGYEDKTEWGLEVGW 719
MS+MNFEK FGQSSIFVTSTLMEEGGVPPSSSPA +LKEAIHVISCGYEDKTEWG E+GW
Sbjct: 659 MSEMNFEKTFGQSSIFVTSTLMEEGGVPPSSSPAVLLKEAIHVISCGYEDKTEWGTELGW 718
Query: 720 IYGSITEDILTGFKMHCRGWRSIYCMPRRAAFKGTAPINLSDRLNQVLRWALGSIEIFFS 779
IYGSITEDILTGFKMHCRGWRSIYCMP+R AFKG+APINLSDRLNQVLRWALGS+EIFFS
Sbjct: 719 IYGSITEDILTGFKMHCRGWRSIYCMPKRPAFKGSAPINLSDRLNQVLRWALGSVEIFFS 778
Query: 780 HHCPLWYGYKEKKLKWLERFAYANTTIYPFTSIPLVAYCVLPAVCLLTDKFIMPPISTFA 839
H PLWYGYK KLKWLERFAYANTTIYPFTSIPL+AYC+LPA+CLLTDKFIMPPISTFA
Sbjct: 779 RHSPLWYGYKGGKLKWLERFAYANTTIYPFTSIPLLAYCILPAICLLTDKFIMPPISTFA 838
Query: 840 GLYFIALFSSIIATGVIELKWSGVSIEEWWRNEQFWVIGGVSAHLFAVIQGLLKVLAGID 899
L+FI+LF SII TG++EL+WSGVSIEEWWRNEQFWVIGG+SAHLFAV+QGLLK+LAGID
Sbjct: 839 SLFFISLFMSIIVTGILELRWSGVSIEEWWRNEQFWVIGGISAHLFAVVQGLLKILAGID 898
Query: 900 TNFTVTSKATDDEDFGELYAIKWTTLLIPPTTILIINIVGVVAGVSDAINNGYQSWGPLF 959
TNFTVTSKATDD+DFGELYA KWTTLLIPPTT+LIINIVGVVAG+SDAINNGYQSWGPLF
Sbjct: 899 TNFTVTSKATDDDDFGELYAFKWTTLLIPPTTVLIINIVGVVAGISDAINNGYQSWGPLF 958
Query: 960 GKLFFSFWVIVHLYPFLKGLMGRQNRTPTIVVIWSVLLASIFSLLWVRIDPFVLKTKGPD 1019
GKLFFSFWVIVHLYPFLKGLMGRQNRTPTIVVIWSVLLASIFSLLWVRIDPFVLKTKGPD
Sbjct: 959 GKLFFSFWVIVHLYPFLKGLMGRQNRTPTIVVIWSVLLASIFSLLWVRIDPFVLKTKGPD 1018
Query: 1020 TKLCGINC 1027
T CGINC
Sbjct: 1019 TSKCGINC 1026
>At5g05170 cellulose synthase catalytic subunit (Ath-B)
Length = 1065
Score = 1466 bits (3795), Expect = 0.0
Identities = 727/1069 (68%), Positives = 840/1069 (78%), Gaps = 71/1069 (6%)
Query: 22 GHEEHKPLKNLDGQVCEICGDGVGLTVDGDLFVACNECGFPVCRPCYEYERREGSQNCPQ 81
G KP+KN+ Q C+IC D VG TVDGD FVAC+ C FPVCRPCYEYER++G+Q+CPQ
Sbjct: 5 GETAGKPMKNIVPQTCQICSDNVGKTVDGDRFVACDICSFPVCRPCYEYERKDGNQSCPQ 64
Query: 82 CKTRYKRLKGSPRVEGDDDEEDV-DDIEHEFNIDDQKNKQVNVVEALLHGKMSYGRGLED 140
CKTRYKRLKGSP + GD DE+ + D+ EFN QK K + E +L ++ G+G E
Sbjct: 65 CKTRYKRLKGSPAIPGDKDEDGLADEGTVEFNYP-QKEK---ISERMLGWHLTRGKGEEM 120
Query: 141 DE--------NSQFPPVISG-GRSGEFPVGSHYGEQMLSSSLHKRIHPYPMSESGKEE-- 189
E ++ P + S SGEF S + S+ + PY +
Sbjct: 121 GEPQYDKEVSHNHLPRLTSRQDTSGEFSAASPERLSVSSTIAGGKRLPYSSDVNQSPNRR 180
Query: 190 ----------GWKDRMDDWKL-QQGNLGPEADE--------DTDAS---------MLDEA 221
WK+R+D WK+ Q+ N GP + + D DAS + DEA
Sbjct: 181 IVDPVGLGNVAWKERVDGWKMKQEKNTGPVSTQAASERGGVDIDASTDILADEALLNDEA 240
Query: 222 RQPLSRKVPIASSKINPYRMVIVARLVILAFFLRYRILNPVHDALGLWLTSIICEIWFAF 281
RQPLSRKV I SS+INPYRMVI+ RLVIL FL YRI NPV +A LWL S+ICEIWFA
Sbjct: 241 RQPLSRKVSIPSSRINPYRMVIMLRLVILCLFLHYRITNPVPNAFALWLVSVICEIWFAL 300
Query: 282 SWILDQFPKWFPIDRETYLDRLSIRYEREGEPNMLAPVDFFVSTVDPMKEPPLVTANTIL 341
SWILDQFPKWFP++RETYLDRL++RY+REGEP+ LA VD FVSTVDP+KEPPLVTANT+L
Sbjct: 301 SWILDQFPKWFPVNRETYLDRLALRYDREGEPSQLAAVDIFVSTVDPLKEPPLVTANTVL 360
Query: 342 SILAVDYPVDKVSCYISDDGASMCTFEALSETAEFARKWVPFCKKFSIEPRAPEMYFSEK 401
SILAVDYPVDKVSCY+SDDGA+M +FE+L+ET+EFARKWVPFCKK+SIEPRAPE YF+ K
Sbjct: 361 SILAVDYPVDKVSCYVSDDGAAMLSFESLAETSEFARKWVPFCKKYSIEPRAPEWYFAAK 420
Query: 402 IDYLKDKVQPTFVKERRAMKREYEEFKVRINALVAKAQKVPPEGWIMQDGTPWPGNNTKD 461
IDYLKDKVQ +FVK+RRAMKREYEEFK+RINALV+KA K P EGW+MQDGTPWPGNNT+D
Sbjct: 421 IDYLKDKVQTSFVKDRRAMKREYEEFKIRINALVSKALKCPEEGWVMQDGTPWPGNNTRD 480
Query: 462 HPGMIQVFLGNSGGVDTEGNQLPRLVYVSREKRPGFQHHKKAGAMNALVRVSAVLTNAPF 521
HPGMIQVFLG +GG+D EGN+LPRLVYVSREKRPGFQHHKKAGAMNALVRVSAVLTN PF
Sbjct: 481 HPGMIQVFLGQNGGLDAEGNELPRLVYVSREKRPGFQHHKKAGAMNALVRVSAVLTNGPF 540
Query: 522 MLNLDCDHYINNSKAVREAMCFLMDPQTGKKVCYVQFPQRFDGIDTHDRYANRNTVFFDI 581
+LNLDCDHYINNSKA+REAMCFLMDP GK+VCYVQFPQRFDGID +DRYANRNTVFFDI
Sbjct: 541 ILNLDCDHYINNSKALREAMCFLMDPNLGKQVCYVQFPQRFDGIDKNDRYANRNTVFFDI 600
Query: 582 NMKGLDGIQGPVYVGTGCVFRRQALYGYNPP--KGPKRPKMVSCDCCPCFGSRKKLKHAK 639
N++GLDGIQGPVYVGTGCVF R ALYGY PP K+P ++S C GSRKK AK
Sbjct: 601 NLRGLDGIQGPVYVGTGCVFNRTALYGYEPPIKVKHKKPSLLS---KLCGGSRKKNSKAK 657
Query: 640 SDVN--------------------GEAASLKGMDDDKEVLMSQMNFEKKFGQSSIFVTST 679
+ + E G DD+K +LMSQM+ EK+FGQS++FV ST
Sbjct: 658 KESDKKKSGRHTDSTVPVFNLDDIEEGVEGAGFDDEKALLMSQMSLEKRFGQSAVFVAST 717
Query: 680 LMEEGGVPPSSSPAGMLKEAIHVISCGYEDKTEWGLEVGWIYGSITEDILTGFKMHCRGW 739
LME GGVPPS++P +LKEAIHVISCGYEDK++WG+E+GWIYGS+TEDILTGFKMH RGW
Sbjct: 718 LMENGGVPPSATPENLLKEAIHVISCGYEDKSDWGMEIGWIYGSVTEDILTGFKMHARGW 777
Query: 740 RSIYCMPRRAAFKGTAPINLSDRLNQVLRWALGSIEIFFSHHCPLWYGYKEKKLKWLERF 799
RSIYCMP+ AFKG+APINLSDRLNQVLRWALGS+EI FS HCP+WYGY +LK+LERF
Sbjct: 778 RSIYCMPKLPAFKGSAPINLSDRLNQVLRWALGSVEILFSRHCPIWYGY-NGRLKFLERF 836
Query: 800 AYANTTIYPFTSIPLVAYCVLPAVCLLTDKFIMPPISTFAGLYFIALFSSIIATGVIELK 859
AY NTTIYP TSIPL+ YC LPAVCL T++FI+P IS A ++F++LF SI ATG++E++
Sbjct: 837 AYVNTTIYPITSIPLLMYCTLPAVCLFTNQFIIPQISNIASIWFLSLFLSIFATGILEMR 896
Query: 860 WSGVSIEEWWRNEQFWVIGGVSAHLFAVIQGLLKVLAGIDTNFTVTSKATD-DEDFGELY 918
WSGV I+EWWRNEQFWVIGGVSAHLFAV QG+LKVLAGIDTNFTVTSKA+D D DF ELY
Sbjct: 897 WSGVGIDEWWRNEQFWVIGGVSAHLFAVFQGILKVLAGIDTNFTVTSKASDEDGDFAELY 956
Query: 919 AIKWTTLLIPPTTILIINIVGVVAGVSDAINNGYQSWGPLFGKLFFSFWVIVHLYPFLKG 978
KWTTLLIPPTT+LI+N+VGVVAGVS AIN+GYQSWGPLFGKLFF+FWVIVHLYPFLKG
Sbjct: 957 LFKWTTLLIPPTTLLIVNLVGVVAGVSYAINSGYQSWGPLFGKLFFAFWVIVHLYPFLKG 1016
Query: 979 LMGRQNRTPTIVVIWSVLLASIFSLLWVRIDPFVLKTKGPDTKLCGINC 1027
LMGRQNRTPTIVV+WSVLLASIFSLLWVRIDPF + GPD CGINC
Sbjct: 1017 LMGRQNRTPTIVVVWSVLLASIFSLLWVRIDPFTSRVTGPDILECGINC 1065
>At4g39350 cellulose synthase catalytic subunit (Ath-A)
Length = 1084
Score = 1466 bits (3794), Expect = 0.0
Identities = 705/1087 (64%), Positives = 848/1087 (77%), Gaps = 65/1087 (5%)
Query: 1 MEASAGLVAGSHNRNELVVIHGHEEHK--PLKNLDGQVCEICGDGVGLTVDGDLFVACNE 58
M L+AGSHNRNE V+I+ E + ++ L GQ C+ICGD + LTV +LFVACNE
Sbjct: 1 MNTGGRLIAGSHNRNEFVLINADESARIRSVQELSGQTCQICGDEIELTVSSELFVACNE 60
Query: 59 CGFPVCRPCYEYERREGSQNCPQCKTRYKRLKGSPRVEGDDDEE-DVDDIEHEFNIDDQK 117
C FPVCRPCYEYERREG+Q CPQCKTRYKR+KGSPRV+GDD+EE D+DD+E+EF D
Sbjct: 61 CAFPVCRPCYEYERREGNQACPQCKTRYKRIKGSPRVDGDDEEEEDIDDLEYEF---DHG 117
Query: 118 NKQVNVVEALLHGKMSYGRGLEDDE--NSQFPPVISGGRSGEFPVGSHYGEQMLSSSLHK 175
+ EA L +++ GRG D SQ P + + H S+
Sbjct: 118 MDPEHAAEAALSSRLNTGRGGLDSAPPGSQIPLLTYCDEDADMYSDRHALIVPPSTGYGN 177
Query: 176 RIHPYPMSESGKEE-------------------GWKDRMDDWKLQQGNL----------- 205
R++P P ++S WKDRM+ WK +QG
Sbjct: 178 RVYPAPFTDSSAPPQARSMVPQKDIAEYGYGSVAWKDRMEVWKRRQGEKLQVIKHEGGNN 237
Query: 206 --GPEADE---DTDASMLDEARQPLSRKVPIASSKINPYRMVIVARLVILAFFLRYRILN 260
G D+ D D M+DE RQPLSRK+PI SS+INPYRM+I+ RL IL F YRIL+
Sbjct: 238 GRGSNDDDELDDPDMPMMDEGRQPLSRKLPIRSSRINPYRMLILCRLAILGLFFHYRILH 297
Query: 261 PVHDALGLWLTSIICEIWFAFSWILDQFPKWFPIDRETYLDRLSIRYEREGEPNMLAPVD 320
PV+DA GLWLTS+ICEIWFA SWILDQFPKW+PI+RETYLDRLS+RYE+EG+P+ LAPVD
Sbjct: 298 PVNDAYGLWLTSVICEIWFAVSWILDQFPKWYPIERETYLDRLSLRYEKEGKPSGLAPVD 357
Query: 321 FFVSTVDPMKEPPLVTANTILSILAVDYPVDKVSCYISDDGASMCTFEALSETAEFARKW 380
FVSTVDP+KEPPL+TANT+LSILAVDYPVDKV+CY+SDDGA+M TFEALS+TAEFARKW
Sbjct: 358 VFVSTVDPLKEPPLITANTVLSILAVDYPVDKVACYVSDDGAAMLTFEALSDTAEFARKW 417
Query: 381 VPFCKKFSIEPRAPEMYFSEKIDYLKDKVQPTFVKERRAMKREYEEFKVRINALVAKAQK 440
VPFCKKF+IEPRAPE YFS+K+DYLK+KV P FV+ERRAMKR+YEEFKV+INALVA AQK
Sbjct: 418 VPFCKKFNIEPRAPEWYFSQKMDYLKNKVHPAFVRERRAMKRDYEEFKVKINALVATAQK 477
Query: 441 VPPEGWIMQDGTPWPGNNTKDHPGMIQVFLGNSGGVDTEGNQLPRLVYVSREKRPGFQHH 500
VP EGW MQDGTPWPGNN +DHPGMIQVFLG+SG DT+GN+LPRLVYVSREKRPGF HH
Sbjct: 478 VPEEGWTMQDGTPWPGNNVRDHPGMIQVFLGHSGVRDTDGNELPRLVYVSREKRPGFDHH 537
Query: 501 KKAGAMNALVRVSAVLTNAPFMLNLDCDHYINNSKAVREAMCFLMDPQTGKKVCYVQFPQ 560
KKAGAMN+L+RVSAVL+NAP++LN+DCDHYINNSKA+RE+MCF+MDPQ+GKKVCYVQFPQ
Sbjct: 538 KKAGAMNSLIRVSAVLSNAPYLLNVDCDHYINNSKAIRESMCFMMDPQSGKKVCYVQFPQ 597
Query: 561 RFDGIDTHDRYANRNTVFFDINMKGLDGIQGPVYVGTGCVFRRQALYGYNPPKGPKRP-K 619
RFDGID HDRY+NRN VFFDINMKGLDGIQGP+YVGTGCVFRRQALYG++ PK K P K
Sbjct: 598 RFDGIDRHDRYSNRNVVFFDINMKGLDGIQGPIYVGTGCVFRRQALYGFDAPKKKKPPGK 657
Query: 620 MVSC---DCCPCFGSRKKLKHAKSD-------VNGEAASLKGMDD---------DKEVLM 660
+C CC C G RKK K D + + +L+ +D+ +K
Sbjct: 658 TCNCWPKWCCLCCGLRKKSKTKAKDKKTNTKETSKQIHALENVDEGVIVPVSNVEKRSEA 717
Query: 661 SQMNFEKKFGQSSIFVTSTLMEEGGVPPSSSPAGMLKEAIHVISCGYEDKTEWGLEVGWI 720
+Q+ EKKFGQS +FV S +++ GGVP ++SPA +L+EAI VISCGYEDKTEWG E+GWI
Sbjct: 718 TQLKLEKKFGQSPVFVASAVLQNGGVPRNASPACLLREAIQVISCGYEDKTEWGKEIGWI 777
Query: 721 YGSITEDILTGFKMHCRGWRSIYCMPRRAAFKGTAPINLSDRLNQVLRWALGSIEIFFSH 780
YGS+TEDILTGFKMHC GWRS+YCMP+RAAFKG+APINLSDRL+QVLRWALGS+EIF S
Sbjct: 778 YGSVTEDILTGFKMHCHGWRSVYCMPKRAAFKGSAPINLSDRLHQVLRWALGSVEIFLSR 837
Query: 781 HCPLWYGYKEKKLKWLERFAYANTTIYPFTSIPLVAYCVLPAVCLLTDKFIMPPISTFAG 840
HCP+WYGY LKWLERF+Y N+ +YP+TS+PL+ YC LPAVCLLT KFI+P IS +AG
Sbjct: 838 HCPIWYGY-GGGLKWLERFSYINSVVYPWTSLPLIVYCSLPAVCLLTGKFIVPEISNYAG 896
Query: 841 LYFIALFSSIIATGVIELKWSGVSIEEWWRNEQFWVIGGVSAHLFAVIQGLLKVLAGIDT 900
+ F+ +F SI TG++E++W GV I++WWRNEQFWVIGG S+HLFA+ QGLLKVLAG++T
Sbjct: 897 ILFMLMFISIAVTGILEMQWGGVGIDDWWRNEQFWVIGGASSHLFALFQGLLKVLAGVNT 956
Query: 901 NFTVTSKATDDEDFGELYAIKWTTLLIPPTTILIINIVGVVAGVSDAINNGYQSWGPLFG 960
NFTVTSKA DD F ELY KWTTLLIPPTT+LIINI+GV+ GVSDAI+NGY SWGPLFG
Sbjct: 957 NFTVTSKAADDGAFSELYIFKWTTLLIPPTTLLIINIIGVIVGVSDAISNGYDSWGPLFG 1016
Query: 961 KLFFSFWVIVHLYPFLKGLMGRQNRTPTIVVIWSVLLASIFSLLWVRIDPFVLKTKGPDT 1020
+LFF+ WVIVHLYPFLKG++G+Q++ PTI+V+WS+LLASI +LLWVR++PFV K GP
Sbjct: 1017 RLFFALWVIVHLYPFLKGMLGKQDKMPTIIVVWSILLASILTLLWVRVNPFVAK-GGPVL 1075
Query: 1021 KLCGINC 1027
++CG+NC
Sbjct: 1076 EICGLNC 1082
>At4g32410 cellulose synthase catalytic subunit (RSW1)
Length = 1081
Score = 1463 bits (3788), Expect = 0.0
Identities = 710/1074 (66%), Positives = 839/1074 (78%), Gaps = 71/1074 (6%)
Query: 1 MEASAGLVAGSHNRNELVVIHGHEEH---KPLKNLDGQVCEICGDGVGLTVDGDLFVACN 57
MEASAGLVAGS+ RNELV I HE KPLKN++GQ+C+ICGD VGL GD+FVACN
Sbjct: 1 MEASAGLVAGSYRRNELVRIR-HESDGGTKPLKNMNGQICQICGDDVGLAETGDVFVACN 59
Query: 58 ECGFPVCRPCYEYERREGSQNCPQCKTRYKRLKGSPRVEGDDDEEDVDDIEHEFNIDDQK 117
EC FPVCRPCYEYER++G+Q CPQCKTR++R +GSPRVEGD+DE+DVDDIE+EFN
Sbjct: 60 ECAFPVCRPCYEYERKDGTQCCPQCKTRFRRHRGSPRVEGDEDEDDVDDIENEFNYAQGA 119
Query: 118 NK----------------QVNVVEALLHGKMSYGRGLEDDENSQFPPVISGGRSGEFPVG 161
NK + + L HG G D S G S +
Sbjct: 120 NKARHQRHGEEFSSSSRHESQPIPLLTHGHTVSGEIRTPDTQSVRTTSGPLGPSDRNAIS 179
Query: 162 SHYGEQMLSSSLH-----KRIHPYPMSESGKEEGWKDRMDDWKLQQ-------------- 202
S Y + + K ++ Y + WK+R++ WKL+Q
Sbjct: 180 SPYIDPRQPVPVRIVDPSKDLNSYGLGNVD----WKERVEGWKLKQEKNMLQMTGKYHEG 235
Query: 203 --GNLGPEADEDTDASMLDEARQPLSRKVPIASSKINPYRMVIVARLVILAFFLRYRILN 260
G + + M D+ R P+SR VPI SS++ PYR+VI+ RL+IL FFL+YR +
Sbjct: 236 KGGEIEGTGSNGEELQMADDTRLPMSRVVPIPSSRLTPYRVVIILRLIILCFFLQYRTTH 295
Query: 261 PVHDALGLWLTSIICEIWFAFSWILDQFPKWFPIDRETYLDRLSIRYEREGEPNMLAPVD 320
PV +A LWLTS+ICEIWFAFSW+LDQFPKW+PI+RETYLDRL+IRY+R+GEP+ L PVD
Sbjct: 296 PVKNAYPLWLTSVICEIWFAFSWLLDQFPKWYPINRETYLDRLAIRYDRDGEPSQLVPVD 355
Query: 321 FFVSTVDPMKEPPLVTANTILSILAVDYPVDKVSCYISDDGASMCTFEALSETAEFARKW 380
FVSTVDP+KEPPLVTANT+LSIL+VDYPVDKV+CY+SDDG++M TFE+LSETAEFA+KW
Sbjct: 356 VFVSTVDPLKEPPLVTANTVLSILSVDYPVDKVACYVSDDGSAMLTFESLSETAEFAKKW 415
Query: 381 VPFCKKFSIEPRAPEMYFSEKIDYLKDKVQPTFVKERRAMKREYEEFKVRINALVAKAQK 440
VPFCKKF+IEPRAPE YF++KIDYLKDK+QP+FVKERRAMKREYEEFKVRINALVAKAQK
Sbjct: 416 VPFCKKFNIEPRAPEFYFAQKIDYLKDKIQPSFVKERRAMKREYEEFKVRINALVAKAQK 475
Query: 441 VPPEGWIMQDGTPWPGNNTKDHPGMIQVFLGNSGGVDTEGNQLPRLVYVSREKRPGFQHH 500
+P EGW MQDGTPWPGNNT+DHPGMIQVFLG+SGG+DT+GN+LPRL+YVSREKRPGFQHH
Sbjct: 476 IPEEGWTMQDGTPWPGNNTRDHPGMIQVFLGHSGGLDTDGNELPRLIYVSREKRPGFQHH 535
Query: 501 KKAGAMNALVRVSAVLTNAPFMLNLDCDHYINNSKAVREAMCFLMDPQTGKKVCYVQFPQ 560
KKAGAMNAL+RVSAVLTN ++LN+DCDHY NNSKA++EAMCF+MDP GKK CYVQFPQ
Sbjct: 536 KKAGAMNALIRVSAVLTNGAYLLNVDCDHYFNNSKAIKEAMCFMMDPAIGKKCCYVQFPQ 595
Query: 561 RFDGIDTHDRYANRNTVFFDINMKGLDGIQGPVYVGTGCVFRRQALYGYNP--PKGPKRP 618
RFDGID HDRYANRN VFFDINMKGLDGIQGPVYVGTGC F RQALYGY+P + P
Sbjct: 596 RFDGIDLHDRYANRNIVFFDINMKGLDGIQGPVYVGTGCCFNRQALYGYDPVLTEEDLEP 655
Query: 619 KMVSCDCCPCFGSRKKLKHAK------------SDVNGEAASL-------KGMDDDKEVL 659
++ CC GSRKK K +K SD N ++ +G DD++ +L
Sbjct: 656 NIIVKSCC---GSRKKGKSSKKYNYEKRRGINRSDSNAPLFNMEDIDEGFEGYDDERSIL 712
Query: 660 MSQMNFEKKFGQSSIFVTSTLMEEGGVPPSSSPAGMLKEAIHVISCGYEDKTEWGLEVGW 719
MSQ + EK+FGQS +F+ +T ME+GG+PP+++PA +LKEAIHVISCGYEDKTEWG E+GW
Sbjct: 713 MSQRSVEKRFGQSPVFIAATFMEQGGIPPTTNPATLLKEAIHVISCGYEDKTEWGKEIGW 772
Query: 720 IYGSITEDILTGFKMHCRGWRSIYCMPRRAAFKGTAPINLSDRLNQVLRWALGSIEIFFS 779
IYGS+TEDILTGFKMH RGW SIYC P R AFKG+APINLSDRLNQVLRWALGSIEI S
Sbjct: 773 IYGSVTEDILTGFKMHARGWISIYCNPPRPAFKGSAPINLSDRLNQVLRWALGSIEILLS 832
Query: 780 HHCPLWYGYKEKKLKWLERFAYANTTIYPFTSIPLVAYCVLPAVCLLTDKFIMPPISTFA 839
HCP+WYGY +L+ LER AY NT +YP TSIPL+AYC+LPA CL+TD+FI+P IS +A
Sbjct: 833 RHCPIWYGY-HGRLRLLERIAYINTIVYPITSIPLIAYCILPAFCLITDRFIIPEISNYA 891
Query: 840 GLYFIALFSSIIATGVIELKWSGVSIEEWWRNEQFWVIGGVSAHLFAVIQGLLKVLAGID 899
++FI LF SI TG++EL+WSGVSIE+WWRNEQFWVIGG SAHLFAV QGLLKVLAGID
Sbjct: 892 SIWFILLFISIAVTGILELRWSGVSIEDWWRNEQFWVIGGTSAHLFAVFQGLLKVLAGID 951
Query: 900 TNFTVTSKATD-DEDFGELYAIKWTTLLIPPTTILIINIVGVVAGVSDAINNGYQSWGPL 958
TNFTVTSKATD D DF ELY KWT LLIPPTT+L++N++G+VAGVS A+N+GYQSWGPL
Sbjct: 952 TNFTVTSKATDEDGDFAELYIFKWTALLIPPTTVLLVNLIGIVAGVSYAVNSGYQSWGPL 1011
Query: 959 FGKLFFSFWVIVHLYPFLKGLMGRQNRTPTIVVIWSVLLASIFSLLWVRIDPFV 1012
FGKLFF+ WVI HLYPFLKGL+GRQNRTPTIV++WSVLLASIFSLLWVRI+PFV
Sbjct: 1012 FGKLFFALWVIAHLYPFLKGLLGRQNRTPTIVIVWSVLLASIFSLLWVRINPFV 1065
>At5g64740 cellulose synthase
Length = 1084
Score = 1436 bits (3716), Expect = 0.0
Identities = 692/1087 (63%), Positives = 834/1087 (76%), Gaps = 64/1087 (5%)
Query: 1 MEASAGLVAGSHNRNELVVIHGHEEHK--PLKNLDGQVCEICGDGVGLTVDGDLFVACNE 58
M L+AGSHNRNE V+I+ E + ++ L GQ C+IC D + LTVDG+ FVACNE
Sbjct: 1 MNTGGRLIAGSHNRNEFVLINADENARIRSVQELSGQTCQICRDEIELTVDGEPFVACNE 60
Query: 59 CGFPVCRPCYEYERREGSQNCPQCKTRYKRLKGSPRVEGDDDEEDVDDIEHEFNIDDQKN 118
C FPVCRPCYEYERREG+Q CPQCKTR+KRLKGSPRVEGD++E+D+DD+++EF +
Sbjct: 61 CAFPVCRPCYEYERREGNQACPQCKTRFKRLKGSPRVEGDEEEDDIDDLDNEFEYGNNGI 120
Query: 119 KQVNVVEALLHGKMSYGRGLEDDEN----SQFPPVISGGRSGEFPVGSH----------Y 164
V E + + + G D ++ SQ P + G E H +
Sbjct: 121 GFDQVSEGMSISRRNSGFPQSDLDSAPPGSQIPLLTYGDEDVEISSDRHALIVPPSLGGH 180
Query: 165 GEQMLSSSLHK---RIHPYPMSESGK-------EEGWKDRMDDWKLQQ----------GN 204
G ++ SL HP PM WKDRM++WK +Q G+
Sbjct: 181 GNRVHPVSLSDPTVAAHPRPMVPQKDLAVYGYGSVAWKDRMEEWKRKQNEKLQVVRHEGD 240
Query: 205 LGPEADEDTDASMLDEARQPLSRKVPIASSKINPYRMVIVARLVILAFFLRYRILNPVHD 264
E +D D M+DE RQPLSRK+PI SSKINPYRM+IV RLVIL F YRIL+PV D
Sbjct: 241 PDFEDGDDADFPMMDEGRQPLSRKIPIKSSKINPYRMLIVLRLVILGLFFHYRILHPVKD 300
Query: 265 ALGLWLTSIICEIWFAFSWILDQFPKWFPIDRETYLDRLSIRYEREGEPNMLAPVDFFVS 324
A LWL S+ICEIWFA SW+LDQFPKW+PI+RETYLDRLS+RYE+EG+P+ L+PVD FVS
Sbjct: 301 AYALWLISVICEIWFAVSWVLDQFPKWYPIERETYLDRLSLRYEKEGKPSGLSPVDVFVS 360
Query: 325 TVDPMKEPPLVTANTILSILAVDYPVDKVSCYISDDGASMCTFEALSETAEFARKWVPFC 384
TVDP+KEPPL+TANT+LSILAVDYPVDKV+CY+SDDGA+M TFEALSETAEFARKWVPFC
Sbjct: 361 TVDPLKEPPLITANTVLSILAVDYPVDKVACYVSDDGAAMLTFEALSETAEFARKWVPFC 420
Query: 385 KKFSIEPRAPEMYFSEKIDYLKDKVQPTFVKERRAMKREYEEFKVRINALVAKAQKVPPE 444
KK+ IEPRAPE YF K+DYLK+KV P FV+ERRAMKR+YEEFKV+INALVA AQKVP +
Sbjct: 421 KKYCIEPRAPEWYFCHKMDYLKNKVHPAFVRERRAMKRDYEEFKVKINALVATAQKVPED 480
Query: 445 GWIMQDGTPWPGNNTKDHPGMIQVFLGNSGGVDTEGNQLPRLVYVSREKRPGFQHHKKAG 504
GW MQDGTPWPGN+ +DHPGMIQVFLG+ G D E N+LPRLVYVSREKRPGF HHKKAG
Sbjct: 481 GWTMQDGTPWPGNSVRDHPGMIQVFLGSDGVRDVENNELPRLVYVSREKRPGFDHHKKAG 540
Query: 505 AMNALVRVSAVLTNAPFMLNLDCDHYINNSKAVREAMCFLMDPQTGKKVCYVQFPQRFDG 564
AMN+L+RVS VL+NAP++LN+DCDHYINNSKA+REAMCF+MDPQ+GKK+CYVQFPQRFDG
Sbjct: 541 AMNSLIRVSGVLSNAPYLLNVDCDHYINNSKALREAMCFMMDPQSGKKICYVQFPQRFDG 600
Query: 565 IDTHDRYANRNTVFFDINMKGLDGIQGPVYVGTGCVFRRQALYGYNPPKGPKRPKMVSCD 624
ID HDRY+NRN VFFDINMKGLDG+QGP+YVGTGCVFRRQALYG++ PK K P+ +C+
Sbjct: 601 IDRHDRYSNRNVVFFDINMKGLDGLQGPIYVGTGCVFRRQALYGFDAPKKKKGPRK-TCN 659
Query: 625 CCP-----CFGSRKKLK-------------------HAKSDVNGEAASLKGMDDDKEVLM 660
C P CFGSRK K HA ++ E KG + ++
Sbjct: 660 CWPKWCLLCFGSRKNRKAKTVAADKKKKNREASKQIHALENIE-EGRVTKGSNVEQSTEA 718
Query: 661 SQMNFEKKFGQSSIFVTSTLMEEGGVPPSSSPAGMLKEAIHVISCGYEDKTEWGLEVGWI 720
QM EKKFGQS +FV S ME GG+ ++SPA +LKEAI VISCGYEDKTEWG E+GWI
Sbjct: 719 MQMKLEKKFGQSPVFVASARMENGGMARNASPACLLKEAIQVISCGYEDKTEWGKEIGWI 778
Query: 721 YGSITEDILTGFKMHCRGWRSIYCMPRRAAFKGTAPINLSDRLNQVLRWALGSIEIFFSH 780
YGS+TEDILTGFKMH GWRS+YC P+ AAFKG+APINLSDRL+QVLRWALGS+EIF S
Sbjct: 779 YGSVTEDILTGFKMHSHGWRSVYCTPKLAAFKGSAPINLSDRLHQVLRWALGSVEIFLSR 838
Query: 781 HCPLWYGYKEKKLKWLERFAYANTTIYPFTSIPLVAYCVLPAVCLLTDKFIMPPISTFAG 840
HCP+WYGY LKWLER +Y N+ +YP+TS+PL+ YC LPA+CLLT KFI+P IS +A
Sbjct: 839 HCPIWYGY-GGGLKWLERLSYINSVVYPWTSLPLIVYCSLPAICLLTGKFIVPEISNYAS 897
Query: 841 LYFIALFSSIIATGVIELKWSGVSIEEWWRNEQFWVIGGVSAHLFAVIQGLLKVLAGIDT 900
+ F+ALFSSI TG++E++W V I++WWRNEQFWVIGGVSAHLFA+ QGLLKVLAG+DT
Sbjct: 898 ILFMALFSSIAITGILEMQWGKVGIDDWWRNEQFWVIGGVSAHLFALFQGLLKVLAGVDT 957
Query: 901 NFTVTSKATDDEDFGELYAIKWTTLLIPPTTILIINIVGVVAGVSDAINNGYQSWGPLFG 960
NFTVTSKA DD +F +LY KWT+LLIPP T+LIIN++GV+ GVSDAI+NGY SWGPLFG
Sbjct: 958 NFTVTSKAADDGEFSDLYLFKWTSLLIPPMTLLIINVIGVIVGVSDAISNGYDSWGPLFG 1017
Query: 961 KLFFSFWVIVHLYPFLKGLMGRQNRTPTIVVIWSVLLASIFSLLWVRIDPFVLKTKGPDT 1020
+LFF+ WVI+HLYPFLKGL+G+Q+R PTI+V+WS+LLASI +LLWVR++PFV K GP
Sbjct: 1018 RLFFALWVIIHLYPFLKGLLGKQDRMPTIIVVWSILLASILTLLWVRVNPFVAK-GGPIL 1076
Query: 1021 KLCGINC 1027
++CG++C
Sbjct: 1077 EICGLDC 1083
>At2g21770 putative cellulose synthase catalytic subunit
Length = 1088
Score = 1428 bits (3697), Expect = 0.0
Identities = 687/1094 (62%), Positives = 841/1094 (76%), Gaps = 75/1094 (6%)
Query: 1 MEASAGLVAGSHNRNELVVIHGHEEHK--PLKNLDGQVCEICGDGVGLTVDGDLFVACNE 58
M L+AGSHNRNE V+I+ + + + L GQ C+IC D + LT +G+ F+ACNE
Sbjct: 1 MNTGGRLIAGSHNRNEFVLINADDTARIRSAEELSGQTCKICRDEIELTDNGEPFIACNE 60
Query: 59 CGFPVCRPCYEYERREGSQNCPQCKTRYKRLKGSPRVEGDDDEEDVDDIEHEFNIDDQKN 118
C FP CRPCYEYERREG+Q CPQC TRYKR+KGSPRVEGD++++D+DD+EHEF D ++
Sbjct: 61 CAFPTCRPCYEYERREGNQACPQCGTRYKRIKGSPRVEGDEEDDDIDDLEHEFYGMDPEH 120
Query: 119 KQVNVVEALLHG-KMSYGRGLED-------DENSQFPPVISGGRSGEFPVGSHYGEQMLS 170
V EA L+ +++ GRG ++ S+ P + + H S
Sbjct: 121 ----VTEAALYYMRLNTGRGTDEVSHLYSASPGSEVPLLTYCDEDSDMYSDRHALIVPPS 176
Query: 171 SSLHKRIHPYPMSESGKE-------------------EGWKDRMDDWKLQQ--------- 202
+ L R+H P ++S WKDRM+ WK QQ
Sbjct: 177 TGLGNRVHHVPFTDSFASIHTRPMVPQKDLTVYGYGSVAWKDRMEVWKKQQIEKLQVVKN 236
Query: 203 ---------GNLGPEADEDTDASMLDEARQPLSRKVPIASSKINPYRMVIVARLVILAFF 253
G + E D D M+DE RQPLSRK+PI SS+INPYRM+I RL IL F
Sbjct: 237 ERVNDGDGDGFIVDELD-DPGLPMMDEGRQPLSRKLPIRSSRINPYRMLIFCRLAILGLF 295
Query: 254 LRYRILNPVHDALGLWLTSIICEIWFAFSWILDQFPKWFPIDRETYLDRLSIRYEREGEP 313
YRIL+PV+DA GLWLTS+ICEIWFA SWILDQFPKW+PI+RETYLDRLS+RYE+EG+P
Sbjct: 296 FHYRILHPVNDAFGLWLTSVICEIWFAVSWILDQFPKWYPIERETYLDRLSLRYEKEGKP 355
Query: 314 NMLAPVDFFVSTVDPMKEPPLVTANTILSILAVDYPVDKVSCYISDDGASMCTFEALSET 373
+ LAPVD FVSTVDP+KEPPL+TANT+LSILAVDYPV+KV+CY+SDDGA+M TFEALS T
Sbjct: 356 SELAPVDVFVSTVDPLKEPPLITANTVLSILAVDYPVEKVACYVSDDGAAMLTFEALSYT 415
Query: 374 AEFARKWVPFCKKFSIEPRAPEMYFSEKIDYLKDKVQPTFVKERRAMKREYEEFKVRINA 433
AEFARKWVPFCKKFSIEPRAPE YFS+K+DYLK KV P FV ERRAMKR+YEEFKV+INA
Sbjct: 416 AEFARKWVPFCKKFSIEPRAPEWYFSQKMDYLKHKVDPAFVMERRAMKRDYEEFKVKINA 475
Query: 434 LVAKAQKVPPEGWIMQDGTPWPGNNTKDHPGMIQVFLGNSGGVDTEGNQLPRLVYVSREK 493
LV+ +QKVP +GW MQDGTPWPGNN +DHPGMIQVFLG+SG D +GN+LPRLVYVSREK
Sbjct: 476 LVSVSQKVPEDGWTMQDGTPWPGNNVRDHPGMIQVFLGHSGVCDMDGNELPRLVYVSREK 535
Query: 494 RPGFQHHKKAGAMNALVRVSAVLTNAPFMLNLDCDHYINNSKAVREAMCFLMDPQTGKKV 553
RPGF HHKKAGAMN+L+RVSAVL+NAP++LN+DCDHYINNSKA+REAMCF+MDPQ+GKK+
Sbjct: 536 RPGFDHHKKAGAMNSLIRVSAVLSNAPYLLNVDCDHYINNSKAIREAMCFMMDPQSGKKI 595
Query: 554 CYVQFPQRFDGIDTHDRYANRNTVFFDINMKGLDGIQGPVYVGTGCVFRRQALYGYNPPK 613
CYVQFPQRFDGID HDRY+NRN VFFDINMKGLDGIQGP+YVGTGCVFRRQALYG++ PK
Sbjct: 596 CYVQFPQRFDGIDRHDRYSNRNVVFFDINMKGLDGIQGPIYVGTGCVFRRQALYGFDAPK 655
Query: 614 GPKRPKMVSCD-----CCPCFGSRKK--------LKHAKSDVNGEAASLKGMDDDKEVL- 659
K+P +C+ CC C G RKK + + + + +L+ +++ +V
Sbjct: 656 -KKQPPGRTCNCWPKWCCLCCGMRKKKTGKVKDNQRKKPKETSKQIHALEHIEEGLQVTN 714
Query: 660 ------MSQMNFEKKFGQSSIFVTSTLMEEGGVPPSSSPAGMLKEAIHVISCGYEDKTEW 713
+Q+ EKKFGQS + V STL+ GGVP + +PA +L+E+I VISCGYE+KTEW
Sbjct: 715 AENNSETAQLKLEKKFGQSPVLVASTLLLNGGVPSNVNPASLLRESIQVISCGYEEKTEW 774
Query: 714 GLEVGWIYGSITEDILTGFKMHCRGWRSIYCMPRRAAFKGTAPINLSDRLNQVLRWALGS 773
G E+GWIYGS+TEDILTGFKMHC GWRS+YCMP+RAAFKG+APINLSDRL+QVLRWALGS
Sbjct: 775 GKEIGWIYGSVTEDILTGFKMHCHGWRSVYCMPKRAAFKGSAPINLSDRLHQVLRWALGS 834
Query: 774 IEIFFSHHCPLWYGYKEKKLKWLERFAYANTTIYPFTSIPLVAYCVLPAVCLLTDKFIMP 833
+EIF S HCP+WYGY LKWLERF+Y N+ +YP+TS+PL+ YC LPA+CLLT KFI+P
Sbjct: 835 VEIFLSRHCPIWYGY-GGGLKWLERFSYINSVVYPWTSLPLLVYCSLPAICLLTGKFIVP 893
Query: 834 PISTFAGLYFIALFSSIIATGVIELKWSGVSIEEWWRNEQFWVIGGVSAHLFAVIQGLLK 893
IS +AG+ F+ +F SI TG++E++W + I++WWRNEQFWVIGGVS+HLFA+ QGLLK
Sbjct: 894 EISNYAGILFLLMFMSIAVTGILEMQWGKIGIDDWWRNEQFWVIGGVSSHLFALFQGLLK 953
Query: 894 VLAGIDTNFTVTSKATDDEDFGELYAIKWTTLLIPPTTILIINIVGVVAGVSDAINNGYQ 953
VLAG+ TNFTVTSKA DD +F ELY KWT+LLIPPTT+LIINIVGV+ GVSDAINNGY
Sbjct: 954 VLAGVSTNFTVTSKAADDGEFSELYIFKWTSLLIPPTTLLIINIVGVIVGVSDAINNGYD 1013
Query: 954 SWGPLFGKLFFSFWVIVHLYPFLKGLMGRQNRTPTIVVIWSVLLASIFSLLWVRIDPFVL 1013
SWGPLFG+LFF+ WVIVHLYPFLKGL+G+Q+R PTI+++WS+LLASI +LLWVR++PFV
Sbjct: 1014 SWGPLFGRLFFALWVIVHLYPFLKGLLGKQDRVPTIILVWSILLASILTLLWVRVNPFVS 1073
Query: 1014 KTKGPDTKLCGINC 1027
K GP ++CG++C
Sbjct: 1074 K-DGPVLEICGLDC 1086
>At5g09870 cellulose synthase catalytic subunit
Length = 1069
Score = 1425 bits (3688), Expect = 0.0
Identities = 689/1087 (63%), Positives = 835/1087 (76%), Gaps = 79/1087 (7%)
Query: 1 MEASAGLVAGSHNRNELVVIHGHEEHK--PLKNLDGQVCEICGDGVGLTVDGDLFVACNE 58
M L+AGSHNRNE V+I+ E + ++ L GQ C+ICGD + L+VDG+ FVACNE
Sbjct: 1 MNTGGRLIAGSHNRNEFVLINADESARIRSVEELSGQTCQICGDEIELSVDGESFVACNE 60
Query: 59 CGFPVCRPCYEYERREGSQNCPQCKTRYKRLKGSPRVEGDDDEEDVDDIEHEFNID---- 114
C FPVCRPCYEYERREG+Q+CPQCKTRYKR+KGSPRVEGD++++ +DD++ EF+
Sbjct: 61 CAFPVCRPCYEYERREGNQSCPQCKTRYKRIKGSPRVEGDEEDDGIDDLDFEFDYSRSGL 120
Query: 115 -----DQKNKQVNVVEALLHGKMSYGRGLEDDENSQFPPVISGGRSGEFPVGSHYGEQML 169
++N + ++ A SQ P + G E SH ++
Sbjct: 121 ESETFSRRNSEFDLASA--------------PPGSQIPLLTYGEEDVEISSDSH--ALIV 164
Query: 170 SSS---LHK---------RIHPYPMSESGK-------EEGWKDRMDDWKLQQ-------- 202
S S +H+ HP PM WKDRM++WK +Q
Sbjct: 165 SPSPGHIHRVHQPHFPDPAAHPRPMVPQKDLAVYGYGSVAWKDRMEEWKRKQNEKYQVVK 224
Query: 203 --GNLGPEADEDTDASMLDEARQPLSRKVPIASSKINPYRMVIVARLVILAFFLRYRILN 260
G+ +D D M+DE RQPLSRKVPI SSKINPYRM+IV RLVIL F YRIL+
Sbjct: 225 HDGDSSLGDGDDADIPMMDEGRQPLSRKVPIKSSKINPYRMLIVLRLVILGLFFHYRILH 284
Query: 261 PVHDALGLWLTSIICEIWFAFSWILDQFPKWFPIDRETYLDRLSIRYEREGEPNMLAPVD 320
PV+DA LWL S+ICEIWFA SW+LDQFPKW+PI+RETYLDRLS+RYE+EG+P+ LA VD
Sbjct: 285 PVNDAYALWLISVICEIWFAVSWVLDQFPKWYPIERETYLDRLSLRYEKEGKPSELAGVD 344
Query: 321 FFVSTVDPMKEPPLVTANTILSILAVDYPVDKVSCYISDDGASMCTFEALSETAEFARKW 380
FVSTVDPMKEPPL+TANT+LSILAVDYPVD+V+CY+SDDGA+M TFEALSETAEFARKW
Sbjct: 345 VFVSTVDPMKEPPLITANTVLSILAVDYPVDRVACYVSDDGAAMLTFEALSETAEFARKW 404
Query: 381 VPFCKKFSIEPRAPEMYFSEKIDYLKDKVQPTFVKERRAMKREYEEFKVRINALVAKAQK 440
VPFCKK++IEPRAPE YF K+DYLK+KV P FV+ERRAMKR+YEEFKV+INALVA AQK
Sbjct: 405 VPFCKKYTIEPRAPEWYFCHKMDYLKNKVHPAFVRERRAMKRDYEEFKVKINALVATAQK 464
Query: 441 VPPEGWIMQDGTPWPGNNTKDHPGMIQVFLGNSGGVDTEGNQLPRLVYVSREKRPGFQHH 500
VP EGW MQDGTPWPGNN +DHPGMIQVFLGN+G D E N+LPRLVYVSREKRPGF HH
Sbjct: 465 VPEEGWTMQDGTPWPGNNVRDHPGMIQVFLGNNGVRDVENNELPRLVYVSREKRPGFDHH 524
Query: 501 KKAGAMNALVRVSAVLTNAPFMLNLDCDHYINNSKAVREAMCFLMDPQTGKKVCYVQFPQ 560
KKAGAMN+L+RVS VL+NAP++LN+DCDHYINNSKA+REAMCF+MDPQ+GKK+CYVQFPQ
Sbjct: 525 KKAGAMNSLIRVSGVLSNAPYLLNVDCDHYINNSKALREAMCFMMDPQSGKKICYVQFPQ 584
Query: 561 RFDGIDTHDRYANRNTVFFDINMKGLDGIQGPVYVGTGCVFRRQALYGYNPPKGPKRPKM 620
RFDGID DRY+NRN VFFDINMKGLDG+QGP+YVGTGCVFRRQALYG++ PK K+ K
Sbjct: 585 RFDGIDKSDRYSNRNVVFFDINMKGLDGLQGPIYVGTGCVFRRQALYGFDAPK-KKKTKR 643
Query: 621 VSCDCCP-----CFGSRKKLKHAKSD---VNGEAA-----------SLKGMDD-DKEVLM 660
++C+C P C G RK K +D N EA+ KG +D K
Sbjct: 644 MTCNCWPKWCLFCCGLRKNRKSKTTDKKKKNREASKQIHALENIEEGTKGTNDAAKSPEA 703
Query: 661 SQMNFEKKFGQSSIFVTSTLMEEGGVPPSSSPAGMLKEAIHVISCGYEDKTEWGLEVGWI 720
+Q+ EKKFGQS +FV S ME GG+ ++SPA +L+EAI VISCGYEDKTEWG E+GWI
Sbjct: 704 AQLKLEKKFGQSPVFVASAGMENGGLARNASPASLLREAIQVISCGYEDKTEWGKEIGWI 763
Query: 721 YGSITEDILTGFKMHCRGWRSIYCMPRRAAFKGTAPINLSDRLNQVLRWALGSIEIFFSH 780
YGS+TEDILTGFKMH GWRS+YC P+ AFKG+APINLSDRL+QVLRWALGS+EIF S
Sbjct: 764 YGSVTEDILTGFKMHSHGWRSVYCTPKIPAFKGSAPINLSDRLHQVLRWALGSVEIFLSR 823
Query: 781 HCPLWYGYKEKKLKWLERFAYANTTIYPFTSIPLVAYCVLPAVCLLTDKFIMPPISTFAG 840
HCP+WYGY LKWLER +Y N+ +YP+TSIPL+ YC LPA+CLLT KFI+P IS +A
Sbjct: 824 HCPIWYGY-GGGLKWLERLSYINSVVYPWTSIPLLVYCSLPAICLLTGKFIVPEISNYAS 882
Query: 841 LYFIALFSSIIATGVIELKWSGVSIEEWWRNEQFWVIGGVSAHLFAVIQGLLKVLAGIDT 900
+ F+ALF SI TG++E++W V I++WWRNEQFWVIGGVSAHLFA+ QGLLKVLAG++T
Sbjct: 883 ILFMALFGSIAVTGILEMQWGKVGIDDWWRNEQFWVIGGVSAHLFALFQGLLKVLAGVET 942
Query: 901 NFTVTSKATDDEDFGELYAIKWTTLLIPPTTILIINIVGVVAGVSDAINNGYQSWGPLFG 960
NFTVTSKA DD +F ELY KWT+LLIPPTT+LIIN++GV+ G+SDAI+NGY SWGPLFG
Sbjct: 943 NFTVTSKAADDGEFSELYIFKWTSLLIPPTTLLIINVIGVIVGISDAISNGYDSWGPLFG 1002
Query: 961 KLFFSFWVIVHLYPFLKGLMGRQNRTPTIVVIWSVLLASIFSLLWVRIDPFVLKTKGPDT 1020
+LFF+FWVI+HLYPFLKGL+G+Q+R PTI+++WS+LLASI +LLWVR++PFV K GP
Sbjct: 1003 RLFFAFWVILHLYPFLKGLLGKQDRMPTIILVWSILLASILTLLWVRVNPFVAK-GGPIL 1061
Query: 1021 KLCGINC 1027
++CG++C
Sbjct: 1062 EICGLDC 1068
>At5g44030 cellulose synthase catalytic subunit-like protein
Length = 1043
Score = 1406 bits (3640), Expect = 0.0
Identities = 693/1055 (65%), Positives = 811/1055 (76%), Gaps = 81/1055 (7%)
Query: 31 NLDGQVCEICGDGVGLTVDGDLFVACNECGFPVCRPCYEYERREGSQNCPQCKTRYKRLK 90
+ ++C++CGD V +G FVAC+ C +PVC+PCYEYER G++ CPQC T YKR K
Sbjct: 12 SFSAKICKVCGDEVKDDDNGQTFVACHVCVYPVCKPCYEYERSNGNKCCPQCNTLYKRHK 71
Query: 91 GSPRVEGDDDEEDVDDIEHEFNIDDQKNKQVNVVEALLHGKMSYGRGLEDDENSQFPPVI 150
GSP++ GD++ DD + E NI +++ + +H +YG L D + Q+ P
Sbjct: 72 GSPKIAGDEENNGPDDSDDELNIKYRQDG------SSIHQNFAYGSVLFDFDKQQWRP-- 123
Query: 151 SGGRSGEFPVGSHYGEQMLSSSLHKRIHPYPMSESGKEEGWKDRMDDWKLQQGNLG---- 206
GR+ GS G+ + + + WK+R+D WK +Q G
Sbjct: 124 -NGRAFS-STGSVLGKDFEAER-----------DGYTDAEWKERVDKWKARQEKRGLVTK 170
Query: 207 ----PEADEDTDASMLD-EARQPLSRKVPIASSKINPYRMVIVARLVILAFFLRYRILNP 261
E ED + LD EARQPL RKVPI+SSKI+PYR+VIV RLVIL FF R+RIL P
Sbjct: 171 GEQTNEDKEDDEEEYLDAEARQPLWRKVPISSSKISPYRIVIVLRLVILVFFFRFRILTP 230
Query: 262 VHDALGLWLTSIICEIWFAFSWILDQFPKWFPIDRETYLDRLSIRYEREGEPNMLAPVDF 321
DA LWL S+ICEIWFA SWILDQFPKWFPI+RETYLDRLS+R+ER+GE N LAPVD
Sbjct: 231 AKDAYPLWLISVICEIWFALSWILDQFPKWFPINRETYLDRLSMRFERDGEKNKLAPVDV 290
Query: 322 FVSTVDPMKEPPLVTANTILSILAVDYPVDKVSCYISDDGASMCTFEALSETAEFARKWV 381
FVSTVDP+KEPP++TANTILSILAVDYPV+KVSCY+SDDGASM F+ LSET+EFAR+WV
Sbjct: 291 FVSTVDPLKEPPIITANTILSILAVDYPVNKVSCYVSDDGASMLLFDTLSETSEFARRWV 350
Query: 382 PFCKKFSIEPRAPEMYFSEKIDYLKDKVQPTFVKERRAMKREYEEFKVRINALVAKAQKV 441
PFCKK+++EPRAPE YFSEKIDYLKDKVQ TFVK+RRAMKREYEEFKVRINALVAKAQK
Sbjct: 351 PFCKKYNVEPRAPEFYFSEKIDYLKDKVQTTFVKDRRAMKREYEEFKVRINALVAKAQKK 410
Query: 442 PPEGWIMQDGTPWPGNNTKDHPGMIQVFLGNSGGVDTEGNQLPRLVYVSREKRPGFQHHK 501
P EGW+MQDGTPWPGNNT+DHPGMIQV+LG G D +GN+LPRLVYVSREKRPG+ HHK
Sbjct: 411 PEEGWVMQDGTPWPGNNTRDHPGMIQVYLGKEGAFDIDGNELPRLVYVSREKRPGYAHHK 470
Query: 502 KAGAMNALVRVSAVLTNAPFMLNLDCDHYINNSKAVREAMCFLMDPQTGKKVCYVQFPQR 561
KAGAMNA+VRVSAVLTNAPFMLNLDCDHYINNSKA+RE+MCFLMDPQ GKK+CYVQFPQR
Sbjct: 471 KAGAMNAMVRVSAVLTNAPFMLNLDCDHYINNSKAIRESMCFLMDPQLGKKLCYVQFPQR 530
Query: 562 FDGIDTHDRYANRNTVFFDINMKGLDGIQGPVYVGTGCVFRRQALYGYNPPKGPKRPKMV 621
FDGID +DRYANRN VFFDINM+GLDGIQGPVYVGTGCVF R ALYGY PP KR KM
Sbjct: 531 FDGIDLNDRYANRNIVFFDINMRGLDGIQGPVYVGTGCVFNRPALYGYEPPVSEKRKKM- 589
Query: 622 SCDC-----CPCFG---------SRKKLKHAKS--------------------------- 640
+CDC C C G S KK KS
Sbjct: 590 TCDCWPSWICCCCGGGNRNHKSDSSKKKSGIKSLFSKLKKKTKKKSDDKTMSSYSRKRSS 649
Query: 641 -----DVNGEAASLKGMDD-DKEVLMSQMNFEKKFGQSSIFVTSTLMEEGGVPPSSSPAG 694
D+ L+G D+ +K LMSQ NFEK+FG S +F+ STLME GG+P +++ +
Sbjct: 650 TEAIFDLEDIEEGLEGYDELEKSSLMSQKNFEKRFGMSPVFIASTLMENGGLPEATNTSS 709
Query: 695 MLKEAIHVISCGYEDKTEWGLEVGWIYGSITEDILTGFKMHCRGWRSIYCMPRRAAFKGT 754
++KEAIHVISCGYE+KTEWG E+GWIYGS+TEDILTGF+MHCRGW+S+YCMP+R AFKG+
Sbjct: 710 LIKEAIHVISCGYEEKTEWGKEIGWIYGSVTEDILTGFRMHCRGWKSVYCMPKRPAFKGS 769
Query: 755 APINLSDRLNQVLRWALGSIEIFFSHHCPLWYGYKEKKLKWLERFAYANTTIYPFTSIPL 814
APINLSDRL+QVLRWALGS+EIFFS HCPLWY + KLK LER AY NT +YPFTSIPL
Sbjct: 770 APINLSDRLHQVLRWALGSVEIFFSRHCPLWYAW-GGKLKILERLAYINTIVYPFTSIPL 828
Query: 815 VAYCVLPAVCLLTDKFIMPPISTFAGLYFIALFSSIIATGVIELKWSGVSIEEWWRNEQF 874
+AYC +PAVCLLT KFI+P I+ FA ++F+ALF SIIAT ++EL+WSGVSI + WRNEQF
Sbjct: 829 LAYCTIPAVCLLTGKFIIPTINNFASIWFLALFLSIIATAILELRWSGVSINDLWRNEQF 888
Query: 875 WVIGGVSAHLFAVIQGLLKVLAGIDTNFTVTSKATDDE--DFGELYAIKWTTLLIPPTTI 932
WVIGGVSAHLFAV QGLLKVL G+DTNFTVTSK DE +FG+LY KWTTLLIPPTT+
Sbjct: 889 WVIGGVSAHLFAVFQGLLKVLFGVDTNFTVTSKGASDEADEFGDLYLFKWTTLLIPPTTL 948
Query: 933 LIINIVGVVAGVSDAINNGYQSWGPLFGKLFFSFWVIVHLYPFLKGLMGRQNRTPTIVVI 992
+I+N+VGVVAGVSDAINNGY SWGPLFGKLFF+FWVIVHLYPFLKGLMGRQNRTPTIVV+
Sbjct: 949 IILNMVGVVAGVSDAINNGYGSWGPLFGKLFFAFWVIVHLYPFLKGLMGRQNRTPTIVVL 1008
Query: 993 WSVLLASIFSLLWVRIDPFVLKTKGPDTKLCGINC 1027
WS+LLASIFSL+WVRIDPF+ K GP K CG++C
Sbjct: 1009 WSILLASIFSLVWVRIDPFLPKQTGPLLKQCGVDC 1043
>At2g25540 cellulose synthase catalytic subunit like protein
Length = 1065
Score = 1388 bits (3592), Expect = 0.0
Identities = 681/1066 (63%), Positives = 815/1066 (75%), Gaps = 68/1066 (6%)
Query: 7 LVAGSHNRNELVVIHGHEEH--KPLKNLDGQVCEICGDGVGLTVDGDLFVACNECGFPVC 64
+VAGS+ R E V + KPLK+L+GQ+C+ICGD VGLT G++FVACNECGFP+C
Sbjct: 1 MVAGSYRRYEFVRNRDDSDDGLKPLKDLNGQICQICGDDVGLTKTGNVFVACNECGFPLC 60
Query: 65 RPCYEYERREGSQNCPQCKTRYKRLKGSPRVEGDDDEEDVDDIEHEFNIDDQKNK----- 119
+ CYEYER++GSQ CPQCK R++R GSPRVE D+ E+DV+DIE+EF+ NK
Sbjct: 61 QSCYEYERKDGSQCCPQCKARFRRHNGSPRVEVDEKEDDVNDIENEFDYTQGNNKARLPH 120
Query: 120 ------------QVNVVEALLHGKMSYGRGLEDDENSQFPPVISGGRSGEFPVGSHYGEQ 167
+ V L HG G D N+ P I G + + +
Sbjct: 121 RAEEFSSSSRHEESLPVSLLTHGHPVSGEIPTPDRNATLSPCIDPQLPGIYQL-LLLPVR 179
Query: 168 MLSSSLHKRIHPYPMSESGKEEGWKDRMDDWKLQQ----------------GNLGPEADE 211
+L S K ++ Y + WK R+ WKL+Q G
Sbjct: 180 ILDPS--KDLNSYGLVNVD----WKKRIQGWKLKQDKNMIHMTGKYHEGKGGEFEGTGSN 233
Query: 212 DTDASMLDEARQPLSRKVPIASSKINPYRMVIVARLVILAFFLRYRILNPVHDALGLWLT 271
+ M+D+AR P+SR V S+++ PYR+VIV RL+IL FL YR +PV DA LWLT
Sbjct: 234 GDELQMVDDARLPMSRVVHFPSARMTPYRIVIVLRLIILGVFLHYRTTHPVKDAYALWLT 293
Query: 272 SIICEIWFAFSWILDQFPKWFPIDRETYLDRLSIRYEREGEPNMLAPVDFFVSTVDPMKE 331
S+ICEIWFAFSW+LDQFPKW+PI+RET+LDRL++RY+R+GEP+ LAPVD FVSTVDPMKE
Sbjct: 294 SVICEIWFAFSWLLDQFPKWYPINRETFLDRLALRYDRDGEPSQLAPVDVFVSTVDPMKE 353
Query: 332 PPLVTANTILSILAVDYPVDKVSCYISDDGASMCTFEALSETAEFARKWVPFCKKFSIEP 391
PPLVTANT+LSILAVDYPVDKV+CY+SDDG++M TFEALSETAEF++KWVPFCKKF+IEP
Sbjct: 354 PPLVTANTVLSILAVDYPVDKVACYVSDDGSAMLTFEALSETAEFSKKWVPFCKKFNIEP 413
Query: 392 RAPEMYFSEKIDYLKDKVQPTFVKERRAMKREYEEFKVRINALVAKAQKVPPEGWIMQDG 451
RAPE YFS+KIDYLKDK+QP+FVKERRAMKREYEEFKVRIN LVAKAQK+P +GW M+DG
Sbjct: 414 RAPEFYFSQKIDYLKDKIQPSFVKERRAMKREYEEFKVRINILVAKAQKIPEDGWTMEDG 473
Query: 452 TPWPGNNTKDHPGMIQVFLGNSGGVDTEGNQLPRLVYVSREKRPGFQHHKKAGAMNALVR 511
T WPGNN +DHPGMIQVFLG+SGG+DT+GN+LPRL+YVSREKRPGFQHHKKAGAMNAL+R
Sbjct: 474 TSWPGNNPRDHPGMIQVFLGHSGGLDTDGNELPRLIYVSREKRPGFQHHKKAGAMNALIR 533
Query: 512 VSAVLTNAPFMLNLDCDHYINNSKAVREAMCFLMDPQTGKKVCYVQFPQRFDGIDTHDRY 571
VSAVLTN ++LN+DCDHY NNSKA++EAMCF+MDP GKK CYVQFPQRFDGID HDRY
Sbjct: 534 VSAVLTNGAYLLNVDCDHYFNNSKAIKEAMCFMMDPAIGKKCCYVQFPQRFDGIDLHDRY 593
Query: 572 ANRNTVFFDINMKGLDGIQGPVYVGTGCVFRRQALYGYNP--PKGPKRPKMVSCDCCPCF 629
ANRNTVFFDIN+KGLDGIQGPVYVGTGC F RQALYGY+P + P ++ CF
Sbjct: 594 ANRNTVFFDINLKGLDGIQGPVYVGTGCCFNRQALYGYDPVLTEEDLEPNII---VKSCF 650
Query: 630 GSRKKLKHAK------------SDVNGEAASLKGMDDDKE-------VLMSQMNFEKKFG 670
GSRKK K K SD N +++ +D+D E +L+SQ EK+FG
Sbjct: 651 GSRKKGKSRKIPNYEDNRSIKRSDSNVPLFNMEDIDEDVEGYEDEMSLLVSQKRLEKRFG 710
Query: 671 QSSIFVTSTLMEEGGVPPSSSPAGMLKEAIHVISCGYEDKTEWGLEVGWIYGSITEDILT 730
QS +F+ +T ME+GG+P +++P +LKEAIHVISCGYE KT+WG E+GWIYGS+TEDILT
Sbjct: 711 QSPVFIAATFMEQGGLPSTTNPLTLLKEAIHVISCGYEAKTDWGKEIGWIYGSVTEDILT 770
Query: 731 GFKMHCRGWRSIYCMPRRAAFKGTAPINLSDRLNQVLRWALGSIEIFFSHHCPLWYGYKE 790
GFKMH RGW SIYC+P R AFKG+APINLSDRLNQVLRWALGSIEI S HCP+WYGY
Sbjct: 771 GFKMHARGWISIYCVPSRPAFKGSAPINLSDRLNQVLRWALGSIEILLSRHCPIWYGY-N 829
Query: 791 KKLKWLERFAYANTTIYPFTSIPLVAYCVLPAVCLLTDKFIMPPISTFAGLYFIALFSSI 850
+LK LER AY NT +YP TSIPL+AYC+LPA CL+T+ FI+P IS A L F+ LF+SI
Sbjct: 830 GRLKLLERIAYINTIVYPITSIPLLAYCMLPAFCLITNTFIIPEISNLASLCFMLLFASI 889
Query: 851 IATGVIELKWSGVSIEEWWRNEQFWVIGGVSAHLFAVIQGLLKVLAGIDTNFTVTSKATD 910
A+ ++ELKWS V++E+WWRNEQFWVIGG SAHLFAV QGLLKV AGIDTNFTVTSKA+D
Sbjct: 890 YASAILELKWSDVALEDWWRNEQFWVIGGTSAHLFAVFQGLLKVFAGIDTNFTVTSKASD 949
Query: 911 -DEDFGELYAIKWTTLLIPPTTILIINIVGVVAGVSDAINNGYQSWGPLFGKLFFSFWVI 969
D DF ELY KWT+LLIPPTTIL++N+VG+VAGVS AIN+GYQSWGPL GKL F+FWV+
Sbjct: 950 EDGDFAELYVFKWTSLLIPPTTILLVNLVGIVAGVSYAINSGYQSWGPLMGKLLFAFWVV 1009
Query: 970 VHLYPFLKGLMGRQNRTPTIVVIWSVLLASIFSLLWVRIDPFVLKT 1015
HLYPFLKGL+GRQNRTPTIV++WS LLASIFSLLWVRI+PFV T
Sbjct: 1010 AHLYPFLKGLLGRQNRTPTIVIVWSALLASIFSLLWVRINPFVSTT 1055
>At4g18780 cellulose synthase catalytic subunit (IRX1)
Length = 985
Score = 1306 bits (3381), Expect = 0.0
Identities = 645/1020 (63%), Positives = 766/1020 (74%), Gaps = 75/1020 (7%)
Query: 36 VCEICGDGVGLTVDGDLFVACNECGFPVCRPCYEYERREGSQNCPQCKTRYKRLKGSPRV 95
+C CG+ +G+ +G+ FVAC+EC FP+C+ C EYE +EG + C +C Y
Sbjct: 8 ICNTCGEEIGVKSNGEFFVACHECSFPICKACLEYEFKEGRRICLRCGNPY--------- 58
Query: 96 EGDDDEEDVDDIEHEFNIDDQKNKQVNVVEALLHGKMSYGRGLEDDENSQFPPVISGGRS 155
DE DD+E K + + S G+ S + S
Sbjct: 59 ----DENVFDDVE-------TKTSKTQSIVPTQTNNTSQDSGIHARHISTVSTIDS---- 103
Query: 156 GEFPVGSHYGEQMLSSSLHKRIHPYPMSESGKEEGWKDRMDDWKLQQGNLGPEAD----- 210
+ YG + + + E WKD+ D K ++ P+A
Sbjct: 104 ---ELNDEYGNPIWKNRV---------------ESWKDKKDK-KSKKKKKDPKATKAEQH 144
Query: 211 ----------EDTDASMLDEARQPLSRKVPIASSKINPYRMVIVARLVILAFFLRYRILN 260
EDT + A LS +PI +KI YR+VI+ RL+ILA F YRI +
Sbjct: 145 EAQIPTQQHMEDTPPNTESGATDVLSVVIPIPRTKITSYRIVIIMRLIILALFFNYRITH 204
Query: 261 PVHDALGLWLTSIICEIWFAFSWILDQFPKWFPIDRETYLDRLSIRYEREGEPNMLAPVD 320
PV A GLWLTS+ICEIWFA SW+LDQFPKW PI+RETY+DRLS R+EREGE + LA VD
Sbjct: 205 PVDSAYGLWLTSVICEIWFAVSWVLDQFPKWSPINRETYIDRLSARFEREGEQSQLAAVD 264
Query: 321 FFVSTVDPMKEPPLVTANTILSILAVDYPVDKVSCYISDDGASMCTFEALSETAEFARKW 380
FFVSTVDP+KEPPL+TANT+LSILA+DYPVDKVSCY+SDDGA+M +FE+L ETA+FARKW
Sbjct: 265 FFVSTVDPLKEPPLITANTVLSILALDYPVDKVSCYVSDDGAAMLSFESLVETADFARKW 324
Query: 381 VPFCKKFSIEPRAPEMYFSEKIDYLKDKVQPTFVKERRAMKREYEEFKVRINALVAKAQK 440
VPFCKK+SIEPRAPE YFS KIDYL+DKVQP+FVKERRAMKR+YEEFK+R+NALVAKAQK
Sbjct: 325 VPFCKKYSIEPRAPEFYFSLKIDYLRDKVQPSFVKERRAMKRDYEEFKIRMNALVAKAQK 384
Query: 441 VPPEGWIMQDGTPWPGNNTKDHPGMIQVFLGNSGGVDTEGNQLPRLVYVSREKRPGFQHH 500
P EGW MQDGT WPGNNT+DHPGMIQVFLG SG D EGN+LPRLVYVSREKRPG+QHH
Sbjct: 385 TPEEGWTMQDGTSWPGNNTRDHPGMIQVFLGYSGARDIEGNELPRLVYVSREKRPGYQHH 444
Query: 501 KKAGAMNALVRVSAVLTNAPFMLNLDCDHYINNSKAVREAMCFLMDPQTGKKVCYVQFPQ 560
KKAGA NALVRVSAVLTNAPF+LNLDCDHY+NNSKAVREAMCFLMDP G+ VC+VQFPQ
Sbjct: 445 KKAGAENALVRVSAVLTNAPFILNLDCDHYVNNSKAVREAMCFLMDPVVGQDVCFVQFPQ 504
Query: 561 RFDGIDTHDRYANRNTVFFDINMKGLDGIQGPVYVGTGCVFRRQALYGYNPPKGPK-RPK 619
RFDGID DRYANRN VFFD+NM+GLDGIQGPVYVGTG VFRRQALYGY+PP P+ P+
Sbjct: 505 RFDGIDKSDRYANRNIVFFDVNMRGLDGIQGPVYVGTGTVFRRQALYGYSPPSKPRILPQ 564
Query: 620 MVSCDCCPCFGSRKK-------LKHAK-SDVNGEAASLKGMDD----DKEVLMSQMNFEK 667
S CC C +K+ K AK +++ +L +D+ D+ +L+SQ +FEK
Sbjct: 565 SSSSSCC-CLTKKKQPQDPSEIYKDAKREELDAAIFNLGDLDNYDEYDRSMLISQTSFEK 623
Query: 668 KFGQSSIFVTSTLMEEGGVPPSSSPAGMLKEAIHVISCGYEDKTEWGLEVGWIYGSITED 727
FG S++F+ STLME GGVP S +P+ ++KEAIHVISCGYE+KTEWG E+GWIYGSITED
Sbjct: 624 TFGLSTVFIESTLMENGGVPDSVNPSTLIKEAIHVISCGYEEKTEWGKEIGWIYGSITED 683
Query: 728 ILTGFKMHCRGWRSIYCMPRRAAFKGTAPINLSDRLNQVLRWALGSIEIFFSHHCPLWYG 787
ILTGFKMHCRGWRSIYCMP R AFKG+APINLSDRL+QVLRWALGS+EIF S HCPLWYG
Sbjct: 684 ILTGFKMHCRGWRSIYCMPLRPAFKGSAPINLSDRLHQVLRWALGSVEIFLSRHCPLWYG 743
Query: 788 YKEKKLKWLERFAYANTTIYPFTSIPLVAYCVLPAVCLLTDKFIMPPISTFAGLYFIALF 847
+LK L+R AY NT +YPFTS+PLVAYC LPA+CLLT KFI+P +S A + F+ LF
Sbjct: 744 CSGGRLKLLQRLAYINTIVYPFTSLPLVAYCTLPAICLLTGKFIIPTLSNLASMLFLGLF 803
Query: 848 SSIIATGVIELKWSGVSIEEWWRNEQFWVIGGVSAHLFAVIQGLLKVLAGIDTNFTVTSK 907
SII T V+EL+WSGVSIE+ WRNEQFWVIGGVSAHLFAV QG LK+LAG+DTNFTVTSK
Sbjct: 804 ISIILTSVLELRWSGVSIEDLWRNEQFWVIGGVSAHLFAVFQGFLKMLAGLDTNFTVTSK 863
Query: 908 ATDDEDFGELYAIKWTTLLIPPTTILIINIVGVVAGVSDAINNGYQSWGPLFGKLFFSFW 967
DD +FGELY +KWTTLLIPPT++LIIN+VGVVAG SDA+N GY++WGPLFGK+FF+FW
Sbjct: 864 TADDLEFGELYIVKWTTLLIPPTSLLIINLVGVVAGFSDALNKGYEAWGPLFGKVFFAFW 923
Query: 968 VIVHLYPFLKGLMGRQNRTPTIVVIWSVLLASIFSLLWVRIDPFVLKTKGPDTKLCGINC 1027
VI+HLYPFLKGLMGRQNRTPTIV++WS+LLAS+FSL+WVRI+PFV KT DT +NC
Sbjct: 924 VILHLYPFLKGLMGRQNRTPTIVILWSILLASVFSLVWVRINPFVSKT---DTTSLSLNC 980
>At1g02730 cellulose synthase catalytic subunit like protein
Length = 1181
Score = 865 bits (2235), Expect = 0.0
Identities = 466/1072 (43%), Positives = 646/1072 (59%), Gaps = 157/1072 (14%)
Query: 34 GQVCEICGDGVGLTVDGDLFVACNECGFPVCRPCYEYERREGSQNCPQCKTRYKRLKGSP 93
GQ+C + G D + ECGF +CR CY G NCP CK Y+ + P
Sbjct: 159 GQICWLKG------CDEKVVHGRCECGFRICRDCYFDCITSGGGNCPGCKEPYRDINDDP 212
Query: 94 RVEGDDDEEDVDDIEHEFNIDDQKNKQVNVVEALLHGKMSYGRGLEDDENSQFPPVISGG 153
E +D+E++ + + + +K+++VV++ +
Sbjct: 213 ETEEEDEEDEAKPLPQMG--ESKLDKRLSVVKSFK----------------------AQN 248
Query: 154 RSGEFPVGSHYGEQMLSSSLHKRIHP---YPMSESGKEEGWKDRMDDWKLQQGNLGPEAD 210
++G+F E + + P Y + G G+
Sbjct: 249 QAGDFDHTRWLFETKGTYGYGNAVWPKDGYGIGSGGGGNGY------------------- 289
Query: 211 EDTDASMLDEARQPLSRKVPIASSKINPYRMVIVARLVILAFFLRYRILNPVHDALGLWL 270
+T + +++PL+RKV ++++ I+PYR++I RLV L FL +R+ +P +A+ LW
Sbjct: 290 -ETPPEFGERSKRPLTRKVSVSAAIISPYRLLIALRLVALGLFLTWRVRHPNREAMWLWG 348
Query: 271 TSIICEIWFAFSWILDQFPKWFPIDRETYLDRLSIRYEREGEPNMLAP--------VDFF 322
S CE+WFA SW+LDQ PK P++R T L L R+E PN+ P +D F
Sbjct: 349 MSTTCELWFALSWLLDQLPKLCPVNRLTDLGVLKERFE---SPNLRNPKGRSDLPGIDVF 405
Query: 323 VSTVDPMKEPPLVTANTILSILAVDYPVDKVSCYISDDGASMCTFEALSETAEFARKWVP 382
VST DP KEPPLVTANTILSILAVDYPV+K++CY+SDDG ++ TFEAL++TA FA WVP
Sbjct: 406 VSTADPEKEPPLVTANTILSILAVDYPVEKLACYLSDDGGALLTFEALAQTASFASTWVP 465
Query: 383 FCKKFSIEPRAPEMYFSEKIDYLKDKVQPTFVKERRAMKREYEEFKVRINALVAKAQK-- 440
FC+K +IEPR PE YF +K ++LK+KV+ FV+ERR +KREY+EFKVRIN+L ++
Sbjct: 466 FCRKHNIEPRNPEAYFGQKRNFLKNKVRLDFVRERRRVKREYDEFKVRINSLPEAIRRRS 525
Query: 441 -----------------------------VPPEGWIMQDGTPWPG--------NNTKDHP 463
VP W M DG+ WPG N+ DH
Sbjct: 526 DAYNVHEELRAKKKQMEMMMGNNPQETVIVPKATW-MSDGSHWPGTWSSGETDNSRGDHA 584
Query: 464 GMIQVFLGNSGGVDTEGNQ---------------LPRLVYVSREKRPGFQHHKKAGAMNA 508
G+IQ L G + LP LVYVSREKRPG+ H+KKAGAMNA
Sbjct: 585 GIIQAMLAPPNAEPVYGAEADAENLIDTTDVDIRLPMLVYVSREKRPGYDHNKKAGAMNA 644
Query: 509 LVRVSAVLTNAPFMLNLDCDHYINNSKAVREAMCFLMDPQTGKKVCYVQFPQRFDGIDTH 568
LVR SA+++N PF+LNLDCDHYI NS A+RE MCF++D + G ++CYVQFPQRF+GID +
Sbjct: 645 LVRTSAIMSNGPFILNLDCDHYIYNSMALREGMCFMLD-RGGDRICYVQFPQRFEGIDPN 703
Query: 569 DRYANRNTVFFDINMKGLDGIQGPVYVGTGCVFRRQALYGYNPPKGPKRPKMVSCDCCPC 628
DRYAN NTVFFD++M+ LDG+QGP+YVGTGC+FRR ALYG++PP+ + +
Sbjct: 704 DRYANHNTVFFDVSMRALDGLQGPMYVGTGCIFRRTALYGFSPPRATEHHGWLGRRKVKI 763
Query: 629 FGSRKKLKHAKSD-----VNGEAASLKGMDDDKEVLMSQMNFEKKFGQSSIFVTSTLMEE 683
R K K D +NGE + D D E L+ K+FG S+ FV S + E
Sbjct: 764 SLRRPKAMMKKDDEVSLPINGEYNEEENDDGDIESLL----LPKRFGNSNSFVASIPVAE 819
Query: 684 ---------GGVPPSSSPAGMLK------------EAIHVISCGYEDKTEWGLEVGWIYG 722
G +S PAG L EAI VISC YEDKTEWG VGWIYG
Sbjct: 820 YQGRLIQDLQGKGKNSRPAGSLAVPREPLDAATVAEAISVISCFYEDKTEWGKRVGWIYG 879
Query: 723 SITEDILTGFKMHCRGWRSIYCMPRRAAFKGTAPINLSDRLNQVLRWALGSIEIFFSHHC 782
S+TED++TG++MH RGWRSIYC+ +R AF+GTAPINL+DRL+QVLRWA GS+EIFFS +
Sbjct: 880 SVTEDVVTGYRMHNRGWRSIYCVTKRDAFRGTAPINLTDRLHQVLRWATGSVEIFFSRNN 939
Query: 783 PLWYGYKEKKLKWLERFAYANTTIYPFTSIPLVAYCVLPAVCLLTDKFIMPPISTFAGLY 842
+ + +++K+L+R AY N +YPFTS+ L+ YC+LPA+ L + +FI+ + +Y
Sbjct: 940 AI---FATRRMKFLQRVAYFNVGMYPFTSLFLIVYCILPAISLFSGQFIVQSLDITFLIY 996
Query: 843 FIALFSSIIATGVIELKWSGVSIEEWWRNEQFWVIGGVSAHLFAVIQGLLKVLAGIDTNF 902
+++ ++ ++E+KWSG+++ EWWRNEQFWVIGG SAH AV+QGLLKV+AG+D +F
Sbjct: 997 LLSITLTLCMLSLLEIKWSGITLHEWWRNEQFWVIGGTSAHPAAVLQGLLKVIAGVDISF 1056
Query: 903 TVTSKAT----DDEDFGELYAIKWTTLLIPPTTILIINIVGVVAGVSDAINNGYQSWGPL 958
T+TSK++ D++F +LY +KW+ L++PP TI+++N++ + G++ + + + W L
Sbjct: 1057 TLTSKSSAPEDGDDEFADLYVVKWSFLMVPPLTIMMVNMIAIAVGLARTLYSPFPQWSKL 1116
Query: 959 FGKLFFSFWVIVHLYPFLKGLMGRQNRTPTIVVIWSVLLASIFSLLWVRIDP 1010
G +FFSFWV+ HLYPF KGLMGR+ R PTIV +WS LL+ I SLLWV I+P
Sbjct: 1117 VGGVFFSFWVLCHLYPFAKGLMGRRGRVPTIVFVWSGLLSIIVSLLWVYINP 1168
>At3g03050 putative cellulose synthase catalytic subunit
Length = 1145
Score = 859 bits (2219), Expect = 0.0
Identities = 480/1081 (44%), Positives = 644/1081 (59%), Gaps = 150/1081 (13%)
Query: 19 VIHGHEEHKPLKNLDGQVCEICGDGVGLTVD--GDLFVACNECGFPVCRPCYEYERREGS 76
VI H + G C + G V + D G + C EC F +CR C+ + G
Sbjct: 112 VIDTETSHPQMAGAKGSSCAVPGCDVKVMSDERGQDLLPC-ECDFKICRDCFMDAVKTGG 170
Query: 77 QNCPQCKTRYKRLKGSPRVEGDDDEEDVDDIEHEFNIDDQKNKQVNVVEALLHGKMSYGR 136
CP CK Y+ + D D D NKQ
Sbjct: 171 M-CPGCKEPYR----------NTDLADFAD----------NNKQ---------------- 193
Query: 137 GLEDDENSQFPPVISGGRSGE-FPVGSHYGEQMLSSSLHKRIHPYPMSESGKEEGWKDRM 195
+ PP G + + ++ S H + E+ G+ +
Sbjct: 194 -----QRPMLPPPAGGSKMDRRLSLMKSTKSGLMRSQTGDFDHNRWLFETSGTYGFGNAF 248
Query: 196 DDWKLQQGNLGPEADEDTDA----SMLDEARQPLSRKVPIASSKINPYRMVIVARLVILA 251
W + GN G + D + ++ +PL+RK+ I ++ I+PYR++I+ R+V+LA
Sbjct: 249 --WT-KDGNFGSDKDGNGHGMGPQDLMSRPWRPLTRKLQIPAAVISPYRLLILIRIVVLA 305
Query: 252 FFLRYRILNPVHDALGLWLTSIICEIWFAFSWILDQFPKWFPIDRETYLDRLSIRYEREG 311
FL +RI + DA+ LW S++CE+WFA SW+LDQ PK PI+R T L+ L ++E
Sbjct: 306 LFLMWRIKHKNPDAIWLWGMSVVCELWFALSWLLDQLPKLCPINRATDLNVLKEKFETPT 365
Query: 312 EPNM-----LAPVDFFVSTVDPMKEPPLVTANTILSILAVDYPVDKVSCYISDDGASMCT 366
N L +D FVST DP KEPPLVT+NTILSILA DYPV+K++CY+SDDG ++ T
Sbjct: 366 PSNPTGKSDLPGLDMFVSTADPEKEPPLVTSNTILSILAADYPVEKLACYVSDDGGALLT 425
Query: 367 FEALSETAEFARKWVPFCKKFSIEPRAPEMYFSEKIDYLKDKVQPTFVKERRAMKREYEE 426
FEA++E A FA WVPFC+K +IEPR P+ YFS K D K+KV+ FVK+RR +KREY+E
Sbjct: 426 FEAMAEAASFANMWVPFCRKHNIEPRNPDSYFSLKRDPYKNKVKADFVKDRRRVKREYDE 485
Query: 427 FKVRINAL------------------------------VAKAQKVPPEGWIMQDGTPWPG 456
FKVRIN+L + + K+P W M DGT WPG
Sbjct: 486 FKVRINSLPDSIRRRSDAYHAREEIKAMKLQRQNRDEEIVEPVKIPKATW-MADGTHWPG 544
Query: 457 --------NNTKDHPGMIQVFLGNSGGVDTEGN------------QLPRLVYVSREKRPG 496
++ DH G+IQV L G +LP LVYVSREKRPG
Sbjct: 545 TWINSGPDHSRSDHAGIIQVMLKPPSDEPLHGVSEGFLDLTDVDIRLPLLVYVSREKRPG 604
Query: 497 FQHHKKAGAMNALVRVSAVLTNAPFMLNLDCDHYINNSKAVREAMCFLMDPQTGKKVCYV 556
+ H+KKAGAMNALVR SA+++N PF+LNLDCDHYI NS+A+RE MCF+MD + G ++CYV
Sbjct: 605 YDHNKKAGAMNALVRASAIMSNGPFILNLDCDHYIYNSQALREGMCFMMD-RGGDRLCYV 663
Query: 557 QFPQRFDGIDTHDRYANRNTVFFDINMKGLDGIQGPVYVGTGCVFRRQALYGYNPPKGPK 616
QFPQRF+GID DRYAN NTVFFD+NM+ LDG+ GPVYVGTGC+FRR ALYG++PP+ +
Sbjct: 664 QFPQRFEGIDPSDRYANHNTVFFDVNMRALDGLMGPVYVGTGCLFRRIALYGFDPPRAKE 723
Query: 617 R-PKMVSCDCCPCFGSRKKLKHAKSDVNGEAASLK---GMDDDKEVLMSQMNFEKKFGQS 672
P SC CF +KK KS V E SL+ DDD+E+ +S + KKFG S
Sbjct: 724 HHPGFCSC----CFSRKKK----KSRVPEENRSLRMGGDSDDDEEMNLSLV--PKKFGNS 773
Query: 673 SIFVTSTLMEE-------------GGVPPSSSP-------AGMLKEAIHVISCGYEDKTE 712
+ + S + E G PP + A + EAI VISC YEDKTE
Sbjct: 774 TFLIDSIPVAEFQGRPLADHPAVQNGRPPGALTIPRELLDASTVAEAIAVISCWYEDKTE 833
Query: 713 WGLEVGWIYGSITEDILTGFKMHCRGWRSIYCMPRRAAFKGTAPINLSDRLNQVLRWALG 772
WG +GWIYGS+TED++TG++MH RGW+S+YC+ +R AF+GTAPINL+DRL+QVLRWA G
Sbjct: 834 WGSRIGWIYGSVTEDVVTGYRMHNRGWKSVYCVTKRDAFRGTAPINLTDRLHQVLRWATG 893
Query: 773 SIEIFFSHHCPLWYGYKEKKLKWLERFAYANTTIYPFTSIPLVAYCVLPAVCLLTDKFIM 832
S+EIFFS + + ++K L+R AY N IYPFTS L+ YC LPA+ L + +FI+
Sbjct: 894 SVEIFFSRNNAF---FASPRMKILQRIAYLNVGIYPFTSFFLIVYCFLPALSLFSGQFIV 950
Query: 833 PPISTFAGLYFIALFSSIIATGVIELKWSGVSIEEWWRNEQFWVIGGVSAHLFAVIQGLL 892
++ +Y + + ++ ++E+KWSG+S+EEWWRNEQFW+IGG SAHL AVIQGLL
Sbjct: 951 QTLNVTFLVYLLIISITLCLLALLEIKWSGISLEEWWRNEQFWLIGGTSAHLAAVIQGLL 1010
Query: 893 KVLAGIDTNFTVTSKATD---DEDFGELYAIKWTTLLIPPTTILIINIVGVVAGVSDAIN 949
KV+AGI+ +FT+TSK+ D++F +LY +KWT+L+IPP TI+++N++ + G S I
Sbjct: 1011 KVVAGIEISFTLTSKSGGEDVDDEFADLYIVKWTSLMIPPITIMMVNLIAIAVGFSRTIY 1070
Query: 950 NGYQSWGPLFGKLFFSFWVIVHLYPFLKGLMGRQNRTPTIVVIWSVLLASIFSLLWVRID 1009
+ W L G +FFSFWV+ HLYPF KGLMGR+ RTPTIV +WS L+A SLLWV I+
Sbjct: 1071 SVIPQWSKLIGGVFFSFWVLAHLYPFAKGLMGRRGRTPTIVYVWSGLVAITISLLWVAIN 1130
Query: 1010 P 1010
P
Sbjct: 1131 P 1131
>At5g16910 cellulose synthase catalytic subunit -like protein
Length = 1145
Score = 855 bits (2210), Expect = 0.0
Identities = 482/1086 (44%), Positives = 638/1086 (58%), Gaps = 158/1086 (14%)
Query: 19 VIHGHEEHKPLKNLDGQVCEICGDGVGLTVD--GDLFVACNECGFPVCRPCYEYERREGS 76
VI H + G C I G + D G + C EC F +CR C+ + G
Sbjct: 110 VIETEPNHPQMAGSKGSSCAIPGCDAKVMSDERGQDLLPC-ECDFKICRDCFIDAVKTGG 168
Query: 77 QNCPQCKTRYKRLKGSPRVEGDDDEEDVDDIEHEFNIDDQKNKQVNVVEALLHGKMSYGR 136
CP CK YK + +V
Sbjct: 169 GICPGCKEPYKNTHLTDQV----------------------------------------- 187
Query: 137 GLEDDENSQFPPVISGGRSGEFP-----VGSHYGEQMLSSSLHKRIHPYPMSESGKEEGW 191
DEN Q P++ GG + V S ++ S H + E+ G+
Sbjct: 188 ----DENGQQRPMLPGGGGSKMERRLSMVKSTNKSALMRSQTGDFDHNRWLFETTGTYGY 243
Query: 192 KDRMDDWKLQQGNLGPEADEDTDAS--------MLDEARQPLSRKVPIASSKINPYRMVI 243
+ W + G+ G D D D ++ +PL+RK+ I + I+PYR++I
Sbjct: 244 GNAF--WT-KDGDFGSGKDGDGDGDGMGMEAQDLMSRPWRPLTRKLKIPAGVISPYRLLI 300
Query: 244 VARLVILAFFLRYRILNPVHDALGLWLTSIICEIWFAFSWILDQFPKWFPIDRETYLDRL 303
R+V+LA FL +R+ + DA+ LW S++CE+WFA SW+LDQ PK PI+R T L L
Sbjct: 301 FIRIVVLALFLTWRVKHQNPDAVWLWGMSVVCELWFALSWLLDQLPKLCPINRATDLQVL 360
Query: 304 SIRYEREGEPNM-----LAPVDFFVSTVDPMKEPPLVTANTILSILAVDYPVDKVSCYIS 358
++E N L D FVST DP KEPPLVTANTILSILA +YPV+K+SCY+S
Sbjct: 361 KEKFETPTASNPTGKSDLPGFDVFVSTADPEKEPPLVTANTILSILAAEYPVEKLSCYVS 420
Query: 359 DDGASMCTFEALSETAEFARKWVPFCKKFSIEPRAPEMYFSEKIDYLKDKVQPTFVKERR 418
DDG ++ TFEA++E A FA WVPFC+K +IEPR P+ YFS K D K+KV+ FVK+RR
Sbjct: 421 DDGGALLTFEAMAEAASFANIWVPFCRKHAIEPRNPDSYFSLKRDPYKNKVKSDFVKDRR 480
Query: 419 AMKREYEEFKVRINAL------------------VAKAQ------------KVPPEGWIM 448
+KRE++EFKVR+N+L K Q K+P W M
Sbjct: 481 RVKREFDEFKVRVNSLPDSIRRRSDAYHAREEIKAMKMQRQNRDDEPMEPVKIPKATW-M 539
Query: 449 QDGTPWPG--------NNTKDHPGMIQVFLGNSGGVDTEGN------------QLPRLVY 488
DGT WPG + DH G+IQV L G +LP LVY
Sbjct: 540 ADGTHWPGTWLTSASDHAKGDHAGIIQVMLKPPSDEPLHGVSEGFLDLTDVDIRLPLLVY 599
Query: 489 VSREKRPGFQHHKKAGAMNALVRVSAVLTNAPFMLNLDCDHYINNSKAVREAMCFLMDPQ 548
VSREKRPG+ H+KKAGAMNALVR SA+++N PF+LNLDCDHYI NS+A+RE MCF+MD +
Sbjct: 600 VSREKRPGYDHNKKAGAMNALVRASAIMSNGPFILNLDCDHYIYNSEALREGMCFMMD-R 658
Query: 549 TGKKVCYVQFPQRFDGIDTHDRYANRNTVFFDINMKGLDGIQGPVYVGTGCVFRRQALYG 608
G ++CYVQFPQRF+GID DRYAN NTVFFD+NM+ LDG+ GPVYVGTGC+FRR ALYG
Sbjct: 659 GGDRLCYVQFPQRFEGIDPSDRYANHNTVFFDVNMRALDGLMGPVYVGTGCLFRRIALYG 718
Query: 609 YNPPKGPKRPKMVSCDCCPCFGSRKKLKHAKSDVNGEAASLKGMD-DDKEVLMSQMNFEK 667
+NPP R K S C C R K K ++ E +L+ D DD+E+ +S + K
Sbjct: 719 FNPP----RSKDFSPSCWSCCFPRSK----KKNIPEENRALRMSDYDDEEMNLSLV--PK 768
Query: 668 KFGQSSIFVTSTLMEE-------------GGVPPSSSP-------AGMLKEAIHVISCGY 707
KFG S+ + S + E G PP + A + EAI VISC Y
Sbjct: 769 KFGNSTFLIDSIPVAEFQGRPLADHPAVKNGRPPGALTIPRELLDASTVAEAIAVISCWY 828
Query: 708 EDKTEWGLEVGWIYGSITEDILTGFKMHCRGWRSIYCMPRRAAFKGTAPINLSDRLNQVL 767
EDKTEWG +GWIYGS+TED++TG++MH RGW+S+YC+ +R AF+GTAPINL+DRL+QVL
Sbjct: 829 EDKTEWGSRIGWIYGSVTEDVVTGYRMHNRGWKSVYCVTKRDAFRGTAPINLTDRLHQVL 888
Query: 768 RWALGSIEIFFSHHCPLWYGYKEKKLKWLERFAYANTTIYPFTSIPLVAYCVLPAVCLLT 827
RWA GS+EIFFS + L K+K L+R AY N IYPFTSI L+ YC LPA+ L +
Sbjct: 889 RWATGSVEIFFSRNNAL---LASSKMKILQRIAYLNVGIYPFTSIFLIVYCFLPALSLFS 945
Query: 828 DKFIMPPISTFAGLYFIALFSSIIATGVIELKWSGVSIEEWWRNEQFWVIGGVSAHLFAV 887
+FI+ ++ +Y + + ++ ++E+KWSG+S+EEWWRNEQFW+IGG SAHL AV
Sbjct: 946 GQFIVQTLNVTFLVYLLIISITLCLLALLEIKWSGISLEEWWRNEQFWLIGGTSAHLAAV 1005
Query: 888 IQGLLKVLAGIDTNFTVTSKATD---DEDFGELYAIKWTTLLIPPTTILIINIVGVVAGV 944
+QGLLKV+AG++ +FT+TSK+ D++F +LY +KWT+L+IPP TI+++N++ + G
Sbjct: 1006 LQGLLKVVAGVEISFTLTSKSGGDDIDDEFADLYMVKWTSLMIPPITIIMVNLIAIAVGF 1065
Query: 945 SDAINNGYQSWGPLFGKLFFSFWVIVHLYPFLKGLMGRQNRTPTIVVIWSVLLASIFSLL 1004
S I + W L G +FFSFWV+ HLYPF KGLMGR+ RTPTIV +WS L+A SLL
Sbjct: 1066 SRTIYSVVPQWSKLIGGVFFSFWVLAHLYPFAKGLMGRRGRTPTIVYVWSGLVAITISLL 1125
Query: 1005 WVRIDP 1010
WV I+P
Sbjct: 1126 WVAINP 1131
>At4g38190 unknown protein
Length = 1111
Score = 843 bits (2178), Expect = 0.0
Identities = 475/1079 (44%), Positives = 636/1079 (58%), Gaps = 170/1079 (15%)
Query: 19 VIHGHEEHKPLKNLDGQVCEI--CGDGVGLTVDGDLFVACNECGFPVCRPCYEYERREGS 76
VI H + G C + C V G + C EC F +CR C+ ++E
Sbjct: 97 VIDSDVTHPQMAGAKGSSCAMPACDGNVMKDERGKDVMPC-ECRFKICRDCFMDAQKETG 155
Query: 77 QNCPQCKTRYKRLKGSPRVEGDDDEEDVDDIEHEFNID----DQK--NKQVNVVEALLHG 130
CP CK +YK GD D++ D + DQ+ N +++++ +G
Sbjct: 156 L-CPGCKEQYKI--------GDLDDDTPDYSSGALPLPAPGKDQRGNNNNMSMMKRNQNG 206
Query: 131 KMSYGRGLEDDENSQFPPVISGGRSGEFPVGSHYGEQMLSSSLHKRIHPYPMSESGKEEG 190
+ + R L + + + G + +P YG+ M +EG
Sbjct: 207 EFDHNRWLFETQGTY------GYGNAYWPQDEMYGDDM-------------------DEG 241
Query: 191 WKDRMDDWKLQQGNLGPEADEDTDASMLDEARQPLSRKVPIASSKINPYRMVIVARLVIL 250
+ M D+ +PLSR++PI ++ I+PYR++IV R V+L
Sbjct: 242 MRGGM-------------------VETADKPWRPLSRRIPIPAAIISPYRLLIVIRFVVL 282
Query: 251 AFFLRYRILNPVHDALGLWLTSIICEIWFAFSWILDQFPKWFPIDRETYLDRLSIRYERE 310
FFL +RI NP DA+ LWL SIICE+WF FSWILDQ PK PI+R T L+ L +++
Sbjct: 283 CFFLTWRIRNPNEDAIWLWLMSIICELWFGFSWILDQIPKLCPINRSTDLEVLRDKFDMP 342
Query: 311 GEPNM-----LAPVDFFVSTVDPMKEPPLVTANTILSILAVDYPVDKVSCYISDDGASMC 365
N L +D FVST DP KEPPLVTANTILSILAVDYPV+KVSCY+SDDG ++
Sbjct: 343 SPSNPTGRSDLPGIDLFVSTADPEKEPPLVTANTILSILAVDYPVEKVSCYLSDDGGALL 402
Query: 366 TFEALSETAEFARKWVPFCKKFSIEPRAPEMYFSEKIDYLKDKVQPTFVKERRAMKREYE 425
+FEA++E A FA WVPFC+K +IEPR P+ YFS KID K+K + FVK+RR +KREY+
Sbjct: 403 SFEAMAEAASFADLWVPFCRKHNIEPRNPDSYFSLKIDPTKNKSRIDFVKDRRKIKREYD 462
Query: 426 EFKVRINALVAKAQ-----------------------------KVPPEGWIMQDGTPWPG 456
EFKVRIN L + KVP W M DGT WPG
Sbjct: 463 EFKVRINGLPDSIRRRSDAFNAREEMKALKQMRESGGDPTEPVKVPKATW-MADGTHWPG 521
Query: 457 --------NNTKDHPGMIQVFL---------GNSGGV-----DTEGNQLPRLVYVSREKR 494
++ DH G++QV L GNS DT+ +LP VYVSREKR
Sbjct: 522 TWAASTREHSKGDHAGILQVMLKPPSSDPLIGNSDDKVIDFSDTD-TRLPMFVYVSREKR 580
Query: 495 PGFQHHKKAGAMNALVRVSAVLTNAPFMLNLDCDHYINNSKAVREAMCFLMDPQTGKKVC 554
PG+ H+KKAGAMNALVR SA+L+N PF+LNLDCDHYI N KAVRE MCF+MD + G+ +C
Sbjct: 581 PGYDHNKKAGAMNALVRASAILSNGPFILNLDCDHYIYNCKAVREGMCFMMD-RGGEDIC 639
Query: 555 YVQFPQRFDGIDTHDRYANRNTVFFDINMKGLDGIQGPVYVGTGCVFRRQALYGYNPPKG 614
Y+QFPQRF+GID DRYAN NTVFFD NM+ LDG+QGPVYVGTG +FRR ALYG++PP
Sbjct: 640 YIQFPQRFEGIDPSDRYANNNTVFFDGNMRALDGVQGPVYVGTGTMFRRFALYGFDPPNP 699
Query: 615 PKRPKMVSCDCCPCFGSRKKLKHAKSDVNGEAASLKGMDDDKEVLMSQMNFEKKFGQSSI 674
K + K + EA + D D +V K+FG S++
Sbjct: 700 DKLLE-------------------KKESETEALTTSDFDPDLDVTQ----LPKRFGNSTL 736
Query: 675 FVTST-LMEEGGVPPSSSPA-------GMLK------------EAIHVISCGYEDKTEWG 714
S + E G P + PA G L+ E++ VISC YEDKTEWG
Sbjct: 737 LAESIPIAEFQGRPLADHPAVKYGRPPGALRVPRDPLDATTVAESVSVISCWYEDKTEWG 796
Query: 715 LEVGWIYGSITEDILTGFKMHCRGWRSIYCMPRRAAFKGTAPINLSDRLNQVLRWALGSI 774
VGWIYGS+TED++TG++MH RGWRS+YC+ +R +F+G+APINL+DRL+QVLRWA GS+
Sbjct: 797 DRVGWIYGSVTEDVVTGYRMHNRGWRSVYCITKRDSFRGSAPINLTDRLHQVLRWATGSV 856
Query: 775 EIFFSHHCPLWYGYKEKKLKWLERFAYANTTIYPFTSIPLVAYCVLPAVCLLTDKFIMPP 834
EIFFS + + K+LK+L+R AY N IYPFTS+ L+ YC LPA L + +FI+
Sbjct: 857 EIFFSRNNAI---LASKRLKFLQRLAYLNVGIYPFTSLFLILYCFLPAFSLFSGQFIVRT 913
Query: 835 ISTFAGLYFIALFSSIIATGVIELKWSGVSIEEWWRNEQFWVIGGVSAHLFAVIQGLLKV 894
+S +Y + + +I V+E+KWSG+ +EEWWRNEQ+W+I G S+HL+AV+QG+LKV
Sbjct: 914 LSISFLVYLLMITICLIGLAVLEVKWSGIGLEEWWRNEQWWLISGTSSHLYAVVQGVLKV 973
Query: 895 LAGIDTNFTVTSKATDDED---FGELYAIKWTTLLIPPTTILIINIVGVVAGVSDAINNG 951
+AGI+ +FT+T+K+ D++ + +LY +KW++L+IPP I ++NI+ +V I
Sbjct: 974 IAGIEISFTLTTKSGGDDNEDIYADLYIVKWSSLMIPPIVIAMVNIIAIVVAFIRTIYQA 1033
Query: 952 YQSWGPLFGKLFFSFWVIVHLYPFLKGLMGRQNRTPTIVVIWSVLLASIFSLLWVRIDP 1010
W L G FFSFWV+ HLYPF KGLMGR+ +TPTIV +W+ L+A SLLW I+P
Sbjct: 1034 VPQWSKLIGGAFFSFWVLAHLYPFAKGLMGRRGKTPTIVFVWAGLIAITISLLWTAINP 1092
>At2g33100 putative cellulose synthase
Length = 1036
Score = 839 bits (2168), Expect = 0.0
Identities = 439/877 (50%), Positives = 579/877 (65%), Gaps = 91/877 (10%)
Query: 215 ASMLDEARQPLSRKVPIASSKINPYRMVIVARLVILAFFLRYRILNPVHDALGLWLTSII 274
+ LD+ +PL+RKV I + ++PYR++IV RLVI+ FFL +RI NP DA+ LW SI+
Sbjct: 158 SDFLDKPWKPLTRKVQIPAKILSPYRLLIVIRLVIVFFFLWWRITNPNEDAMWLWGLSIV 217
Query: 275 CEIWFAFSWILDQFPKWFPIDRETYLDRLSIRYEREGEPNM-----LAPVDFFVSTVDPM 329
CEIWFAFSWILD PK PI+R T L L ++E+ N L VD FVST DP
Sbjct: 218 CEIWFAFSWILDILPKLNPINRATDLAALHDKFEQPSPSNPTGRSDLPGVDVFVSTADPE 277
Query: 330 KEPPLVTANTILSILAVDYPVDKVSCYISDDGASMCTFEALSETAEFARKWVPFCKKFSI 389
KEPPLVTANT+LSILAVDYP++K+S YISDDG ++ TFEA++E FA WVPFC+K I
Sbjct: 278 KEPPLVTANTLLSILAVDYPIEKLSAYISDDGGAILTFEAMAEAVRFAEYWVPFCRKHDI 337
Query: 390 EPRAPEMYFSEKIDYLKDKVQPTFVKERRAMKREYEEFKVRINALVAKAQK--------- 440
EPR P+ YFS K D K+K + FVK+RR +KREY+EFKVRIN L + +K
Sbjct: 338 EPRNPDSYFSIKKDPTKNKKRQDFVKDRRWIKREYDEFKVRINGLPEQIKKRAEQFNMRE 397
Query: 441 ---------------VPPEG-------WIMQDGTPWPG--------NNTKDHPGMIQVF- 469
+PP+G W M DGT WPG ++ DH G++Q+
Sbjct: 398 ELKEKRIAREKNGGVLPPDGVEVVKATW-MADGTHWPGTWFEPKPDHSKGDHAGILQIMS 456
Query: 470 --------LG--NSGGVDTEG--NQLPRLVYVSREKRPGFQHHKKAGAMNALVRVSAVLT 517
+G N G +D G ++P YVSREKRPGF H+KKAGAMN +VR SA+L+
Sbjct: 457 KVPDLEPVMGGPNEGALDFTGIDIRVPMFAYVSREKRPGFDHNKKAGAMNGMVRASAILS 516
Query: 518 NAPFMLNLDCDHYINNSKAVREAMCFLMDPQTGKKVCYVQFPQRFDGIDTHDRYANRNTV 577
N F+LNLDCDHYI NSKA++E MCF+MD + G ++CY+QFPQRF+GID DRYAN NTV
Sbjct: 517 NGAFILNLDCDHYIYNSKAIKEGMCFMMD-RGGDRICYIQFPQRFEGIDPSDRYANHNTV 575
Query: 578 FFDINMKGLDGIQGPVYVGTGCVFRRQALYGYNPPKGPKRPKMVSCDCCPCFGSRKKLKH 637
FFD NM+ LDG+QGPVYVGTGC+FRR ALYG+NPP+ + + + P R + +
Sbjct: 576 FFDGNMRALDGLQGPVYVGTGCMFRRYALYGFNPPRANEYSGVFGQEKAPAMHVRTQSQA 635
Query: 638 AK-SDVNGEAASLKGMDDDKEVLMSQMNFEKKFGQSSIFVTSTLMEE------------- 683
++ S + + + ++DD + + KKFG S++F + + E
Sbjct: 636 SQTSQASDLESDTQPLNDDPD-----LGLPKKFGNSTMFTDTIPVAEYQGRPLADHMSVK 690
Query: 684 GGVPPSS-------SPAGMLKEAIHVISCGYEDKTEWGLEVGWIYGSITEDILTGFKMHC 736
G PP + A + EAI VISC YED TEWG +GWIYGS+TED++TG++MH
Sbjct: 691 NGRPPGALLLPRPPLDAPTVAEAIAVISCWYEDNTEWGDRIGWIYGSVTEDVVTGYRMHN 750
Query: 737 RGWRSIYCMPRRAAFKGTAPINLSDRLNQVLRWALGSIEIFFSHHCPLWYGYKEKKLKWL 796
RGWRS+YC+ +R AF+GTAPINL+DRL+QVLRWA GS+EIFFS + + + ++LK+L
Sbjct: 751 RGWRSVYCITKRDAFRGTAPINLTDRLHQVLRWATGSVEIFFSKNNAM---FATRRLKFL 807
Query: 797 ERFAYANTTIYPFTSIPLVAYCVLPAVCLLTDKFIMPPISTFAGLYFIALFSSIIATGVI 856
+R AY N IYPFTSI LV YC LPA+CL + KFI+ + Y + + ++ ++
Sbjct: 808 QRVAYLNVGIYPFTSIFLVVYCFLPALCLFSGKFIVQSLDIHFLSYLLCITVTLTLISLL 867
Query: 857 ELKWSGVSIEEWWRNEQFWVIGGVSAHLFAVIQGLLKVLAGIDTNFTVTSKAT-DDED-- 913
E+KWSG+ +EEWWRNEQFW+IGG SAHL AV+QGLLKV+AGI+ +FT+TSKA+ +DED
Sbjct: 868 EVKWSGIGLEEWWRNEQFWLIGGTSAHLAAVVQGLLKVIAGIEISFTLTSKASGEDEDDI 927
Query: 914 FGELYAIKWTTLLIPPTTILIINIVGVVAGVSDAINNGYQSWGPLFGKLFFSFWVIVHLY 973
F +LY +KWT L I P TI+I+N+V +V G S I + WG L G +FFS WV+ H+Y
Sbjct: 928 FADLYIVKWTGLFIMPLTIIIVNLVAIVIGASRTIYSVIPQWGKLMGGIFFSLWVLTHMY 987
Query: 974 PFLKGLMGRQNRTPTIVVIWSVLLASIFSLLWVRIDP 1010
PF KGLMGR+ + PTIV +WS L++ SLLW+ I P
Sbjct: 988 PFAKGLMGRRGKVPTIVYVWSGLVSITVSLLWITISP 1024
>At1g32180 cellulose synthase catalytic subunit, putative
Length = 979
Score = 811 bits (2095), Expect = 0.0
Identities = 437/905 (48%), Positives = 568/905 (62%), Gaps = 103/905 (11%)
Query: 192 KDRMDDWKLQQGNLGPEADEDTDASMLDEARQPLSRKVPIASSKINPYRMVIVARLVILA 251
KD D + N+G E ++DT +L + L+R V I+ I YR++IV R+V LA
Sbjct: 77 KDNEPDLTDVRINVGEEEEDDT---LLSKISYSLTRVVKISPIIIALYRILIVVRVVSLA 133
Query: 252 FFLRYRILNPVHDALGLWLTSIICEIWFAFSWILDQFPKWFPIDRETYLDRLSIRYEREG 311
FL +RI NP + AL LWL S+ICE+WFAFSW+LDQ PK FP++ T ++ L +E
Sbjct: 134 LFLFWRIRNPNNKALWLWLLSVICELWFAFSWLLDQIPKLFPVNHATDIEALKATFETPN 193
Query: 312 EPNM-----LAPVDFFVSTVDPMKEPPLVTANTILSILAVDYPVDKVSCYISDDGASMCT 366
N L +D FVST D KEPPLVTANTILSIL+VDYPV+K+S YISDDG S+ T
Sbjct: 194 PDNPTGKSDLPGIDVFVSTADAEKEPPLVTANTILSILSVDYPVEKLSVYISDDGGSLVT 253
Query: 367 FEALSETAEFARKWVPFCKKFSIEPRAPEMYFSEKIDYLKDKVQPTFVKERRAMKREYEE 426
FEA++E A FA+ WVPFC+K IEPR PE YF K D KDKV+ FV+ERR +KR Y+E
Sbjct: 254 FEAIAEAASFAKIWVPFCRKHKIEPRNPESYFGLKRDPYKDKVRHDFVRERRYVKRAYDE 313
Query: 427 FKVRINALVAKAQK------------------------------------VPPEGWIMQD 450
FKVR+NAL ++ V P+ M D
Sbjct: 314 FKVRVNALPHSIRRRSDAFNSKEEIKALEKWKHWKVKVEEDQIKEPRPALVAPKATWMSD 373
Query: 451 GTPWPG--------NNTKDHPGMIQVFLGNSGGVDTEGN--------------QLPRLVY 488
GT WPG ++ DH +IQV L G EG +LP LVY
Sbjct: 374 GTHWPGTWAVSGPHHSRGDHASVIQVLLDPPGDEPVEGKGGEGRALDLEGVDIRLPMLVY 433
Query: 489 VSREKRPGFQHHKKAGAMNALVRVSAVLTNAPFMLNLDCDHYINNSKAVREAMCFLMDPQ 548
VSREKRPG+ H+KKAGAMNALVR SA+++N PF+LNLDCDHY+ NS+A R+ +CF+MD
Sbjct: 434 VSREKRPGYDHNKKAGAMNALVRASAIMSNGPFILNLDCDHYVYNSRAFRDGICFMMD-H 492
Query: 549 TGKKVCYVQFPQRFDGIDTHDRYANRNTVFFDINMKGLDGIQGPVYVGTGCVFRRQALYG 608
G +V YVQFPQRF+GID DRYAN+NTVFFDIN++ LDGIQGP+YVGTGC+FRR ALYG
Sbjct: 493 DGDRVSYVQFPQRFEGIDPSDRYANKNTVFFDINLRALDGIQGPMYVGTGCLFRRTALYG 552
Query: 609 YNPPKGPKRPKMVSCDCCPCFGSRKKLKHAKSDVNGEAASLKGMDDDKEVLMSQMNFEKK 668
+NPP + S C F KK A AS D+E K+
Sbjct: 553 FNPPDVFVVEEEPSGSYC--FPLIKKRSPAT------VASEPEYYTDEEDRFDIGLIRKQ 604
Query: 669 FGQSSIFVTSTLMEEG-------------GVPPSSSPAGM-------LKEAIHVISCGYE 708
FG SS+ V S + E G PP S + EA++VISC YE
Sbjct: 605 FGSSSMLVNSVKVAEFEGRPLATVHSSRLGRPPGSLTGSRKPLDFATVNEAVNVISCWYE 664
Query: 709 DKTEWGLEVGWIYGSITEDILTGFKMHCRGWRSIYCMPRRAAFKGTAPINLSDRLNQVLR 768
DKTEWG VGWIYGS+TED++TGF+MH +GWRS YC+ AF+G+APINL+DRL+QVLR
Sbjct: 665 DKTEWGFNVGWIYGSVTEDVVTGFRMHEKGWRSFYCVTEPDAFRGSAPINLTDRLHQVLR 724
Query: 769 WALGSIEIFFSHHCPLWYGYKEKKLKWLERFAYANTTIYPFTSIPLVAYCVLPAVCLLTD 828
WA GS+EIFFS + ++ G KLK L+R AY N IYPFTSI ++ YC LP + L +
Sbjct: 725 WATGSVEIFFSRNNAIFAG---PKLKLLQRIAYLNVGIYPFTSIFILTYCFLPPLSLFSG 781
Query: 829 KFIMPPISTFAGLYFIALFSSIIATGVIELKWSGVSIEEWWRNEQFWVIGGVSAHLFAVI 888
F++ ++ +Y + + S+ V+E+KWSG+S+EEWWRNEQFW+IGG SAHL AV+
Sbjct: 782 HFVVETLTGSFLIYLLIITLSLCGLAVLEVKWSGISLEEWWRNEQFWLIGGTSAHLVAVL 841
Query: 889 QGLLKVLAGIDTNFTVTSKAT-----DDEDFGELYAIKWTTLLIPPTTILIINIVGVVAG 943
QG+LKV+AG++ +FT+TSK++ +D++F +LY KWT L+IPP TI+I+NIV ++
Sbjct: 842 QGILKVIAGVEISFTLTSKSSTGGDDEDDEFADLYLFKWTALMIPPLTIIILNIVAILFA 901
Query: 944 VSDAINNGYQSWGPLFGKLFFSFWVIVHLYPFLKGLMGRQNRTPTIVVIWSVLLASIFSL 1003
V + + W L G FF+ WV++H+YPF KGLMGR +TPT+V +WS L+A SL
Sbjct: 902 VCRTVFSANPQWSNLLGGTFFASWVLLHMYPFAKGLMGRGGKTPTVVYVWSGLIAICLSL 961
Query: 1004 LWVRI 1008
L++ I
Sbjct: 962 LYITI 966
>At2g32530 cellulose synthase like protein
Length = 755
Score = 397 bits (1019), Expect = e-110
Identities = 264/802 (32%), Positives = 397/802 (48%), Gaps = 92/802 (11%)
Query: 224 PLSRKVPIASSKINPYRMVIVARLVILAFFLRYRILNPVHDALGLWLTSIICEIWFAFSW 283
PL K+ S K R+V + L L L YRIL ++ +W+ + +CE +F+F W
Sbjct: 10 PLCEKI---SYKNYFLRVVDLTILGFLFSLLLYRILL-MNQNNSVWVVAFLCESFFSFIW 65
Query: 284 ILDQFPKWFPIDRETYLDRLSIRYEREGEPNMLAPVDFFVSTVDPMKEPPLVTANTILSI 343
+L KW P ++Y +RL R L VD FV+T DP++EPP++ ANT+LS+
Sbjct: 66 LLITSIKWSPASYKSYPERLDERVHD------LPSVDMFVTTADPVREPPILVANTLLSL 119
Query: 344 LAVDYPVDKVSCYISDDGASMCTFEALSETAEFARKWVPFCKKFSIEPRAPEMYFSEKID 403
LAV+YP +K++CY+SDDG S T+ +L E ++FA+ WVPFCKK++I+ RAP YF
Sbjct: 120 LAVNYPANKLACYVSDDGCSPLTYFSLKEASKFAKIWVPFCKKYNIKVRAPFRYFLNPPA 179
Query: 404 YLKDKVQPTFVKERRAMKREYEEFKVRINALVAKAQKVPPEGWIMQDGTPWPGNNTKDHP 463
+ F K+ KREYE+ R+ + + E D + DH
Sbjct: 180 ATESS---EFSKDWEITKREYEKLSRRVEDATGDSHWLDAE----DDFEDFSNTKPNDHS 232
Query: 464 GMIQVFLGNSGGVDTEGNQLPRLVYVSREKRPGFQHHKKAGAMNALVRVSAVLTNAPFML 523
+++V N GGV E N++P VY+SREKRP + HH KAGAMN LVRVS ++TNAP+ML
Sbjct: 233 TIVKVVWENKGGVGVE-NEVPHFVYISREKRPNYLHHYKAGAMNFLVRVSGLMTNAPYML 291
Query: 524 NLDCDHYINNSKAVREAMCFLMDPQTGKKVC-YVQFPQRFDGIDTHDRYANRNTVFFDIN 582
N+DCD Y N + VR+AMC + C +VQFPQ F +D A+ TV
Sbjct: 292 NVDCDMYANEADVVRQAMCIFLQKSMNSNHCAFVQFPQEF-----YDSNADELTVLQSYL 346
Query: 583 MKGLDGIQGPVYVGTGCVFRRQALYGYNPPKGPKRPKMVSCDCCPCFGSRKKLKHAKSDV 642
+G+ GIQGP Y G+GC R+ +YG +S D GS L K
Sbjct: 347 GRGIAGIQGPTYAGSGCFHTRRVMYG------------LSIDDLEDDGSLSSLATRK--- 391
Query: 643 NGEAASLKGMDDDKEVLMSQMNFEKKFGQSSIFVTSTLMEEGGVP-PSSSPAGMLKEAIH 701
+++ N ++FG S+ VTS + P P ++ A L+ A
Sbjct: 392 ----------------YLAEENLAREFGNSNEMVTSVVEALQRKPNPQNTLANSLEAAQE 435
Query: 702 VISCGYEDKTEWGLEVGWIYGSITEDILTGFKMHCRGWRSIYCMPRRAAFKGTAPINLSD 761
V C +E +T WG +GW+Y S ED T +H RGW S Y P+ AF G P +
Sbjct: 436 VGHCHFEYQTSWGKTIGWLYESTAEDANTSIGIHSRGWTSSYISPKPPAFLGAMPPGGPE 495
Query: 762 RLNQVLRWALGSIEIFFSHHCPLWYGYKEKKLKWLERFAYANTTIYPFTSIPLVAYCVLP 821
+ Q RWA G +E+ F+ PL G +K+++ + AY + SIP + YC+LP
Sbjct: 496 AMLQQRRWATGLLEVLFNKQSPL-IGMFCRKIRFRQSLAYLYIFTWGLRSIPELIYCLLP 554
Query: 822 AVCLLTDKFIMPPISTFAGLYFIALFSSIIATGVIELKWS----GVSIEEWWRNEQFWVI 877
A CLL + + P G+Y + + +++ + W G S++ W+ ++ FW I
Sbjct: 555 AYCLLHNAALFP-----KGVY-LGIVVTLVGMHCLYSLWEFMSLGFSVQSWFASQSFWRI 608
Query: 878 GGVSAHLFAVIQGLLKVLAGIDTNFTVTSKA------------------TDDEDFGELYA 919
+ LF++ +LK+L T F VT K ++D G+ +
Sbjct: 609 KTTCSWLFSIPDIILKLLGISKTVFIVTKKTMPKTMSGSGSEKSQREVDCPNQDSGK-FE 667
Query: 920 IKWTTLLIPPTTILIINIVGVVAGVSDAINNGYQSWGPLFGKLFFSFWVIVHLYPFLKGL 979
+ +P T IL++N+ +AG S + ++ G + V++ PFLKG+
Sbjct: 668 FDGSLYFLPGTFILLVNL-AALAGCSVGLQR-HRGGGSGLAEACGCILVVILFLPFLKGM 725
Query: 980 MGRQNRTPTIVVIWSVLLASIF 1001
+ + WS L + F
Sbjct: 726 FEKGK----YGIPWSTLSKAAF 743
>At2g32620 putative cellulose synthase
Length = 757
Score = 395 bits (1015), Expect = e-110
Identities = 262/769 (34%), Positives = 390/769 (50%), Gaps = 87/769 (11%)
Query: 240 RMVIVARLVILAFFLRYRILNPVHDALGLWLTSIICEIWFAFSWILDQFPKWFPIDRETY 299
R V + L +L L +RIL + + +WL + +CE F+F W+L KW P + + Y
Sbjct: 23 RAVDLTILGLLFSLLLHRILYMSQNGI-IWLVAFLCESCFSFVWLLSTCTKWSPAETKPY 81
Query: 300 LDRLSIR-YEREGEPNMLAPVDFFVSTVDPMKEPPLVTANTILSILAVDYPVDKVSCYIS 358
DRL R Y+ L VD FV T DP++EPP++ NT+LS+LAV+YP +K++CY+S
Sbjct: 82 PDRLDERVYD-------LPSVDMFVPTADPVREPPIMVVNTVLSLLAVNYPANKLACYVS 134
Query: 359 DDGASMCTFEALSETAEFARKWVPFCKKFSIEPRAPEMYFSEKIDYLKDKVQPTFVKERR 418
DDG S T+ +L E ++FA+ WVPFCKK++++ RAP YF + P E
Sbjct: 135 DDGCSPLTYFSLKEASKFAKIWVPFCKKYNLKVRAPFRYF----------LNPFAATEGS 184
Query: 419 AMKREYEEFKVRINALVAKAQKVPPEGWIM-QDGTPWPGNNTK--DHPGMIQVFLGNSGG 475
R++E K L K + + ++ D +NTK DH +I+V N GG
Sbjct: 185 EFSRDWEMTKREYEKLCRKVEDATGDSHLLGTDNELEAFSNTKPNDHSTIIKVVWENKGG 244
Query: 476 VDTEGNQLPRLVYVSREKRPGFQHHKKAGAMNALVRVSAVLTNAPFMLNLDCDHYINNSK 535
V E ++P +VY+SREKRP + HH KAGAMN L RVS ++TNAP+MLN+DCD Y N +
Sbjct: 245 VGDE-KEVPHIVYISREKRPNYLHHYKAGAMNFLARVSGLMTNAPYMLNVDCDMYANEAD 303
Query: 536 AVREAMC-FLMDPQTGKKVCYVQFPQRFDGIDTHDRYANRNTVFFDINMKGLDGIQGPVY 594
VR+AMC FL Q +VQFPQ F +D + TV +G+ GIQGP+
Sbjct: 304 VVRQAMCIFLQKSQNQNHCAFVQFPQEF-----YDSNTIKLTVIKSYMGRGIAGIQGPIN 358
Query: 595 VGTGCVFRRQALYGYNPPKGPKRPKMVSCDCCPCFGSRKKLKHAKSDVNGEAASLKGMDD 654
VG+GC R+ +YG +P + + NG +S+ +
Sbjct: 359 VGSGCFHSRRVMYGLSPD--------------------------ELEDNGSLSSVATRE- 391
Query: 655 DKEVLMSQMNFEKKFGQSSIFVTSTLMEEGGVP-PSSSPAGMLKEAIHVISCGYEDKTEW 713
L+++ + FG S VTS + P P + ++ A V C YE +T W
Sbjct: 392 ----LLAEDSLSSGFGNSKEMVTSVVEALQRKPNPQNILTNSIEAAQEVGHCDYESQTSW 447
Query: 714 GLEVGWIYGSITEDILTGFKMHCRGWRSIYCMPRRAAFKGTAPINLSDRLNQVLRWALGS 773
G +GW+Y S++ED+ T +H RGW S Y P AF G+ P + + Q RWA GS
Sbjct: 448 GKTIGWLYDSMSEDMNTSIGIHSRGWTSSYIAPDPPAFLGSMPPGGLEAMIQQRRWATGS 507
Query: 774 IEIFFSHHCPLWYGYKEKKLKWLERFAYANTTIYPFTSIPLVAYCVLPAVCLLTDKFIMP 833
IE+ F+ PL G +KL++ +R AY +I SIP + YC+LPA CLL + + P
Sbjct: 508 IEVLFNKQSPL-LGLFCRKLRFRQRVAYLCVSIC-VRSIPELIYCLLPAYCLLHNSALFP 565
Query: 834 PISTFAGLYFIALFSSIIATGVIELKWSGVSIEEWWRNEQFWVIGGVSAHLFAVIQGLLK 893
G+ + + T + E G SI+ W+ ++ FW I S+ LF++ +LK
Sbjct: 566 K-GLCLGITMLLAGMHCLYT-LWEFMCLGHSIQSWYVSQSFWRIVATSSWLFSIFDIILK 623
Query: 894 VLAGIDTNFTVTSKAT-----------------DDEDFGELYAIKWTTLLIPPTTILIIN 936
+L G+ N + SK T DD + +P T I+++N
Sbjct: 624 LL-GLSKNVFLVSKKTMPVETMSGSGIGPSQREDDGPNSGKTEFDGSLYFLPGTFIVLVN 682
Query: 937 ---IVGVVAGVSDAINNGYQSWGPLFGKLFFSFWVIVHLYPFLKGLMGR 982
+VGV G+ + + + G G+ V++ +PFLKGL +
Sbjct: 683 LAALVGVFVGLQRS-SYSHGGGGSGLGEACACILVVMLFFPFLKGLFAK 730
>At2g32540 putative cellulose synthase
Length = 755
Score = 393 bits (1009), Expect = e-109
Identities = 261/811 (32%), Positives = 394/811 (48%), Gaps = 84/811 (10%)
Query: 220 EARQPLSRKVPIASSKINPYRMVIVARLVILAFFLRYRILNPVHDALGLWLTSIICEIWF 279
E+ PL S K R V + L +L L YRIL+ V+ +W+ + +CE F
Sbjct: 3 ESSSPLPPLCERISHKSYFLRAVDLTILGLLLSLLLYRILH-VNQKDTVWIVAFLCETCF 61
Query: 280 AFSWILDQFPKWFPIDRETYLDRLSIRYEREGEPNMLAPVDFFVSTVDPMKEPPLVTANT 339
F W+L KW P D +TY +RL R L PVD FV+T DP++EPPL+ NT
Sbjct: 62 TFVWLLITNIKWSPADYKTYPERLDERVHE------LPPVDMFVTTADPVREPPLIVVNT 115
Query: 340 ILSILAVDYPVDKVSCYISDDGASMCTFEALSETAEFARKWVPFCKKFSIEPRAPEMYFS 399
+LS+LAV+YP +K++CY+SDDG S T+ +L E ++FA+ WVPFCKK+++ RAP MYF
Sbjct: 116 VLSLLAVNYPANKLACYVSDDGCSPLTYFSLKEASKFAKIWVPFCKKYNVRVRAPFMYFR 175
Query: 400 EKIDYLKDKVQPTFVKERRAMKREYEEFKVRINALVAKAQKVPPEGWIMQDGTPWPGNNT 459
+ + F K+ KREYE+ ++ + + E D + +
Sbjct: 176 NSPEAAEGS---EFSKDWEMTKREYEKLSQKVEDATGSSHWLDAE----DDFEAFLNTKS 228
Query: 460 KDHPGMIQVFLGNSGGVDTEGNQLPRLVYVSREKRPGFQHHKKAGAMNALVRVSAVLTNA 519
DH +++V N GGV E ++P +VY+SREKRP HH KAGAMN LVRVS ++TNA
Sbjct: 229 NDHSTIVKVVWENKGGVGDE-KEVPHVVYISREKRPNHFHHYKAGAMNFLVRVSGLMTNA 287
Query: 520 PFMLNLDCDHYINNSKAVREAMCFLMDPQTGKKVC-YVQFPQRFDGIDTHDRYANRNTVF 578
P+MLN+DCD Y+N + VR+AMC + C +VQ+PQ F +D TV
Sbjct: 288 PYMLNVDCDMYVNEADVVRQAMCIFLQKSMDSNHCAFVQYPQDF-----YDSNVGELTVL 342
Query: 579 FDINMKGLDGIQGPVYVGTGCVFRRQALYGYNPPKGPKRPKMVSCDCCPCFGSRKKLKHA 638
+G+ GIQGP Y G+GC R+ +YG +
Sbjct: 343 QLYLGRGIAGIQGPQYAGSGCFHTRRVMYGLS---------------------------- 374
Query: 639 KSDVNGEAASLKGMDDDKEVLMSQMNFEKKFGQSSIFVTSTLMEEGGVP-PSSSPAGMLK 697
D G+ SL + K +++ + ++FG S V S + P P + L+
Sbjct: 375 -LDDLGDDGSLSSIATRK--YLAEESLTREFGNSKEMVKSVVDALQRKPFPQKNLKDSLE 431
Query: 698 EAIHVISCGYEDKTEWGLEVGWIYGSITEDILTGFKMHCRGWRSIYCMPRRAAFKGTAPI 757
A + C YE +T WG +GW+Y S TED+ T +H RGW S Y P AF G P
Sbjct: 432 TAQEMGHCHYEYQTSWGKNIGWLYDSTTEDVNTSIGIHSRGWTSSYIFPDPPAFLGCMPQ 491
Query: 758 NLSDRLNQVLRWALGSIEIFFSHHCPLWYGYKEKKLKWLERFAYANTTIYPFTSIPLVAY 817
+ + Q RWA G +EI F+ PL G +K+++ + AY + SIP + Y
Sbjct: 492 GGPEVMVQQRRWATGLLEILFNKQSPL-IGMFCRKIRFRQSLAYLYVFSWGLRSIPELFY 550
Query: 818 CVLPAVCLLTDKFIMPPISTFAGLYFIALFSSIIATGVIELKWS----GVSIEEWWRNEQ 873
C+LPA CLL + + P G+Y + + +++ + W G SI+ W+ +
Sbjct: 551 CLLPAYCLLHNSALFP-----KGVY-LGIIITLVGIHCLYTLWEFMNLGFSIQSWYVTQS 604
Query: 874 FWVIGGVSAHLFAVIQGLLKVLAGIDTNFTVTSKATDDEDFGE----------------- 916
F I + LF+V+ +LK+L T F VT K + G
Sbjct: 605 FGRIKTTCSWLFSVLDVILKLLGISKTVFIVTKKTMPETKSGSGSKKSQREVDCPNQDSG 664
Query: 917 LYAIKWTTLLIPPTTILIINIVGVVAGVSDAINNGYQSWGPLFGKLFFSFWVIVHLYPFL 976
+ + +P T I+++N+ + + + G G + V++ PFL
Sbjct: 665 KFEFDGSLYFLPGTFIVLVNLAALAGCLVGLQSRG--GGGSGLAEACGCILVVILFLPFL 722
Query: 977 KGLMGR-QNRTPTIVVIWSVLLASIFSLLWV 1006
KG+ + + P + + LA++F +L V
Sbjct: 723 KGMFEKGKYGIPFSTLSKAAFLAALFVVLSV 753
>At1g55850 putative cellulose synthase catalytic subunit (At1g55850)
Length = 729
Score = 283 bits (724), Expect = 4e-76
Identities = 159/386 (41%), Positives = 223/386 (57%), Gaps = 30/386 (7%)
Query: 233 SSKINPYRMVIVARLVILAFFLRYRI-----LNPVHDALGLWLTSIICEIWFAFSWILDQ 287
+ ++ YR + V + YRI V D L +W I EIWF W++ Q
Sbjct: 26 TGRVIAYRFFSASVFVCICLIWFYRIGEIGDNRTVLDRL-IWFVMFIVEIWFGLYWVVTQ 84
Query: 288 FPKWFPIDRETYLDRLSIRYEREGEPNMLAPVDFFVSTVDPMKEPPLVTANTILSILAVD 347
+W P+ R + DRLS RY + L +D FV T DP+ EPPL+ NT+LS+ A+D
Sbjct: 85 SSRWNPVWRFPFSDRLSRRYGSD-----LPRLDVFVCTADPVIEPPLLVVNTVLSVTALD 139
Query: 348 YPVDKVSCYISDDGASMCTFEALSETAEFARKWVPFCKKFSIEPRAPEMYFSEKIDYLKD 407
YP +K++ Y+SDDG S TF AL+E AEFA+ WVPFCKKF++EP +P Y S K + L
Sbjct: 140 YPPEKLAVYLSDDGGSELTFYALTEAAEFAKTWVPFCKKFNVEPTSPAAYLSSKANCLDS 199
Query: 408 KVQPTFVKERRAMKREYEEFKVRINALVAKAQKVPPEGWIMQ-DG-TPWPGNNTK-DHPG 464
+ + + Y E RI A+ ++P E + DG + W + T+ +H
Sbjct: 200 AAEE--------VAKLYREMAARIET-AARLGRIPEEARVKYGDGFSQWDADATRRNHGT 250
Query: 465 MIQVFLGNSGGVDTEGNQL--PRLVYVSREKRPGFQHHKKAGAMNALVRVSAVLTNAPFM 522
++QV + EGN + P LVY+SREKRP H+ KAGAMNAL+RVS+ +T +
Sbjct: 251 ILQVLVDGR-----EGNTIAIPTLVYLSREKRPQHHHNFKAGAMNALLRVSSKITCGKII 305
Query: 523 LNLDCDHYINNSKAVREAMCFLMDPQTGKKVCYVQFPQRFDGIDTHDRYANRNTVFFDIN 582
LNLDCD Y NNSK+ R+A+C L+D + GK++ +VQFPQ FD + +D Y + V D+
Sbjct: 306 LNLDCDMYANNSKSTRDALCILLDEKEGKEIAFVQFPQCFDNVTRNDLYGSMMRVGIDVE 365
Query: 583 MKGLDGIQGPVYVGTGCVFRRQALYG 608
GLDG GP+Y+GTGC RR + G
Sbjct: 366 FLGLDGNGGPLYIGTGCFHRRDVICG 391
Score = 145 bits (367), Expect = 9e-35
Identities = 86/303 (28%), Positives = 151/303 (49%), Gaps = 11/303 (3%)
Query: 704 SCGYEDKTEWGLEVGWIYGSITEDILTGFKMHCRGWRSIYCMPRRAAFKGTAPINLSDRL 763
SC YE+ T+WG E+G YG ED++TG + CRGW+S Y P + AF G AP NL L
Sbjct: 420 SCTYEENTQWGKEMGVKYGCPVEDVITGLTIQCRGWKSAYLNPEKQAFLGVAPTNLHQML 479
Query: 764 NQVLRWALGSIEIFFSHHCPLWYGYKEKKLKWLERFAYANTTIYPFTSIPLVAYCVLPAV 823
Q RW+ G +I S + P+WYG + L + Y ++ +S+P++ Y VL ++
Sbjct: 480 VQQRRWSEGDFQIMLSKYSPVWYGKGKISLGLI--LGYCCYCLWAPSSLPVLIYSVLTSL 537
Query: 824 CLLTDKFIMPPISTFAGLYFIALFSSIIATGVIELKWSGVSIEEWWRNEQFWVIGGVSAH 883
CL + P +S+ + F + + A + E W G + WW ++ W+ S+
Sbjct: 538 CLFKGIPLFPKVSSSWFIPFGYVTVAATAYSLAEFLWCGGTFRGWWNEQRMWLYRRTSSF 597
Query: 884 LFAVIQGLLKVLAGIDTNFTVTSKATDDEDFGELYAIKWTTLLIPPTTILIINIVGVV-- 941
LF + + K+L ++ F +T+K +E+ E Y + + L++ +G++
Sbjct: 598 LFGFMDTIKKLLGVSESAFVITAKVA-EEEAAERYKEEVMEFGVESPMFLVLGTLGMLNL 656
Query: 942 ----AGVSDAINNGYQSWGPLFGKLFFSFWVIVHLYPFLKGLMGRQN--RTPTIVVIWSV 995
A V+ ++ + + + ++V +P KG++ RQ+ + P V + SV
Sbjct: 657 FCFAAAVARLVSGDGGDLKTMGMQFVITGVLVVINWPLYKGMLLRQDKGKMPMSVTVKSV 716
Query: 996 LLA 998
+LA
Sbjct: 717 VLA 719
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.140 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,188,634
Number of Sequences: 26719
Number of extensions: 1155974
Number of successful extensions: 3579
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 32
Number of HSP's that attempted gapping in prelim test: 3360
Number of HSP's gapped (non-prelim): 99
length of query: 1027
length of database: 11,318,596
effective HSP length: 109
effective length of query: 918
effective length of database: 8,406,225
effective search space: 7716914550
effective search space used: 7716914550
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0037a.7


