

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0037a.4
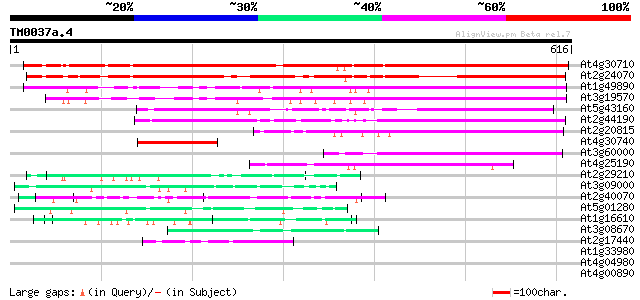
(616 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g30710 putative protein 585 e-167
At2g24070 unknown protein 458 e-129
At1g49890 unknown protein 283 2e-76
At3g19570 unknown protein 280 2e-75
At5g43160 putative protein 133 3e-31
At2g44190 unknown protein 124 2e-28
At2g20815 putative protein 115 9e-26
At4g30740 putative protein 114 1e-25
At3g60000 putative protein 114 1e-25
At4g25190 putative protein 101 1e-21
At2g29210 proline-rich protein like 59 8e-09
At3g09000 unknown protein 58 2e-08
At2g40070 En/Spm-like transposon protein 57 3e-08
At5g01280 putative protein 50 4e-06
At1g16610 arginine/serine-rich protein 45 9e-05
At3g08670 hypothetical protein 44 3e-04
At2g17440 unknown protein 43 6e-04
At1g33980 unknown protein 40 0.003
At4g04980 39 0.007
At4g00890 39 0.007
>At4g30710 putative protein
Length = 644
Score = 585 bits (1509), Expect = e-167
Identities = 353/642 (54%), Positives = 438/642 (67%), Gaps = 63/642 (9%)
Query: 16 AVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTA 75
A +T R L+ ++KNNA AT RR RT EV+SRY+SPTP T +G RCPSP++TR
Sbjct: 4 ATDTTRRRLLPSDKNNAVVAT---RRPRTMEVSSRYRSPTP--TKNG--RCPSPSVTRPT 56
Query: 76 TPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHN 135
++S + KR++SAERKRPSTPPSP S STP+ D +ID SSR+++ RLPE +
Sbjct: 57 VSSSSQSVAAKRAVSAERKRPSTPPSPTSP--STPIRDLSIDLPASSRRLSTGRLPE--S 112
Query: 136 LWPSTMRSLSVSFQSDIISIPVSKKERPVTSAS-DRTLRPASNVAHR-QGETPSTIRKAT 193
LWPSTMRSLSVSFQSD +S+PVSKKERPV+S+S DRTLRP+SN+A + + ET S RK T
Sbjct: 113 LWPSTMRSLSVSFQSDSVSVPVSKKERPVSSSSGDRTLRPSSNIAQKHKAETTSVSRKPT 172
Query: 194 PERKRSPLKGKN-ASDQSENSKPVDSLPSRLIDQHRWPSRIGGKVSSSALNRSVDFGDAR 252
PERKRSPLKGKN SD SENSKPVD SRLI+QHRWPSRIGGK++S++LNRS+D GD
Sbjct: 173 PERKRSPLKGKNNVSDLSENSKPVDGPHSRLIEQHRWPSRIGGKITSNSLNRSLDLGDKA 232
Query: 253 MLKTPASGTGFS-SLRRLSLS-EEASKPLQRASSDSVRLLSLVASGRTGSEVKSVDDCSL 310
P SG G SLRR+SL +S+PL + SS++ LV S KS D+
Sbjct: 233 SRGIPTSGPGMGPSLRRMSLPLSSSSRPLHKTSSNTSSYGGLV------SPTKSEDNNIA 286
Query: 311 HDLRPHRS-STPHTDRAGLSIAGVRSQSLSAPGSRLPSPSRISVQSTSS-------SRGV 362
R S DRA L+ A R L APGSR SPSR S S+SS SRGV
Sbjct: 287 RTSGAQRLLSAGSLDRATLATAVARLHPLPAPGSRPASPSRTSFLSSSSISRGMSTSRGV 346
Query: 363 SPSR-------------------------------SRPSTPPSRGGVSPSRIRPTSSSIQ 391
SPSR +RPSTPPSRG +SPSRIR T++S Q
Sbjct: 347 SPSRGLSPSRGLSPTRGLSPSRGLSPSRGTNTSCFARPSTPPSRG-ISPSRIRQTTTSTQ 405
Query: 392 SNDSVSVLSFIADFRKGKKGAAFIEDAHQLRLLYNRFMQWRFANARAEAVLYIQNAIVQK 451
S+ + SVLSFI D +KGKK A++IED HQLRLL+NR++QWRFA ARAE+V+YIQ ++
Sbjct: 406 SSTTTSVLSFITDVKKGKK-ASYIEDVHQLRLLHNRYLQWRFAIARAESVMYIQRLTSEE 464
Query: 452 TLYNVWIATLSLWESVTRKKINLQQLKLELKLNSVLNDQMAYLDEWATLETDHVDALSGA 511
TL+NVW A L + VTR++I LQQLKLE+KLNS+LNDQM L++WATLE DHV +L GA
Sbjct: 465 TLFNVWHAISELQDHVTRQRIGLQQLKLEIKLNSLLNDQMVSLEDWATLERDHVSSLVGA 524
Query: 512 VEDLEASTLRLPVTRGAMVDIEHLKVAICQAVDVMQAMGSAIRSLFSQVEGMNDLISGVA 571
+ DLEA+TLRLP T G D E LK A+ A+DVMQAMGS+I SL S+VE MN +++ +A
Sbjct: 525 ISDLEANTLRLPATGGTKADTESLKAAMSSALDVMQAMGSSIWSLLSKVEEMNIMVTELA 584
Query: 572 VVATKEKSILDECEMLLASVAALQVEESSLQTHLIQFKQVLG 613
VV TKE S+ +CE LLAS A +Q+EE SL+THLIQ ++ G
Sbjct: 585 VVVTKESSMQGKCEDLLASTAIMQIEECSLRTHLIQTRREEG 626
>At2g24070 unknown protein
Length = 560
Score = 458 bits (1178), Expect = e-129
Identities = 304/619 (49%), Positives = 379/619 (61%), Gaps = 120/619 (19%)
Query: 19 TPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPA 78
+PRPPL +EKNN + T RR+RT EV+SRY+SPTP T RRCPSP +TRTA P+
Sbjct: 13 SPRPPLAPSEKNNVGSVT---RRARTMEVSSRYRSPTPTKT----RRCPSPIVTRTA-PS 64
Query: 79 ASLQLLPKRSLSAERKR-PSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLW 137
+S + KR++SAER R PSTP +TPV+D +D +SSR+++ RLPE +LW
Sbjct: 65 SSPESFLKRAVSAERNRGPSTP--------TTPVSDVLVDLPVSSRRLSTGRLPE--SLW 114
Query: 138 PSTMRSLSVSFQSDIISIPVSKKERP-VTSASDRTLRPAS-NVAHRQ-GETPSTIRKATP 194
PSTMRSLSVSFQSD +S+PVSKKE+P VTS++DRTLRP+S N+AH+Q ET S RK TP
Sbjct: 115 PSTMRSLSVSFQSDSVSVPVSKKEKPLVTSSTDRTLRPSSSNIAHKQQSETTSVTRKQTP 174
Query: 195 ERKRSPLKGKNASD-QSENSKPVDSLPSRLID-QHRWPSRIGGKVSSSALNRSVDFGDAR 252
ERKRSPLKGKN S QSENSKP+D S LI QHRW RI G NRS D GD
Sbjct: 175 ERKRSPLKGKNVSPGQSENSKPMDGSHSMLIPPQHRWSGRIRG-------NRSFDLGD-- 225
Query: 253 MLKTPASGTGFSSLRRLSLS-EEASKPLQRASSDSVRLLSLVASGR--TGSEVKSVDDCS 309
++RR+SL S +++SSD RL S +GR S S D S
Sbjct: 226 -----------KAVRRVSLPLSNKSSRHKKSSSDITRLFSCYDNGRLEVSSSTTSEDSSS 274
Query: 310 LHDLRPHRSSTPHTDRAGLSIAGVRSQSLSAPGSRLPSPSRISVQSTSSS--RGVSPSRS 367
L+ H + L R +SAPGSR SPSR S S+SSS RG+SPSR
Sbjct: 275 TESLK-------HFSTSSLP----RLHPMSAPGSRTASPSRSSFSSSSSSNSRGMSPSRG 323
Query: 368 ----------------RPSTPPSRGGVSPSRIRPTSSSIQSNDSVSVLSFIADFRKGKKG 411
R STPPSRG VSPSRIR T+ S +N SVLSFIAD +KGKK
Sbjct: 324 VSPMRGLSPVGNRSLVRSSTPPSRG-VSPSRIRQTAQSSSTN--TSVLSFIADVKKGKK- 379
Query: 412 AAFIEDAHQLRLLYNRFMQWRFANARAEAVLYIQNAIVQKTLYNVWIATLSLWESVTRKK 471
A +IED HQLRLLYNR+ QWRFANARAE V Y+Q+ I +
Sbjct: 380 ATYIEDVHQLRLLYNRYSQWRFANARAEGVSYVQSLIAK--------------------- 418
Query: 472 INLQQLKLELKLNSVLNDQMAYLDEWATLETDHVDALSGAVEDLEASTLRLPVTRGAMVD 531
M L++WA +E +H+ +L+GA+ DLEA+TLRLP+ G D
Sbjct: 419 -------------------MVCLEDWAMVEREHISSLAGAIGDLEANTLRLPLAGGTKAD 459
Query: 532 IEHLKVAICQAVDVMQAMGSAIRSLFSQVEGMNDLISGVAVVATKEKSILDECEMLLASV 591
+ LK+A+ A+DVMQ+MGS+I SL SQ+E MN L+S +AV+A E +LD+CE LLAS
Sbjct: 460 LGSLKLAMSSALDVMQSMGSSIWSLHSQMEEMNKLVSDLAVIAKTENFLLDKCENLLAST 519
Query: 592 AALQVEESSLQTHLIQFKQ 610
A +++EE SL+THLIQ KQ
Sbjct: 520 AVMEIEERSLKTHLIQKKQ 538
>At1g49890 unknown protein
Length = 659
Score = 283 bits (724), Expect = 2e-76
Identities = 237/703 (33%), Positives = 336/703 (47%), Gaps = 165/703 (23%)
Query: 16 AVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSG------------- 62
A+ T P ++ + T RR R ++V SRY SP+P + S
Sbjct: 5 AISTTDPRNPPRDRPQSLTNNGGQRRPRGKQVPSRYLSPSPSHSVSSTTTTTTTTTTTTS 64
Query: 63 -------------SRRCPSPN--LTRTATPAASLQ-----LLPKRSLSAERKRPSTPPSP 102
S+R PSP+ L+R+ T +AS LLPKRS S +R+RPS
Sbjct: 65 SSSSSSSSAILRTSKRYPSPSPLLSRSTTNSASNSIKTPSLLPKRSQSVDRRRPS----- 119
Query: 103 PSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQSDIISIPVSKKER 162
A+ + + AA+++ L ST RSLSVSFQ + S+P+SKK+
Sbjct: 120 ------------AVSVTVGTEMSAATKM-----LITST-RSLSVSFQGEAFSLPISKKKE 161
Query: 163 PVTSASDRTLRPASNVAHRQGETPSTIRKATPERKRS-PLKGKNASDQSENSKPVDSLPS 221
+ TP + RK+TPER+RS P++ DQ ENSKPVD
Sbjct: 162 TTS-------------------TPVSHRKSTPERRRSTPVR-----DQRENSKPVD---- 193
Query: 222 RLIDQHRWP--SRIGGKVS--SSALNRSVDFGDARMLKTPASGTGFSSLRRLSLSE---- 273
Q RWP SR G S ++L+RS+D G R K + G S L + E
Sbjct: 194 ----QQRWPGASRRGNSESVVPNSLSRSLDCGSDRG-KLGSGFVGRSMLHNSMIDESPRV 248
Query: 274 -----------------EASKPLQRASSDSVRLLSLVASGRTGSEVKSVDDCSLHDLRPH 316
+ +QR ++ L S V+ T S+ SV S + ++
Sbjct: 249 SVNGRLSLDLGGRDEYLDIGDDIQRRPNNG--LTSSVSCDFTASDTDSVSSGSTNGVQEC 306
Query: 317 RS----------STPHTDRAGLSI---AGVRSQSLSAPGSRLPSPSRISVQSTSSSRGVS 363
S S P A R + L PGS L S + S SS G+S
Sbjct: 307 GSGVNGEISKSKSLPRNIMASARFWQETNSRLRRLQDPGSPLSSSPGLKTSSISSKFGLS 366
Query: 364 PSRSRPSTP---------PSRGGV----SPSRIRPTSSSIQS------------------ 392
S + P P RG SPS++ T++S +
Sbjct: 367 KRFSSDAVPLSSPRGMASPVRGSAIRSASPSKLWATTTSSPARALSSPSRARNGVSDQMN 426
Query: 393 ----NDSVSVLSFIADFRKGKKGAAFIEDAHQLRLLYNRFMQWRFANARAEAVLYIQNAI 448
N++ S+LSF AD R+GK G + DAH LRLLYNR +QWRF NARA++ + +Q
Sbjct: 427 AYNRNNTPSILSFSADIRRGKIGEDRVMDAHLLRLLYNRDLQWRFVNARADSTVMVQRLN 486
Query: 449 VQKTLYNVWIATLSLWESVTRKKINLQQLKLELKLNSVLNDQMAYLDEWATLETDHVDAL 508
+K L+N W++ L SVT K+I L L+ +LKL S+L QM +L+EW+ L+ DH +L
Sbjct: 487 AEKNLWNAWVSISELRHSVTLKRIKLLLLRQKLKLASILRGQMGFLEEWSLLDRDHSSSL 546
Query: 509 SGAVEDLEASTLRLPVTRGAMVDIEHLKVAICQAVDVMQAMGSAIRSLFSQVEGMNDLIS 568
SGA E L+ASTLRLP+ +VDI+ LK A+ AVDVMQAM S+I SL S+V+ MN ++
Sbjct: 547 SGATESLKASTLRLPIVGKTVVDIQDLKHAVSSAVDVMQAMSSSIFSLTSKVDEMNSVMV 606
Query: 569 GVAVVATKEKSILDECEMLLASVAALQVEESSLQTHLIQFKQV 611
V KEK +L+ C+ L+ VAA+QV + S++TH+IQ ++
Sbjct: 607 ETVNVTAKEKVLLERCQGCLSRVAAMQVTDCSMKTHIIQLSRI 649
>At3g19570 unknown protein
Length = 644
Score = 280 bits (716), Expect = 2e-75
Identities = 234/672 (34%), Positives = 333/672 (48%), Gaps = 166/672 (24%)
Query: 40 RRSRT-REVTSRYKSPTPP-------------STPSGS-------RRCPSPNLTRTATPA 78
RR R + V SRY SP+P ST S S +R PSP L+RT A
Sbjct: 39 RRPRAAKNVPSRYLSPSPSHSTTTTTTTATSTSTSSSSSVILRSSKRYPSPLLSRTTNSA 98
Query: 79 ASL----QLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGH 134
++L LPKRS S +R+RPS V+D+ + +++ + S
Sbjct: 99 SNLVYTPSSLPKRSQSVDRRRPSA------------VSDTRTEMSAATKMLITST----- 141
Query: 135 NLWPSTMRSLSVSFQSDIISIPVSKKERPVTSASDRTLRPASNVAHRQGETPSTIRKATP 194
RSLSVSFQ + S P+SKK ++ TP + RK TP
Sbjct: 142 -------RSLSVSFQGEAFSFPISKK--------------------KETATPVSHRKCTP 174
Query: 195 ERKRS-PLKGKNASDQSENSKPVDSLPSRLIDQHRWP--SRIGGKVS--SSALNRSVDF- 248
ER+R+ P++ DQ ENSKPVD Q WP SR G S ++L+RSVD
Sbjct: 175 ERRRATPVR-----DQRENSKPVD--------QQLWPGASRRGSSESVVPNSLSRSVDSD 221
Query: 249 --------------------------GDARMLKTPASGTGFSSLRRLSLSEEASKPLQRA 282
GD R+ G G +R + + +++ P
Sbjct: 222 SDDGRKLGSGFVGRSMLQHSQSSRVSGDGRLNLGFVGGDGMLEMRDENKARQSTHP---- 277
Query: 283 SSDSVRLLSLVASGRTGSEVKSVD--------DCSLHDLRPHRS-------STPHTDRAG 327
RL S V+ T S+ SV +C ++ RS ST
Sbjct: 278 -----RLASSVSCDFTASDTDSVSSGSTNGAHECGSGEVSKTRSLPRNGMASTKFWQETN 332
Query: 328 LSIAGVR---SQSLSAPGSRLPSPSRISVQS---------TSSSRGV-SPSR--SRPSTP 372
+ ++ S S+P SR+ S S QS TSS RG+ SP R +RP++P
Sbjct: 333 SRLRRMQDPGSPQCSSPSSRISSISSKFSQSKRFSSDSPLTSSPRGMTSPIRGATRPASP 392
Query: 373 ----------PSRGGVSPSRIRPTSS---SIQSNDSVSVLSFIADFRKGKKGAAFIEDAH 419
P+R SPSR+R S + + S+L F AD R+GK G + DAH
Sbjct: 393 SKLWATATSAPARTSSSPSRVRNGVSEQMNAYNRTLPSILCFSADIRRGKIGEDRVMDAH 452
Query: 420 QLRLLYNRFMQWRFANARAEAVLYIQNAIVQKTLYNVWIATLSLWESVTRKKINLQQLKL 479
LRLLYNR +QWRFANARA++ L +Q +K L+N W++ L SVT K+I L ++
Sbjct: 453 LLRLLYNRDLQWRFANARADSTLMVQRLSAEKILWNAWVSISELRHSVTLKRIKLLLMRQ 512
Query: 480 ELKLNSVLNDQMAYLDEWATLETDHVDALSGAVEDLEASTLRLPVTRGAMVDIEHLKVAI 539
+LKL S+L +QM YL+EW+ L+ +H ++LSGA E L+ASTLRLPV+ A+VDI+ LK A+
Sbjct: 513 KLKLASILKEQMCYLEEWSLLDRNHSNSLSGATEALKASTLRLPVSGKAVVDIQDLKHAV 572
Query: 540 CQAVDVMQAMGSAIRSLFSQVEGMNDLISGVAVVATKEKSILDECEMLLASVAALQVEES 599
AVDVM AM S+I SL S+VE MN +++ + + KE+ +L++C+ L VAA+QV +
Sbjct: 573 SSAVDVMHAMVSSIFSLTSKVEEMNSVMAEMVNITGKEEVLLEQCQGFLTRVAAMQVTDC 632
Query: 600 SLQTHLIQFKQV 611
S++TH+IQ ++
Sbjct: 633 SMKTHIIQLSRL 644
>At5g43160 putative protein
Length = 445
Score = 133 bits (334), Expect = 3e-31
Identities = 138/479 (28%), Positives = 224/479 (45%), Gaps = 83/479 (17%)
Query: 140 TMRSLSVSFQSDIISIPVSKKERPVTSASDRTLR--PASNVAHRQ-GETPSTIRKATPER 196
T ++S SF +++ K+ +P + S+ + R +VA R G T S +++P+R
Sbjct: 5 TAATISPSFNANV------KQNKPPSFPSESSNRRPKTRDVASRYLGGTSSFFHQSSPKR 58
Query: 197 KRSPLKGKNASDQSENSKPVDSLPSRLIDQHRWPSRIGGKVSSSALNRSVDF-------G 249
+SP+ + + S + S P R R S+ + +S + F G
Sbjct: 59 CQSPIVTRPVTPSSVATNRPQSTPRRESLDRREVSKAERMLLTSGRSLFASFQADSFTPG 118
Query: 250 DARMLKTPASGT----GFSSLRRLSLSEEASKPLQRASSDSVRLLSLVASGRTGSEVKSV 305
KT +S T G +L LS++ + LQ + S +SV
Sbjct: 119 TLERRKTTSSATISKSGGGKQEKLKLSDQWPRSLQPSCLSS----------------RSV 162
Query: 306 DDCSLHDLRPHRSSTPHTDRAGLSIAGVRSQSLSAPGSRLPSPSRISVQSTSSSRGVSPS 365
D D R + + L + V ++ +S R+ S + +++ S S G S
Sbjct: 163 D---FTDTRKKLIGSGNGVARALQDSMVSNRPVSR--ERITS---VDLETESVSSGSSNG 214
Query: 366 RSRPSTPPSRGGV-----SPSRIRPTSSSIQ--SNDSVSVLSFIADFRKGKKGAAFIEDA 418
R + P+RG V S R+ P+S ++ S DS SVLS K A +
Sbjct: 215 RGKML--PARGNVVKARVSQDRLEPSSHGLRKISVDS-SVLS--------PKEANCFQKV 263
Query: 419 HQLRLLYNRFMQWRFANARAEAVLYIQNAIVQKTLYNVWIATLSLWESVTRKKINLQQLK 478
+ LL+ + + LYN W + +L+ SV+ K+I +Q LK
Sbjct: 264 SRFDLLH---------------------LVNLRRLYNAWRSISNLYNSVSMKRIEMQHLK 302
Query: 479 LELKLNSVLNDQMAYLDEWATLETDHVDALSGAVEDLEASTLRLPVTRGAMVDIEHLKVA 538
LKL S+LN QM +L+EW ++ +++ +L GA E L+ STL LPV GAMV+++ +K A
Sbjct: 303 QNLKLISILNMQMGHLEEWLVIDRNYMGSLVGAAEALKGSTLCLPVDCGAMVNVQSVKDA 362
Query: 539 ICQAVDVMQAMGSAIRSLFSQVEGMNDLISGVAVVATKEKSILDECEMLLASVAALQVE 597
IC AVDVMQAM S+I L +V ++ L + + V K++ +LD C LL +++ALQV+
Sbjct: 363 ICSAVDVMQAMASSICLLLPKVGKISSLAAELGRVNAKDEGMLDVCRDLLNTISALQVQ 421
Score = 41.6 bits (96), Expect = 0.001
Identities = 77/289 (26%), Positives = 110/289 (37%), Gaps = 90/289 (31%)
Query: 17 VETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGS-RRCPSPNLTRTA 75
V+ +PP +E +N RR +TR+V SRY T S +RC SP +TR
Sbjct: 17 VKQNKPPSFPSESSN--------RRPKTRDVASRYLGGTSSFFHQSSPKRCQSPIVTRPV 68
Query: 76 TPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHN 135
TP+ S RP STP +S L R+V+ +
Sbjct: 69 TPS-----------SVATNRPQ----------STPRRES-----LDRREVS-----KAER 97
Query: 136 LWPSTMRSLSVSFQSDIISIPVSKKERPVTSASDRTLRPASNVAHRQGETPSTIRKATPE 195
+ ++ RSL SFQ+D + P + + R TS+ +TI
Sbjct: 98 MLLTSGRSLFASFQADSFT-PGTLERRKTTSS-------------------ATI------ 131
Query: 196 RKRSPLKGKNASDQSENSKPVDSLPSRLIDQHRWPSRIGGKVSSSALNRSVDFGDARMLK 255
K+ + E K D +WP + SS RSVDF D R K
Sbjct: 132 -------SKSGGGKQEKLKLSD----------QWPRSLQPSCLSS---RSVDFTDTRK-K 170
Query: 256 TPASGTGFSSLRRLSLSEEASKPLQRASSDSVRL-LSLVASGRTGSEVK 303
SG G + R L S +++P+ R SV L V+SG + K
Sbjct: 171 LIGSGNGVA--RALQDSMVSNRPVSRERITSVDLETESVSSGSSNGRGK 217
>At2g44190 unknown protein
Length = 474
Score = 124 bits (310), Expect = 2e-28
Identities = 132/478 (27%), Positives = 213/478 (43%), Gaps = 45/478 (9%)
Query: 138 PSTMRSLSVSFQSDIISIPVSKKERPVTSASDRTLRPASNVAHRQGETPSTIRKATPERK 197
PST R S +S P+S +S+S L ++ + R R + +R
Sbjct: 26 PSTRRPRVREVSSRFMS-PISSSSSSSSSSSAGDLHQLTSNSPRHHHQHQNQRSTSAQRM 84
Query: 198 RSPLKGKNASDQ--SENSKPVDS-LPSRLIDQHRWPSRIGGKVSSSALNRSVDFGDARML 254
R LK + + SE ++ +DS P + +D + P + +D ML
Sbjct: 85 RRQLKMQEGDENRPSETARSLDSPFPLQQVDGGKNPKQHIRSKPLKENGHRLDTPTTAML 144
Query: 255 KTPASGTGFSSLRRLSLSEEASKPLQRASSDSVRLLSLVASGRTGSEVKSVDDCSLHDLR 314
P S + + R L+ S A+ L R+S S LS G + + + + DL
Sbjct: 145 PPP-SRSRLNQQRLLTAS--AATRLLRSSGIS---LSSSTDGEEDNNNREIFKSNGPDLL 198
Query: 315 PHRSSTPHTDRAGLSI--AGVRSQSLSAPGSRLPSPSRISVQSTSSSRGVSPSRSRPSTP 372
P T T + A S+SLS+ + + R S+ S GV S P
Sbjct: 199 P----TIRTQAKAFNTPTASPLSRSLSSDDASMFRDVRASL---SLKNGVGLS-----LP 246
Query: 373 PSRGGVSPSRIRPTSSSIQSNDSVSVLSFIADFRKGKKGAAFIEDAHQLRLLYNRFMQWR 432
P V+P+ S IQ AD +K KK D H L+LL+NR++QWR
Sbjct: 247 P----VAPN------SKIQ-----------ADTKKQKKALGQQADVHSLKLLHNRYLQWR 285
Query: 433 FANARAEAVLYIQNAIVQKTLYNVWIATLSLWESVTRKKINLQQLKLELKLNSVLNDQMA 492
FANA AE Q A ++ Y++ + L +SV RK+I LQ L+ + ++ Q
Sbjct: 286 FANANAEVKTQSQKAQAERMFYSLGLKMSELSDSVQRKRIELQHLQRVKAVTEIVESQTP 345
Query: 493 YLDEWATLETDHVDALSGAVEDLEASTLRLPVTRGAMVDIEHLKVAICQAVDVMQAMGSA 552
L++WA LE + +L E L ++LRLP+ V+ + L A+ A M+ +
Sbjct: 346 SLEQWAVLEDEFSTSLLETTEALLNASLRLPLDSKIKVETKELAEALVVASKSMEGIVQN 405
Query: 553 IRSLFSQVEGMNDLISGVAVVATKEKSILDECEMLLASVAALQVEESSLQTHLIQFKQ 610
I +L + + M L+S +A V+ EK+ +++C + L + Q+EE L++ LIQ ++
Sbjct: 406 IGNLVPKTQEMETLMSELARVSGIEKASVEDCRVALLKTHSSQMEECYLRSQLIQHQK 463
Score = 39.3 bits (90), Expect = 0.007
Identities = 46/170 (27%), Positives = 71/170 (41%), Gaps = 19/170 (11%)
Query: 15 KAVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRT 74
+++E P P + A A + RR R REV+SR+ SP S+ S S T
Sbjct: 7 RSMEHPSTPAINAP---APVPPPSTRRPRVREVSSRFMSPISSSSSSSSSSSAGDLHQLT 63
Query: 75 A-TPAASLQLLPKRSLSAERKR--------PSTPPSPPSRRLSTPVNDSAIDGRLSSRKV 125
+ +P Q +RS SA+R R PS +R L +P +DG + ++
Sbjct: 64 SNSPRHHHQHQNQRSTSAQRMRRQLKMQEGDENRPSETARSLDSPFPLQQVDGGKNPKQH 123
Query: 126 AASR--LPEGHNLWPSTMRSLSVSFQSDIISIPVSKKERPVTSASDRTLR 173
S+ GH L T L +S + +++ SA+ R LR
Sbjct: 124 IRSKPLKENGHRLDTPTTAMLPPPSRSRL-----NQQRLLTASAATRLLR 168
>At2g20815 putative protein
Length = 438
Score = 115 bits (287), Expect = 9e-26
Identities = 113/373 (30%), Positives = 173/373 (46%), Gaps = 39/373 (10%)
Query: 268 RLSLSEEASKPLQRASS--DSVRLLSLVASGRTGSEVKSVDDCSLHDLRPHRSSTPHTDR 325
R S+ E A L RASS +S LL + T SE+ V S L +RSS H +
Sbjct: 42 RFSVDECA---LYRASSRRNSCSLLYESFNDETDSELSDVSCAS--SLSTNRSSWNH--K 94
Query: 326 AGLSIAGVRSQSLSAPGSRLPSPSRISVQS----TSSSRGV------SPSRSRPSTPPSR 375
G+ ++ L+A S+ + +++ Q T+SS+G+ + S SR + S+
Sbjct: 95 PGIKVSSKYLHDLTAKPSKGNNKTKLRSQDDSQRTNSSKGIENRLQRNNSVSRYGSSMSQ 154
Query: 376 GGVSPSRIRPT------SSSIQSNDSVSVLSFIA----DFRKGKKGAAFI---------- 415
+SP R T SS ++ V I FR K + F
Sbjct: 155 WALSPGRSLDTQAVTVPSSKLKPPRGKGVGKLINLGFDFFRSKNKSSPFTSPLKPKTCDT 214
Query: 416 EDAHQLRLLYNRFMQWRFANARAEAVLYIQNAIVQKTLYNVWIATLSLWESVTRKKINLQ 475
E AHQL+L+ NR +QWRF NARA V + + L W + L V +++I LQ
Sbjct: 215 ESAHQLKLMNNRLLQWRFVNARACDVNKNVASQEKNQLLCAWDTLIKLNNLVLQERIKLQ 274
Query: 476 QLKLELKLNSVLNDQMAYLDEWATLETDHVDALSGAVEDLEASTLRLPVTRGAMVDIEHL 535
+ LE+KLN V Q+ +L+ W +E H+ +LS + L + RLP+ GA V++E
Sbjct: 275 KKNLEMKLNYVFLSQVKHLEAWEDMEIQHLSSLSIIRDSLHSVLSRLPLKEGAKVNLESA 334
Query: 536 KVAICQAVDVMQAMGSAIRSLFSQVEGMNDLISGVAVVATKEKSILDECEMLLASVAALQ 595
I A V A+ S + +EG+ L S +A V +EK +L++C LL ++ L+
Sbjct: 335 VSIIKNAEAVTDAIISTVDDYAPTMEGIVPLASQLAEVVVQEKLMLEKCHDLLRMISELE 394
Query: 596 VEESSLQTHLIQF 608
Q+ QF
Sbjct: 395 DCVFPCQSFATQF 407
>At4g30740 putative protein
Length = 91
Score = 114 bits (286), Expect = 1e-25
Identities = 65/91 (71%), Positives = 74/91 (80%), Gaps = 3/91 (3%)
Query: 141 MRSLSVSFQSDIISIPVSKKERPVTSAS-DRTLRPASNVAHR-QGETPSTIRKATPERKR 198
MR LSVSFQSD +S+PVSKKER V+S+S DRTLRP+SN+A + + ET S RK TPERKR
Sbjct: 1 MRILSVSFQSDSVSVPVSKKERLVSSSSGDRTLRPSSNIAQKHKAETTSVSRKPTPERKR 60
Query: 199 SPLKGK-NASDQSENSKPVDSLPSRLIDQHR 228
SPLKGK N SD SENSKPVD SRLI+QHR
Sbjct: 61 SPLKGKNNVSDLSENSKPVDGPHSRLIEQHR 91
>At3g60000 putative protein
Length = 451
Score = 114 bits (286), Expect = 1e-25
Identities = 76/263 (28%), Positives = 129/263 (48%), Gaps = 19/263 (7%)
Query: 345 LPSPSRISVQSTSSSRGVSPSRSRPSTPPSRGGVSPSRIRPTSSSIQSNDSVSVLSFIAD 404
+PS SV S SSS V R S+ SR G+ + P AD
Sbjct: 198 VPSSLNRSVSSPSSSCNV-----RESSSFSRLGLPLPPMAPKVP--------------AD 238
Query: 405 FRKGKKGAAFIEDAHQLRLLYNRFMQWRFANARAEAVLYIQNAIVQKTLYNVWIATLSLW 464
+K +K +ED H L+LL+NR++QWRFANA A+ + +++ L
Sbjct: 239 TKKQRKVTEQLEDVHSLKLLHNRYLQWRFANANAQVKTQTHKTQTETMIHSFGSKISELH 298
Query: 465 ESVTRKKINLQQLKLELKLNSVLNDQMAYLDEWATLETDHVDALSGAVEDLEASTLRLPV 524
+SV RK+I LQ+L L ++ Q L++W+ +E ++ ++S ++ ++LRLP+
Sbjct: 299 DSVQRKRIELQRLLKTKALLAITESQTPCLEQWSAIEEEYSTSVSQTIQAFSNASLRLPL 358
Query: 525 TRGAMVDIEHLKVAICQAVDVMQAMGSAIRSLFSQVEGMNDLISGVAVVATKEKSILDEC 584
MVD + L + A ++ + + + + + M L+S + VA E+S+ + C
Sbjct: 359 DGDIMVDSKQLGDGLVAASKIVDGITQNVGNYMPKAKEMESLLSELTRVARSERSLTENC 418
Query: 585 EMLLASVAALQVEESSLQTHLIQ 607
+ L A Q+EE S+++ LIQ
Sbjct: 419 VVALLKTQASQIEECSMRSQLIQ 441
>At4g25190 putative protein
Length = 443
Score = 101 bits (251), Expect = 1e-21
Identities = 85/318 (26%), Positives = 141/318 (43%), Gaps = 36/318 (11%)
Query: 264 SSLRRLSLSEEASKPLQRASSDSVRLLSLVASGRTGSEVKSVDDCSLHDLRPHRSSTPHT 323
+SL + S +S P SS V + ++ R+ + S D S ++ P R+S +
Sbjct: 33 ASLSSSTSSSSSSSPSN--SSKRVMITRSQSTTRSSRPIGSSDSKSGENIIPARNSASRS 90
Query: 324 DRAGLSIAGVRSQSLSAPGSRLPSPSRISVQSTSSSRGVSPSRSRPS------------- 370
+S + L +R S +S +SSRGV P S PS
Sbjct: 91 QEIN------NGRSRESFARYLEQRTRGSPRSNASSRGVKPGASSPSAWALSPGRLSTMK 144
Query: 371 TPPSRGG------VSPSRIRPTSSSIQSNDSVSVLSFIADFRKGKKGAAFIE-DAHQLRL 423
TP S ++P + + I+S +V + F KK + E D H+ R+
Sbjct: 145 TPLSSSAPTTSMCMTPPESPVSKAKIRSGGGGAVAGVLKYFMAQKKVSPVQEEDYHRFRI 204
Query: 424 LYNRFMQWRFANARAEAVLYIQNAIVQKTLYNVWIATLSLWESVTRKKINLQQLKLELKL 483
NR +QWRF NAR EA + V+ L+ VW+ + V I +Q+L+ ++K+
Sbjct: 205 FQNRLLQWRFVNARTEATMANLKINVEDQLFWVWLRIYKMRNYVVENLIEIQRLRQDIKV 264
Query: 484 NSVLNDQMAYLDEWATLETDHVDALSGAVEDLEASTLRLPVTRGA--------MVDIEHL 535
VL+ QM L+EW+ ++ + +ALS L A ++RLP+ GA +D+ +
Sbjct: 265 REVLSLQMPLLNEWSKIDAKNSEALSKLTRKLHALSVRLPLVHGATKLIIWKLQIDMVSI 324
Query: 536 KVAICQAVDVMQAMGSAI 553
+ A++VM + I
Sbjct: 325 HEEMVIAIEVMDEIEDVI 342
Score = 33.5 bits (75), Expect = 0.36
Identities = 31/120 (25%), Positives = 52/120 (42%), Gaps = 7/120 (5%)
Query: 65 RCPSPNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRK 124
R PSPN R+ T ++S+ L + S S+ S PS + + SSR
Sbjct: 9 RPPSPNNNRSRTISSSISLPVSLNASLSSSTSSSSSSSPSNSSKRVMITRSQSTTRSSRP 68
Query: 125 VAASRLPEGHNLWP---STMRSLSV----SFQSDIISIPVSKKERPVTSASDRTLRPASN 177
+ +S G N+ P S RS + S +S + + P ++AS R ++P ++
Sbjct: 69 IGSSDSKSGENIIPARNSASRSQEINNGRSRESFARYLEQRTRGSPRSNASSRGVKPGAS 128
>At2g29210 proline-rich protein like
Length = 878
Score = 58.9 bits (141), Expect = 8e-09
Identities = 101/376 (26%), Positives = 148/376 (38%), Gaps = 64/376 (17%)
Query: 41 RSRTREVTSRYKSPTPPS-----TPSGSRRCPSPNLTRTATPAASLQLLPKRSLSAERKR 95
RSR+R R++ PT PS RR PSP R +P+ + S A R R
Sbjct: 282 RSRSRSPIRRHRRPTHEGRRQSPAPSRRRRSPSPPARRRRSPSPPARRRRSPSPPARRHR 341
Query: 96 PSTP-------PSPPSRR-LSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPSTM----RS 143
TP PSPP+RR S P +R+ + P PS + RS
Sbjct: 342 SPTPPARQRRSPSPPARRHRSPPPARRRRSPSPPARRRRSPSPPARRRRSPSPLYRRNRS 401
Query: 144 LSVSFQSDIISIPVSKKER---------PVTSASDRT--LRPASNVAHRQGETPSTIRKA 192
S ++ + P++K+ R PV D T P+ ++ R P R
Sbjct: 402 PSPLYRRNRSRSPLAKRGRSDSPGRSPSPVARLRDPTGARLPSPSIEQRLPSPPVAQRLP 461
Query: 193 TPERKRSPLKGKNASDQSENSKPVDS-LPSRLIDQHRWPSRIGGKVSSSALNRSVDFGDA 251
+P +R+ L + + + P + LPS P RIGG S A N
Sbjct: 462 SPPPRRAGLPSPPPAQRLPSPPPRRAGLPS--------PMRIGG---SHAANH------- 503
Query: 252 RMLKTPASGTGFSSLRRLSLSEEASKPLQRASSDSVRLLSLVASGRTGSEVKSVDDCSLH 311
L++P+ + R+ L + + + D R +SL GR S D S+
Sbjct: 504 --LESPSPSSLSPPGRKKVLPSPPVRRRRSLTPDEER-VSLSQGGRHTSPSHIKQDGSMS 560
Query: 312 DLRPHRSSTPHTDRAGLSIAGVRSQSLSAP-GSRLPSP-SRISVQSTSSSRGVSPSRSRP 369
+R S+P + R Q +P R P+P +R S +S+S+SR SP R R
Sbjct: 561 PVRGRGKSSPSS----------RHQKARSPVRRRSPTPVNRRSRRSSSASR--SPDRRRR 608
Query: 370 STPPSRGGVSPSRIRP 385
+P S S SR P
Sbjct: 609 RSPSSSRSPSRSRSPP 624
Score = 48.5 bits (114), Expect = 1e-05
Identities = 86/358 (24%), Positives = 132/358 (36%), Gaps = 80/358 (22%)
Query: 19 TPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTP-------PSTPSGSRRCPSPNL 71
+P PP + + ARR R+ R +SP+P PS P+ RR PSP
Sbjct: 342 SPTPPA-----RQRRSPSPPARRHRSPPPARRRRSPSPPARRRRSPSPPARRRRSPSPLY 396
Query: 72 TRTATPAASLQLLPKRSLSAERKRPSTPPSPPS--RRLSTPVN----DSAIDGRLSSRKV 125
R +P+ + RS A+R R +P PS RL P +I+ RL S V
Sbjct: 397 RRNRSPSPLYRRNRSRSPLAKRGRSDSPGRSPSPVARLRDPTGARLPSPSIEQRLPSPPV 456
Query: 126 A------------------ASRLPE-------------------GHNLWPSTMRSLSVSF 148
A A RLP ++L + SLS
Sbjct: 457 AQRLPSPPPRRAGLPSPPPAQRLPSPPPRRAGLPSPMRIGGSHAANHLESPSPSSLSPPG 516
Query: 149 QSDIISIPVSKKERPVTSASDRTLRPASNVAHRQGETPSTIRKATPERKRSPLKGKNASD 208
+ ++ P ++ R +T +R V+ QG ++ + SP++G+ S
Sbjct: 517 RKKVLPSPPVRRRRSLTPDEER-------VSLSQGGRHTSPSHIKQDGSMSPVRGRGKSS 569
Query: 209 QSENSKPVDSLPSRLIDQHRWPSRIGGKV-SSSALNRSVDFGDARMLKTPASGTGFSSLR 267
S + S P R R P+ + + SS+ +RS D R ++P+S S R
Sbjct: 570 PSSRHQKARS-PVR----RRSPTPVNRRSRRSSSASRS---PDRRRRRSPSSSRSPSRSR 621
Query: 268 RLSLSEEASKPLQRASSDSVRLLSLVASGRTGSEVKSVDDCSLHDLRPHRSSTPHTDR 325
+ + P R R + GR E V + L R+S P TD+
Sbjct: 622 SPPVLHRSPSPRGRKHQRERR-----SPGRLSEEQDRVQNSKL----LKRTSVPDTDK 670
Score = 32.0 bits (71), Expect = 1.0
Identities = 55/205 (26%), Positives = 78/205 (37%), Gaps = 19/205 (9%)
Query: 192 ATPERKRSPLKGKNASDQSE-NSKPVDSLPSRLIDQHRWPSRIGGKVSSSALNRSVDF-- 248
A P R R P G+ A +++ + D +P R D R P R S S N
Sbjct: 173 AKPSRDR-PEDGRRADEKNGVKERRRDLIPPRRGDASRSPLRGSRSRSISKTNSGSKSYS 231
Query: 249 GDARMLKTPASGTGFSSLRRLSLSEEA----SKPLQRASSDSVRLLSLVASGRTGSEVKS 304
G+ + T S S R+ LS S+ ++R+ S R + R+ S ++
Sbjct: 232 GERKSRSTSQSSDASISPRKRRLSNSRRRSRSRSVRRSLSPRRRRIHSPFRSRSRSPIRR 291
Query: 305 ----VDDCSLHDLRPHRS----STPHTDRAGLSIAGVRSQSLSAPGSRLPSPSRISVQST 356
+ P R S P R S R +S S P R SP+ + Q
Sbjct: 292 HRRPTHEGRRQSPAPSRRRRSPSPPARRRRSPSPPARRRRSPSPPARRHRSPTPPARQRR 351
Query: 357 SSSRGVSPSRSRPSTPPSRGGVSPS 381
S S P+R S PP+R SPS
Sbjct: 352 SPS---PPARRHRSPPPARRRRSPS 373
>At3g09000 unknown protein
Length = 541
Score = 57.8 bits (138), Expect = 2e-08
Identities = 99/389 (25%), Positives = 146/389 (37%), Gaps = 72/389 (18%)
Query: 6 SQQASSRRLKAVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRR 65
S SSR TP + + T+T+R TR SR +PT +T + +R
Sbjct: 148 SSSGSSRSTSRPATPT-------RRSTTPTTSTSRPVTTRASNSRSSTPTSRATLTAARA 200
Query: 66 CPSPNLTRTATPAA--SLQLLPKRS----LSAERKRPSTPPSPPSRRLSTPVNDSAIDGR 119
S RT T ++ + P RS SA K+P + P+ P+RR STP S + +
Sbjct: 201 TTSTAAPRTTTTSSGSARSATPTRSNPRPSSASSKKPVSRPATPTRRPSTPTGPSIVSSK 260
Query: 120 LSSRKVAASRLPEGHNLWPSTMRSLSVSFQSDIISIPVSKKERP----------VTSASD 169
SR + S + PS S S + S S P E P T+ +D
Sbjct: 261 APSRGTSPSPTVNSLSKAPSRGTSPSPTLNS---SRPWKPPEMPGFSLEAPPNLRTTLAD 317
Query: 170 RTL---RPASNVAHRQGETPSTIRK----------------ATPERKRSPLKGKNASDQS 210
R + R VA G +I + +P R R+P+ N S
Sbjct: 318 RPVSASRGRPGVASAPGSRSGSIERGGGPTSGGSGNARRQSCSPSRGRAPIGNTNGSLTG 377
Query: 211 ENSKPVDSLPSRLIDQHRWPSRIGGKVSSSALNRSVDFGDARMLKTPASGTGFSSLRRLS 270
+ S D + P +G K+ +N G R+ + G+G SS S
Sbjct: 378 VRGRAKASNGGSGCD-NLSPVAMGNKMVERVVNMR-KLGPPRLTENGGRGSGKSSSAFNS 435
Query: 271 LSEEASKPLQRASSDSVRLLSLVASGRTGSEVKSVDDCSLHDLRPHRSSTPHTDRAGLSI 330
L + L ++S D + G TG +LRP + P + S+
Sbjct: 436 LG--YGRNLSKSSIDMAIRHMDIRRGMTG------------NLRPLVTKVPAS-----SM 476
Query: 331 AGVRSQSLSAPGSRLPSPSRISVQSTSSS 359
VRS+ PGS SP ++ ST SS
Sbjct: 477 YSVRSR----PGSVSSSP--VATSSTVSS 499
Score = 38.9 bits (89), Expect = 0.009
Identities = 98/435 (22%), Positives = 167/435 (37%), Gaps = 54/435 (12%)
Query: 6 SQQASSRRLKAVETPRPPLVLAEKNNA-------ATATATARRSRTREVTSRYKSPTPPS 58
S+ ASS+R T + +E + T + R V +++ +P
Sbjct: 53 SETASSQRYPLRRTAAENFLYSENEKSDYDWLLTPPGTPQFEKESHRSVMNQHDAPNSRP 112
Query: 59 TPSGSR--RCP----SPNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVN 112
T SR C S N + T ++S+ L +R S+ R ++ P+ P+RR +TP
Sbjct: 113 TVLKSRLGNCREDIVSGNNNKPQTSSSSVAGL-RRPSSSGSSRSTSRPATPTRRSTTPTT 171
Query: 113 DSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQSDIISIPVSKKERPVTSASDRTL 172
S+ + +R + P++ +L+ + + + P + +S S R+
Sbjct: 172 --------STSRPVTTRASNSRSSTPTSRATLTAARATTSTAAP---RTTTTSSGSARSA 220
Query: 173 RPA-SNVAHRQGETPSTI-RKATPERKRSPLKGKNASDQSENSKPVDSLPSRLIDQHRWP 230
P SN + + R ATP R+ S G + S+ P+ + + P
Sbjct: 221 TPTRSNPRPSSASSKKPVSRPATPTRRPSTPTGPSIVSSKAPSRGTSPSPT-VNSLSKAP 279
Query: 231 SRIGGKVSSSALNRSVDFGDARM----LKTPASGTGFSSLRRLSLSE----EASKPLQRA 282
SR G S LN S + M L+ P + + R +S S AS P R
Sbjct: 280 SR--GTSPSPTLNSSRPWKPPEMPGFSLEAPPNLRTTLADRPVSASRGRPGVASAPGSR- 336
Query: 283 SSDSVRLLSLVASGRTGSEVKSVDDCSLHDLRPHRSSTPHTDRAGLSIAGVRSQS-LSAP 341
S S+ SG +G+ + CS P R P + G S+ GVR ++ S
Sbjct: 337 -SGSIERGGGPTSGGSGNARR--QSCS-----PSRGRAPIGNTNG-SLTGVRGRAKASNG 387
Query: 342 GSRLPSPSRISVQSTSSSRGVSPSRSRPSTPPSRGGVSPSRIRPTSSSI-----QSNDSV 396
GS + S +++ + R V+ + P GG + +S+ S S+
Sbjct: 388 GSGCDNLSPVAMGNKMVERVVNMRKLGPPRLTENGGRGSGKSSSAFNSLGYGRNLSKSSI 447
Query: 397 SVLSFIADFRKGKKG 411
+ D R+G G
Sbjct: 448 DMAIRHMDIRRGMTG 462
>At2g40070 En/Spm-like transposon protein
Length = 510
Score = 57.0 bits (136), Expect = 3e-08
Identities = 108/397 (27%), Positives = 160/397 (40%), Gaps = 54/397 (13%)
Query: 29 KNNAATATATARRSRTRE-----VTSRYKSPTPP-STPSGSRRCPSPNL---TRTATPAA 79
K+ AT T+ S T +TSR ++ +P S+ SG+ R PS + +R ATP
Sbjct: 105 KSRPATLTSRLANSSTESAARNHLTSRQQTSSPGLSSSSGASRRPSSSGGPGSRPATPTG 164
Query: 80 SLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPS 139
L S S+ RPSTP S + +S+ S + R + V+A+ P
Sbjct: 165 RSSTLTANSKSS---RPSTPTSRAT--VSSATRPSLTNSRST---VSATTKP-------- 208
Query: 140 TMRSLSVSFQSDIISIPVSKKERPVTSASDRTLRPASNVAHRQGETPSTIRK-ATPERKR 198
T S S S S ++ SK P TS T R A +V TPST K A P R
Sbjct: 209 TPMSRSTSLSSSRLTPTASK---PTTS----TARSAGSVTR---STPSTTTKSAGPSRST 258
Query: 199 SPLKGKNASDQSENSKPVDSLPSRLIDQHRWPSRIGGKVSSSALNRSVDFGDARMLKTPA 258
+PL A + S+P PS+ I + P+R +S+A + +PA
Sbjct: 259 TPLSRSTARSSTPTSRPT-LPPSKTISRSSTPTRRPIASASAATTTANPTISQIKPSSPA 317
Query: 259 SGTGFSSLRRLSLSEEASKPLQRASSDSVRLLSLVASGRTGSEVKSVDDCSLHDLRPHRS 318
+ + L RA+S +VR S G +++ +L P R
Sbjct: 318 PAKPMP-------TPSKNPALSRAASPTVRSRPWKPSDMPGFSLETPP--NLRTTLPERP 368
Query: 319 STPHTDRAGLSIAGVRSQSLSAPGSRLPSPSRISVQSTSSSRGVSPSRSRPSTPP--SRG 376
+ R G + RS S+ G P R QS S SRG +P S S+ P +RG
Sbjct: 369 LSATRGRPGAPSS--RSGSVEPGG---PPGGRPRRQSCSPSRGRAPMYSSGSSVPAVNRG 423
Query: 377 -GVSPSRIRPTSSSIQSNDSVSVLSFIADFRKGKKGA 412
+ + P + + V + +A R KG+
Sbjct: 424 YSKASDNVSPVMMGTKMVERVINMRKLAPPRSDDKGS 460
Score = 48.9 bits (115), Expect = 8e-06
Identities = 78/322 (24%), Positives = 128/322 (39%), Gaps = 42/322 (13%)
Query: 71 LTRTATPA-ASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASR 129
LT TP SL++ R++ ++ + P+ + RL+ +SA L+SR+ +S
Sbjct: 78 LTPPGTPLFPSLEMESHRTMMSQTGDSKSRPATLTSRLANSSTESAARNHLTSRQQTSS- 136
Query: 130 LPEGHNLWPSTMRSLSVSFQSDIISIPVSKKERPVTSASDRTLRPASNVAHRQGETPST- 188
P S S + P S+ P +S T A+ + PST
Sbjct: 137 --------PGLSSSSGASRRPSSSGGPGSRPATPTGRSSTLT-------ANSKSSRPSTP 181
Query: 189 IRKATPERKRSPLKGKNASDQSENSKPVDSLPSRLIDQHRWPSRIGGKVSSSALNRSVDF 248
+AT P + S S +KP S + R +S+A +
Sbjct: 182 TSRATVSSATRPSLTNSRSTVSATTKPTPMSRSTSLSSSRLTPTASKPTTSTARSAG--- 238
Query: 249 GDARMLKTPASGTGFSSLRRLSLSEEASKPLQRASSDSVRLLSLVASGRTGSEVKSVDDC 308
+ TP++ T + R ++ PL R+++ S S S T K++
Sbjct: 239 --SVTRSTPSTTTKSAGPSR------STTPLSRSTARS----STPTSRPTLPPSKTISRS 286
Query: 309 SLHDLRPHRSSTPHTDRAGLSIAGVRSQSLSAPGSRLPSPSRISVQSTSSSRGVSPS-RS 367
S RP S++ T A +I+ ++ S AP +P+PS+ + + SR SP+ RS
Sbjct: 287 STPTRRPIASASAATTTANPTISQIKPSS-PAPAKPMPTPSK----NPALSRAASPTVRS 341
Query: 368 RPSTPPSRGGVS---PSRIRPT 386
RP P G S P +R T
Sbjct: 342 RPWKPSDMPGFSLETPPNLRTT 363
Score = 43.5 bits (101), Expect = 3e-04
Identities = 53/207 (25%), Positives = 84/207 (39%), Gaps = 24/207 (11%)
Query: 10 SSRRLKAVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSP 69
+SR + T P+ + +++ T TA + T T+R STPS + + P
Sbjct: 197 NSRSTVSATTKPTPMSRSTSLSSSRLTPTASKPTTS--TARSAGSVTRSTPSTTTKSAGP 254
Query: 70 NLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASR 129
+R+ TP L RS S RP+ PPS R STP +R+ AS
Sbjct: 255 --SRSTTP---LSRSTARS-STPTSRPTLPPSKTISRSSTP-----------TRRPIASA 297
Query: 130 LPEGHNLWPSTMRSLSVSFQSDIISIPVSKKERPVTSASDRTLR----PASNVAHRQGET 185
P T+ + S + +P K ++ A+ T+R S++ ET
Sbjct: 298 SAATTTANP-TISQIKPSSPAPAKPMPTPSKNPALSRAASPTVRSRPWKPSDMPGFSLET 356
Query: 186 PSTIRKATPERKRSPLKGKNASDQSEN 212
P +R PER S +G+ + S +
Sbjct: 357 PPNLRTTLPERPLSATRGRPGAPSSRS 383
>At5g01280 putative protein
Length = 460
Score = 50.1 bits (118), Expect = 4e-06
Identities = 100/398 (25%), Positives = 159/398 (39%), Gaps = 74/398 (18%)
Query: 6 SQQASSRRLKAVETP-RPPLVLAEKNNAATATATARRSR----TREVTSRYKSPTPPSTP 60
S +SSR TP R A++ + T+ AT+ +R + TS +S + PS+
Sbjct: 86 SSSSSSRSTSRPPTPTRKSKTPAKRPSTPTSRATSTTTRATLTSSSTTSSTRSWSRPSSS 145
Query: 61 SGSRRC-----------PSPNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLST 109
SG+ P+ + + T +A+ R +SA +P + S P+RR ST
Sbjct: 146 SGTGTSRVTLTAARATRPTTSTDQQTTGSATSTRSNNRPMSAPNSKPGSRSSTPTRRPST 205
Query: 110 PVNDSAIDGRLSSRKVA-------ASRLPEGHNLWPSTMRSLSVSFQSDIISIPVSKKER 162
P S + ++ ++ AS + P M SV S ++ + +R
Sbjct: 206 PNGSSTVLRSKPTKPLSKPALSLEASPIVRSRPWEPYEMPGFSVEAPS---NLRTTLPDR 262
Query: 163 PVTSASDRTLRPASNVAHRQGETPSTIRK---ATPERKRSPLKGKNASDQSENSKPVDSL 219
P T++S RT ++ + R T + K +P R R+P N ++
Sbjct: 263 PQTASSSRTRAFDASSSSRSASTERDVAKRQSCSPSRSRAP-----------NGNVNGAV 311
Query: 220 PSRLIDQHRWPSRIGGKVSSSAL--NRSVD-FGDARMLKTPASGTGFSSLRRLSLSEEAS 276
PS L Q + G++ S A N+ V+ + R L TP L+E S
Sbjct: 312 PS-LRGQRAKTNNDDGRLISHAAKGNQKVEKVVNMRKLATP------------RLTESGS 358
Query: 277 KPLQRASSDSVRLLSLVASGRTG---SEVKSVDDCSLHDLRPHRSSTPHTDRAGLSIAGV 333
+ L DS S SG G + KS D +L H D S+AG
Sbjct: 359 RRLGGGGGDSSAGKSSSGSGGFGFGRNLSKSSIDMALR----------HMDVRKGSMAGN 408
Query: 334 RSQSLSAPGSRLPSPSRISVQSTSSSRGVSPSRSRPST 371
S+ ++ P+ S SV+S +R VS SRS S+
Sbjct: 409 FRHSV----TKAPATSVYSVRS-CRNRPVSSSRSSESS 441
Score = 36.2 bits (82), Expect = 0.055
Identities = 73/352 (20%), Positives = 127/352 (35%), Gaps = 48/352 (13%)
Query: 86 KRSLSAERKRPSTPPSPPSRRLSTPVN-----DSAIDGRLSSRKVAASRLPEGHNLWPST 140
+R S+ R ++ P P+R+ TP S + + +S W +
Sbjct: 83 RRPSSSSSSRSTSRPPTPTRKSKTPAKRPSTPTSRATSTTTRATLTSSSTTSSTRSW--S 140
Query: 141 MRSLSVSFQSDIISIPVSKKERPVTSASDRTLRPASNVA--HRQGETPST---IRKATPE 195
S S + +++ ++ RP TS +T A++ +R P++ R +TP
Sbjct: 141 RPSSSSGTGTSRVTLTAARATRPTTSTDQQTTGSATSTRSNNRPMSAPNSKPGSRSSTPT 200
Query: 196 RKRSPLKGKNASDQSENSKPVDSLPSRLIDQHRWPSRIGGKVSSSALNRSVDFGDARMLK 255
R+ S G + +S+ +KP+ S+ + +S + RS +
Sbjct: 201 RRPSTPNGSSTVLRSKPTKPL--------------SKPALSLEASPIVRSRPW------- 239
Query: 256 TPASGTGFSSLRRLSLSEEASKPLQRASSDSVRLLSLVASGRTGSEVKSVDDCSLHDLRP 315
P GFS +L Q ASS R +S R+ S + V P
Sbjct: 240 EPYEMPGFSVEAPSNLRTTLPDRPQTASSSRTRAFDASSSSRSASTERDV--AKRQSCSP 297
Query: 316 HRSSTPHTDRAGL--SIAGVRSQSLSAPGSRLPSPSR--ISVQSTSSSRGVSPSRSRPST 371
RS P+ + G S+ G R+++ + G + ++ V+ + R ++ R S
Sbjct: 298 SRSRAPNGNVNGAVPSLRGQRAKTNNDDGRLISHAAKGNQKVEKVVNMRKLATPRLTESG 357
Query: 372 PPSRGGVSPSRIRPTSSSIQ---------SNDSVSVLSFIADFRKGKKGAAF 414
GG SSS S S+ + D RKG F
Sbjct: 358 SRRLGGGGGDSSAGKSSSGSGGFGFGRNLSKSSIDMALRHMDVRKGSMAGNF 409
Score = 33.9 bits (76), Expect = 0.27
Identities = 25/78 (32%), Positives = 40/78 (51%), Gaps = 10/78 (12%)
Query: 316 HRSSTPHTDRAGLSIAGVRSQSLSAPGSRLPSPSRISVQSTSSSRGVSPSRSRPSTPPSR 375
H++++ H+ + I S S S SR P+P+R + +P++ RPSTP SR
Sbjct: 68 HQTTSLHSSSSVSGIRRPSSSSSSRSTSRPPTPTR---------KSKTPAK-RPSTPTSR 117
Query: 376 GGVSPSRIRPTSSSIQSN 393
+ +R TSSS S+
Sbjct: 118 ATSTTTRATLTSSSTTSS 135
>At1g16610 arginine/serine-rich protein
Length = 414
Score = 45.4 bits (106), Expect = 9e-05
Identities = 61/211 (28%), Positives = 82/211 (37%), Gaps = 19/211 (9%)
Query: 27 AEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCP-SPNLTRTATPAASLQLLP 85
AEK+ R ++ R +SP P S RR P SP+ R +P P
Sbjct: 206 AEKDGGPRRPRETSPQRKTGLSPRRRSPLPRRGLSPRRRSPDSPHRRRPGSPIRRRGDTP 265
Query: 86 KRSLSAERKRPSTPPSPPSRRLSTPVNDS--AIDGRLSSRKVAASRLPEGHNLWPSTMRS 143
R A R +P SPP RR +P S I G R+ S LP P R
Sbjct: 266 PRRRPASPSRGRSPSSPPPRRYRSPPRGSPRRIRGSPVRRR---SPLPLRRRS-PPPRRL 321
Query: 144 LSVSFQSDIISIPVSKKERPVTS-ASDRTLRPASNVAHRQGETPSTIRKATPER------ 196
S +S I S RP S +S + R A R+G + S +P R
Sbjct: 322 RSPPRRSPIRRRSRSPIRRPGRSRSSSISPRKGRGPAGRRGRSSSYSSSPSPRRIPRKIS 381
Query: 197 ----KRSPLKGK-NASDQSENSKPVDSLPSR 222
+ PL+GK ++S+ S +S P P R
Sbjct: 382 RSRSPKRPLRGKRSSSNSSSSSSPPPPPPPR 412
Score = 45.4 bits (106), Expect = 9e-05
Identities = 100/418 (23%), Positives = 156/418 (36%), Gaps = 90/418 (21%)
Query: 39 ARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPAASLQLL--PKRSLSAERKRP 96
A+ SR R S S + S+ S S PS +++R+ + + SL P RS+S+ + P
Sbjct: 2 AKPSRGRRSPSVSGSSSRSSSRSRSGSSPSRSISRSRSRSRSLSSSSSPSRSVSSGSRSP 61
Query: 97 --------------STPPSPPSRRLSTP-------------------VNDSAID------ 117
+PP PPS+ S+P VN++ +
Sbjct: 62 PRRGKSPAGPARRGRSPPPPPSKGASSPSKKAVQESLVLHVDSLSRNVNEAHLKEIFGNF 121
Query: 118 GRLSSRKVAASR---LPEGHNLWPSTMRSLSVSFQ-------------SDIISIP----V 157
G + ++A R LP GH R+ + Q ++P V
Sbjct: 122 GEVIHVEIAMDRAVNLPRGHGYVEFKARADAEKAQLYMDGAQIDGKVVKATFTLPPRQKV 181
Query: 158 SKKERPVTSASDRTLRPASNVA---------HRQGETPSTIRKATPERKRSPLKGKNASD 208
S +PV++A R + N A R ET + R+RSPL + S
Sbjct: 182 SSPPKPVSAAPKRDAPKSDNAAADAEKDGGPRRPRETSPQRKTGLSPRRRSPLPRRGLSP 241
Query: 209 QSEN-SKPVDSLPSRLIDQHRWPSRIGGKVSSSALNRSVDFGDARMLKTPASGTGFSSLR 267
+ + P P I + R + + +S + RS R ++P G+ +R
Sbjct: 242 RRRSPDSPHRRRPGSPI-RRRGDTPPRRRPASPSRGRSPSSPPPRRYRSPPRGSP-RRIR 299
Query: 268 RLSLSEEASKPLQRASSDSVRLLSLVASG----RTGSEVKSVDDCSLHDLRPHRS-STPH 322
+ + PL+R S RL S R+ S ++ RP RS S+
Sbjct: 300 GSPVRRRSPLPLRRRSPPPRRLRSPPRRSPIRRRSRSPIR----------RPGRSRSSSI 349
Query: 323 TDRAGLSIAGVRSQSLSAPGSRLPSPSRISVQSTSSSRGVSPSRSRPSTPPSRGGVSP 380
+ R G AG R +S S S PSP RI + + S P R + S+ S SP
Sbjct: 350 SPRKGRGPAGRRGRSSSYSSS--PSPRRIPRKISRSRSPKRPLRGKRSSSNSSSSSSP 405
Score = 42.4 bits (98), Expect = 8e-04
Identities = 96/405 (23%), Positives = 152/405 (36%), Gaps = 89/405 (21%)
Query: 48 TSRYKSPTPPSTPSGSRRCPSPNLTRTATPAASL----QLLPKRSLS--AERKRPSTPPS 101
+SR +S + PS R S +L+ +++P+ S+ + P+R S +R +PP
Sbjct: 21 SSRSRSGSSPSRSISRSRSRSRSLSSSSSPSRSVSSGSRSPPRRGKSPAGPARRGRSPPP 80
Query: 102 PPSRRLSTP-------------------VNDSAID------GRLSSRKVAASR---LPEG 133
PPS+ S+P VN++ + G + ++A R LP G
Sbjct: 81 PPSKGASSPSKKAVQESLVLHVDSLSRNVNEAHLKEIFGNFGEVIHVEIAMDRAVNLPRG 140
Query: 134 HNLWPSTMRSLSVSFQ-------------SDIISIP----VSKKERPVTSASDRTLRPAS 176
H R+ + Q ++P VS +PV++A R +
Sbjct: 141 HGYVEFKARADAEKAQLYMDGAQIDGKVVKATFTLPPRQKVSSPPKPVSAAPKRDAPKSD 200
Query: 177 NVAH--RQGETPSTIRKATPERK-------RSPLKGKNASDQSEN-SKPVDSLPSRLIDQ 226
N A + P R+ +P+RK RSPL + S + + P P I +
Sbjct: 201 NAAADAEKDGGPRRPRETSPQRKTGLSPRRRSPLPRRGLSPRRRSPDSPHRRRPGSPI-R 259
Query: 227 HRWPSRIGGKVSSSALNRSVDFGDARMLKTPASGTGFSSLRRLSLSEEASKPLQRASSDS 286
R + + +S + RS R ++P G+ +R + + PL+R S
Sbjct: 260 RRGDTPPRRRPASPSRGRSPSSPPPRRYRSPPRGSP-RRIRGSPVRRRSPLPLRRRSPPP 318
Query: 287 VRLLSLVASG----RTGSEVKSVDDCSLHDLRPHRS-STPHTDRAGLSIAGVRSQSLSAP 341
RL S R+ S ++ RP RS S+ + R G AG R +S S
Sbjct: 319 RRLRSPPRRSPIRRRSRSPIR----------RPGRSRSSSISPRKGRGPAGRRGRSSSYS 368
Query: 342 GSRLP-----------SPSRISVQSTSSSRGVSPSRSRPSTPPSR 375
S P SP R SSS S S P PP +
Sbjct: 369 SSPSPRRIPRKISRSRSPKRPLRGKRSSSNSSSSSSPPPPPPPRK 413
Score = 37.4 bits (85), Expect = 0.025
Identities = 72/271 (26%), Positives = 102/271 (37%), Gaps = 30/271 (11%)
Query: 139 STMRSLSVSFQSDIISIPVSKKERPVTSAS-DRTLRPASNVAHRQGETPSTIRKATPERK 197
S+ RS S S S IS S+ +S+S R++ S R+G++P+ A R
Sbjct: 20 SSSRSRSGSSPSRSISRSRSRSRSLSSSSSPSRSVSSGSRSPPRRGKSPAG--PARRGRS 77
Query: 198 RSPLKGKNASDQSENSKP------VDSLPSRLIDQHRWPSRIGGKVSSSALNRSVDFGDA 251
P K AS S+ + VDSL + + H I G V+
Sbjct: 78 PPPPPSKGASSPSKKAVQESLVLHVDSLSRNVNEAHL--KEIFGNFGEVI---HVEIAMD 132
Query: 252 RMLKTPASGTGFSSLRRLSLSEEASKPLQRASSDSVRL---LSLVASGRTGSEVKSVDDC 308
R + P G G+ + + +E+A + A D + +L + S K V
Sbjct: 133 RAVNLPR-GHGYVEFKARADAEKAQLYMDGAQIDGKVVKATFTLPPRQKVSSPPKPVSAA 191
Query: 309 SLHDLRPHRSSTPHTDRAG-----LSIAGVRSQSLSAPGSRLP------SPSRISVQSTS 357
D ++ ++ G + R LS P R P SP R S S
Sbjct: 192 PKRDAPKSDNAAADAEKDGGPRRPRETSPQRKTGLS-PRRRSPLPRRGLSPRRRSPDSPH 250
Query: 358 SSRGVSPSRSRPSTPPSRGGVSPSRIRPTSS 388
R SP R R TPP R SPSR R SS
Sbjct: 251 RRRPGSPIRRRGDTPPRRRPASPSRGRSPSS 281
Score = 36.6 bits (83), Expect = 0.042
Identities = 29/74 (39%), Positives = 37/74 (49%), Gaps = 5/74 (6%)
Query: 329 SIAGVRSQSLSAPGSRLPSPSRISVQSTSSSRGVSPS----RSR-PSTPPSRGGVSPSRI 383
SI+ RS+S S S PS S S + RG SP+ R R P PPS+G SPS+
Sbjct: 33 SISRSRSRSRSLSSSSSPSRSVSSGSRSPPRRGKSPAGPARRGRSPPPPPSKGASSPSKK 92
Query: 384 RPTSSSIQSNDSVS 397
S + DS+S
Sbjct: 93 AVQESLVLHVDSLS 106
Score = 31.2 bits (69), Expect = 1.8
Identities = 50/214 (23%), Positives = 80/214 (37%), Gaps = 30/214 (14%)
Query: 192 ATPERKRSPLKGKNASDQSENSKPVDSLPSRLIDQHRWPSRIGGKVSSSALNRSVDFGD- 250
A P R R +S +S + S PSR I + R SR SSS+ +RSV G
Sbjct: 2 AKPSRGRRSPSVSGSSSRSSSRSRSGSSPSRSISRSR--SRSRSLSSSSSPSRSVSSGSR 59
Query: 251 --ARMLKTPASGTGFSSLRRLSLSEEASKPLQRASSDSVRLLSLVASGRTGSEVKSVDDC 308
R K+PA S+ AS P ++A +S+ L S ++V++
Sbjct: 60 SPPRRGKSPAGPARRGRSPPPPPSKGASSPSKKAVQESLVL-------HVDSLSRNVNEA 112
Query: 309 SLHDLRPHRSSTPHTDRAGLSIAGVRSQSLSAPGSRLPSPSRISVQSTS----------- 357
L ++ + H + IA R+ +L + +R +
Sbjct: 113 HLKEIFGNFGEVIHVE-----IAMDRAVNLPRGHGYVEFKARADAEKAQLYMDGAQIDGK 167
Query: 358 --SSRGVSPSRSRPSTPPSRGGVSPSRIRPTSSS 389
+ P R + S+PP +P R P S +
Sbjct: 168 VVKATFTLPPRQKVSSPPKPVSAAPKRDAPKSDN 201
Score = 28.9 bits (63), Expect = 8.8
Identities = 25/53 (47%), Positives = 27/53 (50%), Gaps = 2/53 (3%)
Query: 346 PSPSRISVQSTSSSRGVSPSRSRPSTPPSRG-GVSPSRIRPTSSSIQSNDSVS 397
PS R S S S S S SRSR + PSR S SR R SSS + SVS
Sbjct: 4 PSRGRRS-PSVSGSSSRSSSRSRSGSSPSRSISRSRSRSRSLSSSSSPSRSVS 55
>At3g08670 hypothetical protein
Length = 567
Score = 43.9 bits (102), Expect = 3e-04
Identities = 59/237 (24%), Positives = 92/237 (37%), Gaps = 21/237 (8%)
Query: 174 PASNVAHRQGETP--STIRKATPERKRSPLKGKNASDQSENSKPV--DSLPSRLIDQHRW 229
P N +H P ++ +A+ K S L + +S+P S+ I ++
Sbjct: 108 PLGNDSHSSLAAPKIASSARASSASKASRLSVSQSESGYHSSRPARSSSVTRPSISTSQY 167
Query: 230 PSRIGGKVSSSALNRSVDFGDARMLKTPASGTGFSSLRRLSLSEEASKPLQRASSDSVRL 289
S G+ SS LN S + + + S SS R S P + +S+
Sbjct: 168 SSFTSGRSPSSILNTSSASVSSYIRPSSPSSRSSSSAR-------PSTPTRTSSASRSST 220
Query: 290 LSLVASGRTGSEVKSVDDCSLHDLRPHRSSTPHTDRAGLSIAGVRSQSL-SAPGSRLPSP 348
S + G + S S+ RP SS P T + ++ + S P SR +P
Sbjct: 221 PSRIRPGSSSS--------SMDKARPSLSSRPSTPTSRPQLSASSPNIIASRPNSRPSTP 272
Query: 349 SRISVQSTSSSRGVSPSRSRPSTPPSRGGVSPSRIRPTSSSIQSNDSVSVLSFIADF 405
+R S STS S P+ S S G PS RP+S + ++ +ADF
Sbjct: 273 TRRSPSSTSLSATSGPTIS-GGRAASNGRTGPSLSRPSSPGPRVRNTPQQPIVLADF 328
Score = 39.7 bits (91), Expect = 0.005
Identities = 91/409 (22%), Positives = 151/409 (36%), Gaps = 60/409 (14%)
Query: 9 ASSRRLKAVETPRPPLVLAEKNNAATATATARRS---RTREVTSRYKSPTPPSTPSGSRR 65
+S+R A + R L +++ + ++ AR S R TS+Y S T +PS
Sbjct: 124 SSARASSASKASR--LSVSQSESGYHSSRPARSSSVTRPSISTSQYSSFTSGRSPSSILN 181
Query: 66 CPSPNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPS-RRLSTP--VNDSAIDGRLSS 122
S +++ P++ S S+ RPSTP S R STP + + +
Sbjct: 182 TSSASVSSYIRPSSP------SSRSSSSARPSTPTRTSSASRSSTPSRIRPGSSSSSMDK 235
Query: 123 RKVAASRLPEGHNLWPSTMRSLSVSFQSDIISIPVSKKERPVTSASDRTLRPASNVAHRQ 182
+ + S P P++ LS S + I S P S+ P + T A++
Sbjct: 236 ARPSLSSRPS----TPTSRPQLSASSPNIIASRPNSRPSTPTRRSPSSTSLSATS----- 286
Query: 183 GETPSTIRKATPERKRSPLKGKNASDQSENSKPVDSLPSRLIDQHRWPSRIGGKVSSSAL 242
G T S R A+ R G + S S V + P + I +P + +S
Sbjct: 287 GPTISGGRAASNGR-----TGPSLSRPSSPGPRVRNTPQQPIVLADFPLDTPPNLRTSLP 341
Query: 243 NRSVDFGDARMLKTPASGTGFSSLRRLSLSEEASKPLQRASSDSV----RLLSLVASGRT 298
+R + G +R P G+ + S E P+ R +S + RL GR
Sbjct: 342 DRPISAGRSR----PVGGSSMA-----KASPEPKGPITRRNSSPIVTRGRLTETQGKGRF 392
Query: 299 GSEVKSVDDC----------SLHDLRPHRSSTPHTDRAGLSIAGVRSQSLSAPGSRLPSP 348
G + + D + R ++ST TD SL +
Sbjct: 393 GGNGQHLTDAPEPRRISNVSDITSRRTVKTSTTVTDNNNGLGRSFSKSSLDMAIRHMDIR 452
Query: 349 SRISVQSTSSSRGVSPSRSRPSTPPSRGGVSPSRIRPTSSSIQSNDSVS 397
+ + S+ + P RP++ S+I+P S +DS+S
Sbjct: 453 NGKTNGCALSTTTLFPQSIRPAS---------SKIQPIRSGNNHSDSIS 492
Score = 39.7 bits (91), Expect = 0.005
Identities = 49/184 (26%), Positives = 83/184 (44%), Gaps = 13/184 (7%)
Query: 32 AATATATARRSRTREVTSRYKSPTP----PSTPSGSRRCPSPNLTRTATPAASLQLLPKR 87
++ ++++AR S +S +S TP P + S S P+L+ + S L
Sbjct: 197 SSRSSSSARPSTPTRTSSASRSSTPSRIRPGSSSSSMDKARPSLSSRPSTPTSRPQLSAS 256
Query: 88 SLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNL-WPST--MRSL 144
S + RP++ PS P+RR + + SA G S AAS G +L PS+ R
Sbjct: 257 SPNIIASRPNSRPSTPTRRSPSSTSLSATSGPTISGGRAASNGRTGPSLSRPSSPGPRVR 316
Query: 145 SVSFQSDIIS-IPVSKKERPVTSASDRTLRPASNVAHRQGETPSTIRKATPERKRSPLKG 203
+ Q +++ P+ TS DR + + + S++ KA+PE K P+
Sbjct: 317 NTPQQPIVLADFPLDTPPNLRTSLPDRPI----SAGRSRPVGGSSMAKASPEPK-GPITR 371
Query: 204 KNAS 207
+N+S
Sbjct: 372 RNSS 375
>At2g17440 unknown protein
Length = 526
Score = 42.7 bits (99), Expect = 6e-04
Identities = 49/165 (29%), Positives = 71/165 (42%), Gaps = 20/165 (12%)
Query: 147 SFQSDIISIPVSKKERPVTSASDRTLRPASNVAHRQGETPSTIRKATPERKRSPLKGKNA 206
SF SD + P SK + D TL A NVA GE S I+ A+ + K A
Sbjct: 156 SFYSDGLLAP-SKPQ-----VLDSTLHQAKNVAGNDGEKLSLIKLAS----LIEVSAKKA 205
Query: 207 SDQSENSKPVDSLPSRLIDQHRWPSRIGGKVSSSALNRSVDFGDARMLKTPASGTGFSSL 266
+ + +L RL+DQ W GK+SS +D + ++ PA+ G SL
Sbjct: 206 TQEL-------NLQHRLMDQLEWLPDSLGKLSSLV---RLDLSENCIMVLPATIGGLISL 255
Query: 267 RRLSLSEEASKPLQRASSDSVRLLSLVASGRTGSEVKSVDDCSLH 311
RL L L + D + L++L SG S + S + +H
Sbjct: 256 TRLDLHSNRIGQLPESIGDLLNLVNLNLSGNQLSSLPSSFNRLIH 300
>At1g33980 unknown protein
Length = 482
Score = 40.4 bits (93), Expect = 0.003
Identities = 41/168 (24%), Positives = 72/168 (42%), Gaps = 11/168 (6%)
Query: 90 SAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQ 149
S +R P PP P + + T ++ ++ D R + + RL +G L + S S +F
Sbjct: 274 SKDRDNPDNPPPQPEQHIDTNLSRNSTDSRQNQKSDVGGRLIKGILLRNDSRPSQSSTFV 333
Query: 150 SDIISIPVSKKERPVTSASDRTLRPASNVAHRQGETPSTIRKATPERKRSPLKGKNA--- 206
+ S+ E R RPA+ A + T TI + R R+ +
Sbjct: 334 QSEQRVEPSEAEN-----YKRPSRPANTRAGKDYHTSGTISEKQERRTRNKDRPDRVMWA 388
Query: 207 --SDQSENSKPVDSLPSRLIDQHRWPSRIGGKVSSSALNRSVDFGDAR 252
D SE+ +P+ S + + R S+ G+V +S+ +++ G AR
Sbjct: 389 PRRDGSED-QPLSSAGNNGEVKDRMFSQRSGEVVNSSGGHTLENGSAR 435
>At4g04980
Length = 681
Score = 39.3 bits (90), Expect = 0.007
Identities = 24/81 (29%), Positives = 32/81 (38%)
Query: 28 EKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPAASLQLLPKR 87
E N + S T S K PP P S + PSP ++ T ++ P
Sbjct: 269 EHENETEDHSETTTSETDSTESSPKEDVPPPPPLTSPQTPSPTVSTFNTKSSLRSQPPPP 328
Query: 88 SLSAERKRPSTPPSPPSRRLS 108
S E K P+ PP PP + S
Sbjct: 329 PPSPEHKAPAPPPPPPMSKAS 349
>At4g00890
Length = 431
Score = 39.3 bits (90), Expect = 0.007
Identities = 77/349 (22%), Positives = 132/349 (37%), Gaps = 60/349 (17%)
Query: 54 PTPPSTPSG--SRRCPSPNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPV 111
P P S P S + ++T TP S + R+L + S P SPP S
Sbjct: 55 PEPKSLPLSPLSPKSKEEPESQTMTPLMSKHVKTHRNLLIQ----SPPKSPPESTESQQQ 110
Query: 112 NDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQSDIISIPVSKKERPVTSASDRT 171
+++ R + + P ST+++ S S+ P+ S +
Sbjct: 111 FLASVPLRPLTTEPKTPLSPS------STLKATEES---------QSQPSPPLESIVETW 155
Query: 172 LRPASNVAHRQGETPSTIRKATPERKR---SPLKGKNASDQSENSKPVDSLPSRLIDQHR 228
RP+ + G+ E K SP NA+++ E+ LPS+ ID+ R
Sbjct: 156 FRPSPPTSTETGDETQLPIPPPQEAKTPPSSPSMMLNATEEFESQPKPPLLPSKSIDETR 215
Query: 229 WPSRIGGKVSS--SALNRSVDFGDARMLKTPASGTGFSSLRRLSLSEEASKPLQRASSDS 286
S + + SS ++S+D + R P S S++ ++ +S
Sbjct: 216 LRSPLMSQASSPPPLPSKSIDENETRSQSPPISP---------PKSDKQARSQTHSSPSP 266
Query: 287 VRLLSLVASGRTGSEVKSVDDCSLHDLRPHRSSTPHTDRAGLSIAGVRSQSLSAPGSRLP 346
LLS AS H+S +P + + SLS+ S +P
Sbjct: 267 PPLLSPKAS------------------ENHQSKSPMPPPSPTA-----QISLSSLKSPIP 303
Query: 347 SPSRISVQSTSSSRGVSPSRSRPSTPPSRGGVSPSRIRPTSSSIQSNDS 395
SP+ I+ S + S++ PS PS + P++I+ S + + +S
Sbjct: 304 SPATITAPPPPFSSPL--SQTTPSPKPSLPQIEPNQIKSPSPKLTNTES 350
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.125 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,709,369
Number of Sequences: 26719
Number of extensions: 544222
Number of successful extensions: 3093
Number of sequences better than 10.0: 189
Number of HSP's better than 10.0 without gapping: 49
Number of HSP's successfully gapped in prelim test: 143
Number of HSP's that attempted gapping in prelim test: 2437
Number of HSP's gapped (non-prelim): 522
length of query: 616
length of database: 11,318,596
effective HSP length: 105
effective length of query: 511
effective length of database: 8,513,101
effective search space: 4350194611
effective search space used: 4350194611
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0037a.4


