

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0037a.3
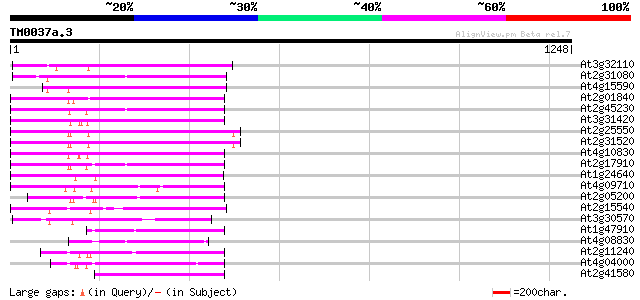
(1248 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g32110 non-LTR reverse transcriptase, putative 342 8e-94
At2g31080 putative non-LTR retroelement reverse transcriptase 339 5e-93
At4g15590 reverse transcriptase like protein 284 3e-76
At2g01840 putative non-LTR retroelement reverse transcriptase 280 3e-75
At2g45230 putative non-LTR retroelement reverse transcriptase 278 1e-74
At3g31420 hypothetical protein 273 3e-73
At2g25550 putative non-LTR retroelement reverse transcriptase 271 2e-72
At2g31520 putative non-LTR retroelement reverse transcriptase 269 8e-72
At4g10830 putative protein 257 3e-68
At2g17910 putative non-LTR retroelement reverse transcriptase 253 4e-67
At1g24640 hypothetical protein 249 9e-66
At4g09710 RNA-directed DNA polymerase -like protein 247 3e-65
At2g05200 putative non-LTR retroelement reverse transcriptase 238 2e-62
At2g15540 putative non-LTR retroelement reverse transcriptase 234 2e-61
At3g30570 putative reverse transcriptase 226 6e-59
At1g47910 reverse transcriptase, putative 216 7e-56
At4g08830 putative protein 215 1e-55
At2g11240 pseudogene 213 6e-55
At4g04000 putative reverse transcriptase 200 5e-51
At2g41580 putative non-LTR retroelement reverse transcriptase 196 9e-50
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 342 bits (877), Expect = 8e-94
Identities = 199/511 (38%), Positives = 291/511 (56%), Gaps = 22/511 (4%)
Query: 7 YGSPNLSIRNFLWQELRRIAANTGEPWVVMGDFNTYLNAPDKWGGDPPNLLAMGKFRDCL 66
Y +P S R+ LW L + + P V+ GDFNT + ++ GG+ F + +
Sbjct: 671 YAAPTASRRSGLWDRLGDVIRSMDGPVVIGGDFNTIVRLDERSGGNGRLSSDSLAFGEWI 730
Query: 67 DDCYLSDLGFKGPPFTWEGRGVKERLDWALGNDQWL------RSFPEASIFHLPQLKSDH 120
+D L DLGFKG FTW+ RG +ER A D+ L + EAS+ HLP L SDH
Sbjct: 731 NDHSLIDLGFKGNKFTWK-RGREERFFVAKRLDRVLCCAHARLKWQEASVLHLPFLASDH 789
Query: 121 KPLLLQLSP-IDIDHSQRPNR---QVFGEIGRRKRHLMRRLEGINS--RLRMQHIPYLE- 173
PL +QL+P + + +RP R G ++ L + I++ L++Q + L
Sbjct: 790 APLYVQLTPEVSGNRGRRPFRFEAAWLSHPGFKELLLTSWNKDISTPEALKVQELLDLHQ 849
Query: 174 -----KLQKRLWKEYQTTLIQEELLWRQKSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEA 228
K ++ L K++ L QEE++W QKSR W HGD+NT+FFHTST++RR+RN+IE
Sbjct: 850 SDDLLKKEEELLKDFDVVLEQEEVVWMQKSREKWFVHGDRNTKFFHTSTIIRRRRNQIEM 909
Query: 229 LTDEHGVSVTDPITLRSMAIDFFQHLYTSPGTDVLYHV--RNAFPTLPRDTLLEISNPLE 286
L D G +++ L + AID+++ LY+ D + + F L ++ P
Sbjct: 910 LQDNDGRWLSNAQELETHAIDYYKRLYSLDDLDAVVEQLPQEGFTALSEADFSSLTKPFS 969
Query: 287 ADEIKAAVFSMRALKAPGPDGLNTLFFQSQWDTIGSSVVQFIQDCIQRPEKIREVNDTLI 346
E++ A+ SM KAPGPDG +F+Q W+ +G SV +F+ D +E ND L+
Sbjct: 970 PLEVEGAIRSMGKYKAPGPDGFQPVFYQQGWEVVGESVTKFVMDFFSSGSFPQETNDVLV 1029
Query: 347 VLIPKIDRPNLLSQYRLIGICNVIYKTLTKALANRLKGVLGDLISPNQCSFIPGRQSSDN 406
VLI K+ +P ++Q+R I +CNV++KT+TK + RLKGV+ LI P Q SFIPGR S+DN
Sbjct: 1030 VLIAKVLKPEKITQFRPISLCNVLFKTITKVMVGRLKGVINKLIGPAQTSFIPGRLSTDN 1089
Query: 407 IIIAQEVFHSMRYLKRKKGWIAIKIDLEKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCV 466
I++ QEV HSMR K KGW+ +K+DLEKAYDR+ W L+DTL GL + I+ CV
Sbjct: 1090 IVVVQEVVHSMRRKKGVKGWMLLKLDLEKAYDRIRWDLLEDTLKAAGLPGTWVQWIMKCV 1149
Query: 467 SLSSFQVLFNGSKTE-FPSFQGHPPR*PIIP 496
S ++L+NG KT+ F +G P+ P
Sbjct: 1150 EGPSMRLLWNGEKTDAFKPLRGLRQGDPLSP 1180
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 339 bits (870), Expect = 5e-93
Identities = 195/488 (39%), Positives = 285/488 (57%), Gaps = 19/488 (3%)
Query: 7 YGSPNLSIRNFLWQELRRIAANTGEPWVVMGDFNTYLNAPDKWGGD---PPNLLAMGKFR 63
Y +P++S R+ LW EL+ + P ++ GDFNT L ++ GG+ P+ LA G
Sbjct: 7 YAAPSVSRRSGLWGELKDVVNGLEGPLLIGGDFNTILWVDERMGGNGRLSPDSLAFG--- 63
Query: 64 DCLDDCYLSDLGFKGPPFTW-----EGRGVKERLDWALGNDQWLRSFPEASIFHLPQLKS 118
D +++ L DLGFKG FTW E V +RLD + EA + HLP + S
Sbjct: 64 DWINELSLIDLGFKGNKFTWRRGRQESTVVAKRLDRVFVCAHARLKWQEAVVSHLPFMAS 123
Query: 119 DHKPLLLQLSPIDIDHSQRPNRQVFGEIGRRKRHLMRRLEGINSRLRMQHIPYLEKLQKR 178
DH PL +QL P+ ++ NR+VFG+I RK L+ ++ + L + L ++
Sbjct: 124 DHAPLYVQLEPLQQRKLRKWNREVFGDIHVRKEKLVADIKEVQDLLGVVLSDDLLAKEEV 183
Query: 179 LWKEYQTTLIQEELLWRQKSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVT 238
L KE L QEE LW QKSR ++ GD+NT FFHTST++RR+RN+IE+L + VT
Sbjct: 184 LLKEMDLVLEQEETLWFQKSREKYIELGDRNTTFFHTSTIIRRRRNRIESLKGDDDRWVT 243
Query: 239 DPITLRSMAIDFFQHLYTSPGTDVLYHVRNAFPTLPRDTLLEISNP--LEA---DEIKAA 293
D + L +MA+ +++ LY+ + + VRN PT ++ E L+A E+ +A
Sbjct: 244 DKVELEAMALTYYKRLYS---LEDVSEVRNMLPTGGFASISEAEKAALLQAFTKAEVVSA 300
Query: 294 VFSMRALKAPGPDGLNTLFFQSQWDTIGSSVVQFIQDCIQRPEKIREVNDTLIVLIPKID 353
V SM KAPGPDG +F+Q W+T+G SV +F+ + + ND L+VLI K+
Sbjct: 301 VKSMGRFKAPGPDGYQPVFYQQCWETVGPSVTRFVLEFFETGVLPASTNDALLVLIAKVA 360
Query: 354 RPNLLSQYRLIGICNVIYKTLTKALANRLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEV 413
+P + Q+R + +CNV++K +TK + RLK V+ LI P Q SFIPGR S DNI++ QE
Sbjct: 361 KPERIQQFRPVSLCNVLFKIITKMMVTRLKNVISKLIGPAQASFIPGRLSIDNIVLVQEA 420
Query: 414 FHSMRYLKRKKGWIAIKIDLEKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCVSLSSFQV 473
HSMR K +KGW+ +K+DLEKAYDR+ W FL++TL GL + I+ V+ S V
Sbjct: 421 VHSMRRKKGRKGWMLLKLDLEKAYDRVRWDFLQETLEAAGLSEGWTSRIMAGVTDPSMSV 480
Query: 474 LFNGSKTE 481
L+NG +T+
Sbjct: 481 LWNGERTD 488
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 284 bits (726), Expect = 3e-76
Identities = 160/440 (36%), Positives = 238/440 (53%), Gaps = 32/440 (7%)
Query: 74 LGFKGPPFTW-----EGRGVKERLDWALGNDQWLRSFPEASIFHLPQLKSDHKPLLLQLS 128
+GFKG FTW E V +RLD L + EA + + + +P + +
Sbjct: 1 MGFKGNRFTWRRGLVESTFVAKRLDRVLFCAHARLKWQEALLCPAQNVDARRRPFRFEAA 60
Query: 129 ---------------------PIDIDHSQ----RPNRQVFGEIGRRKRHLMRRLEGINSR 163
P+ ++ + + N++VFG I RK ++ L+ +
Sbjct: 61 WLSHEGFKELLTASWDTGLSTPVALNRLRWQLKKWNKEVFGNIHVRKEKVVSDLKAVQDL 120
Query: 164 LRMQHIPYLEKLQKRLWKEYQTTLIQEELLWRQKSRVAWLNHGDKNTRFFHTSTMVRRKR 223
L + L + L KE+ L QEE LW QKSR L GD+NT FFHTST++RR+R
Sbjct: 121 LEVVQTDDLLMKEDTLLKEFDVLLHQEETLWFQKSREKLLALGDRNTTFFHTSTVIRRRR 180
Query: 224 NKIEALTDEHGVSVTDPITLRSMAIDFFQHLYTSPGTDVLYHV--RNAFPTLPRDTLLEI 281
N+IE L D VT+ L +A+D+++ LY+ V+ FP L R+ +
Sbjct: 181 NRIEMLKDSEDRWVTEKEALEKLAMDYYRKLYSLEDVSVVRGTLPTEGFPRLTREEKNNL 240
Query: 282 SNPLEADEIKAAVFSMRALKAPGPDGLNTLFFQSQWDTIGSSVVQFIQDCIQRPEKIREV 341
+ P DE+ AV SM KAPGPDG +F+Q W+T+G SV +F+ + + +
Sbjct: 241 NRPFTRDEVVVAVRSMGRFKAPGPDGYQPVFYQQCWETVGESVSKFVMEFFESGVLPKST 300
Query: 342 NDTLIVLIPKIDRPNLLSQYRLIGICNVIYKTLTKALANRLKGVLGDLISPNQCSFIPGR 401
ND L+VL+ K+ +P ++Q+R + +CNV++K +TK + RLK V+ LI P Q SFIPGR
Sbjct: 301 NDVLLVLLAKVAKPERITQFRPVSLCNVLFKIITKMMVIRLKNVISKLIGPAQASFIPGR 360
Query: 402 QSSDNIIIAQEVFHSMRYLKRKKGWIAIKIDLEKAYDRLEWSFLKDTLLEVGLDRNFCDL 461
S DNI++ QE HSMR K +KGW+ +K+DLEKAYDR+ W FL +TL GL +
Sbjct: 361 LSFDNIVVVQEAVHSMRRKKGRKGWMLLKLDLEKAYDRIRWDFLAETLEAAGLSEGWIKR 420
Query: 462 ILDCVSLSSFQVLFNGSKTE 481
I++CV+ +L+NG KT+
Sbjct: 421 IMECVAGPEMSLLWNGEKTD 440
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 280 bits (717), Expect = 3e-75
Identities = 168/526 (31%), Positives = 273/526 (50%), Gaps = 50/526 (9%)
Query: 1 FLVSFTYGSPNLSIRNFLWQELRRIAANTGEPWVVMGDFNTYLNAPDKWGGDPPNLLAMG 60
F +S YG P S R+ LW++L+R++A+ PW++ GDFN LN +K GG ++ ++
Sbjct: 461 FYLSCIYGHPIPSERHHLWEKLQRVSAHRSGPWMMCGDFNEILNLNEKKGGRRRSIGSLQ 520
Query: 61 KFRDCLDDCYLSDLGFKGPPFTWEGRGVKERLDWALG----NDQWLRSFPEASIFHLPQL 116
F + ++ C + DL KG P++W G+ E ++ L N W SFP LP
Sbjct: 521 NFTNMINCCNMKDLKSKGNPYSWVGKRQNETIESCLDRVFINSDWQASFPAFETEFLPIA 580
Query: 117 KSDHKPLLLQLSP----------IDIDHSQRPN--------------------------- 139
SDH P+++ ++ D H Q +
Sbjct: 581 GSDHAPVIIDIAEEVCTKRGQFRYDRRHFQFEDFVDSVQRGWNRGRSDSHGGYYEKLHCC 640
Query: 140 RQVFGEIGRR-KRHLMRRLEGINSRL----RMQHIPYLEKLQKRLWKEYQTTLIQEELLW 194
RQ + RR K + ++E + R+ R +P+ L RL ++ EEL W
Sbjct: 641 RQELAKWKRRTKTNTAEKIETLKYRVDAAERDHTLPHQTIL--RLRQDLNQAYRDEELYW 698
Query: 195 RQKSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVTDPITLRSMAIDFFQHL 254
KSR W+ GD+NT FF+ ST +R+ RN+I+A+TD G+ T+ +A ++F L
Sbjct: 699 HLKSRNRWMLLGDRNTMFFYASTKLRKSRNRIKAITDAQGIENFRDDTIGKVAENYFADL 758
Query: 255 YTSPGTDVLYHVRNAF-PTLPRDTLLEISNPLEADEIKAAVFSMRALKAPGPDGLNTLFF 313
+T+ T + + P + E+ + E++ AVF++ A +APG DG F+
Sbjct: 759 FTTTQTSDWEEIISGIAPKVTEQMNHELLQSVTDQEVRDAVFAIGADRAPGFDGFTAAFY 818
Query: 314 QSQWDTIGSSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVIYKT 373
WD IG+ V ++ + ++N T I LIPKI P +S YR I +C YK
Sbjct: 819 HHLWDLIGNDVCLMVRHFFESDVMDNQINQTQICLIPKIIDPKHMSDYRPISLCTASYKI 878
Query: 374 LTKALANRLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYLKR-KKGWIAIKID 432
++K L RLK LGD+IS +Q +F+PG+ SDN+++A E+ HS++ + + G++A+K D
Sbjct: 879 ISKILIKRLKQCLGDVISDSQAAFVPGQNISDNVLVAHELLHSLKSRRECQSGYVAVKTD 938
Query: 433 LEKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCVSLSSFQVLFNGS 478
+ KAYDR+EW+FL+ ++++G + I+ CV+ S++VL NGS
Sbjct: 939 ISKAYDRVEWNFLEKVMIQLGFAPRWVKWIMTCVTSVSYEVLINGS 984
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 278 bits (712), Expect = 1e-74
Identities = 171/526 (32%), Positives = 267/526 (50%), Gaps = 50/526 (9%)
Query: 1 FLVSFTYGSPNLSIRNFLWQELRRIAANTGEPWVVMGDFNTYLNAPDKWGGDPPNLLAMG 60
F ++ YG P + R LW+ L R+ + PW++ GDFN ++ +K GG +
Sbjct: 99 FYLTCIYGEPVQAERGELWERLTRLGLSRSGPWMLTGDFNELVDPSEKIGGPARKESSCL 158
Query: 61 KFRDCLDDCYLSDLGFKGPPFTWEGRGVKE----RLDWALGNDQWLRSFPEASIFHLPQL 116
+FR L+ C L ++ G F+W G E RLD + N W+ FP+A +L ++
Sbjct: 159 EFRQMLNSCGLWEVNHSGYQFSWYGNRNDELVQCRLDRTVANQAWMELFPQAKATYLQKI 218
Query: 117 KSDHKPLLLQLSPID---------------------------IDHSQRPNRQVFGEIGRR 149
SDH PL+ L + S + N + +I
Sbjct: 219 CSDHSPLINNLVGDNWRKWAGFKYDKRWVQREGFKDLLCNFWSQQSTKTNALMMEKIASC 278
Query: 150 KRHLMR--RLEGINSRLRMQH-----------IPYLEKLQKRLWKEYQTTLIQEELLWRQ 196
+R + + R+ +S +R+Q IP+ + RL KE EE W++
Sbjct: 279 RREISKWKRVSKPSSAVRIQELQFKLDAATKQIPFDRRELARLKKELSQEYNNEEQFWQE 338
Query: 197 KSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVTDPITLRSMAIDFFQHLYT 256
KSR+ W+ +GD+NT++FH +T RR +N+I+ L DE G T L +A +F+ L+
Sbjct: 339 KSRIMWMRNGDRNTKYFHAATKNRRAQNRIQKLIDEEGREWTSDEDLGRVAEAYFKKLFA 398
Query: 257 SPGTDVLYHVR---NAFPTLPRDTLLEISNPLEADEIKAAVFSMRALKAPGPDGLNTLFF 313
S DV Y V N P + + P+ +E++ A FS+ K PGPDG+N +
Sbjct: 399 SE--DVGYTVEELENLTPLVSDQMNNNLLAPITKEEVQRATFSINPHKCPGPDGMNGFLY 456
Query: 314 QSQWDTIGSSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVIYKT 373
Q W+T+G + + +Q + +N T I LIPKI + ++ +R I +CNVIYK
Sbjct: 457 QQFWETMGDQITEMVQAFFRSGSIEEGMNKTNICLIPKILKAEKMTDFRPISLCNVIYKV 516
Query: 374 LTKALANRLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSM-RYLKRKKGWIAIKID 432
+ K +ANRLK +L LIS Q +F+ GR SDNI+IA E+ H++ K + +IAIK D
Sbjct: 517 IGKLMANRLKKILPSLISETQAAFVKGRLISDNILIAHELLHALSSNNKCSEEFIAIKTD 576
Query: 433 LEKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCVSLSSFQVLFNGS 478
+ KAYDR+EW FL+ + +G ++ LI++CV +QVL NG+
Sbjct: 577 ISKAYDRVEWPFLEKAMRGLGFADHWIRLIMECVKSVRYQVLINGT 622
>At3g31420 hypothetical protein
Length = 1491
Score = 273 bits (699), Expect = 3e-73
Identities = 170/529 (32%), Positives = 274/529 (51%), Gaps = 51/529 (9%)
Query: 1 FLVSFTYGSPNLSIRNFLWQELRRI-AANTGEPWVVMGDFNTYLNAPDKWGGDPPNLLAM 59
F +SF YG PN S R+ LW+ L R+ G W +MGDFN L+ +K GG
Sbjct: 279 FYLSFVYGHPNPSHRHHLWERLERLNTTRQGTAWFIMGDFNEILSNREKRGGRLRPERTF 338
Query: 60 GKFRDCLDDCYLSDLGFKGPPFTWEGRG----VKERLDWALGNDQWLRSFPEASIFHLPQ 115
FR+ + C +DL G F+W G+ V LD + N++W FPE+ +
Sbjct: 339 QDFRNMVRGCNFTDLKSVGDRFSWAGKRGDHHVTCSLDRTMANNEWHTLFPESETVFMEY 398
Query: 116 LKSDHKPLLLQLSPIDIDH-------SQRPNRQVFGEIGRRKRH-----------LMRRL 157
+SDH+PL+ +S + S+ +++ F ++ + H L R+L
Sbjct: 399 GESDHRPLVTNISAQKEERRGFFSYDSRLTHKEGFKQVVLNQWHRRNGSFEGDSSLNRKL 458
Query: 158 -------------EGINSRLRMQHIPY-----------LEKLQKRLWKEYQTTLIQEELL 193
+N+ R++ I + + ++++ ++ EE+
Sbjct: 459 VECRQAISRWKKNNRVNAEERIKIIRHKLDRAIATGTATPRERRQMRQDLNQAYADEEIY 518
Query: 194 WRQKSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVTDPITLRSMAIDFFQH 253
W+ KSR WLN GD+NTR+FH++T RR RN++ ++ D G + +AI++F
Sbjct: 519 WQTKSRSRWLNAGDRNTRYFHSTTKTRRCRNRLLSVQDSDGDICRGDENIAKVAINYFDD 578
Query: 254 LYTS-PGTDVLY-HVRNAFPTLPRDTLLE-ISNPLEADEIKAAVFSMRALKAPGPDGLNT 310
LY S P T + Y V F D + E + P+ EI+ +VFS+ + P PDG
Sbjct: 579 LYKSTPNTSLRYADVFQGFQQKITDEINEDLIRPVTELEIEESVFSVAPSRTPDPDGFTA 638
Query: 311 LFFQSQWDTIGSSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVI 370
F+Q W I V+ + +R E N T + LIPK++ P ++++R I +CNV
Sbjct: 639 DFYQQFWPDIKQKVIDEVTRFFERSELDERHNHTNLCLIPKVETPTTIAKFRPIALCNVS 698
Query: 371 YKTLTKALANRLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYLKRK-KGWIAI 429
YK ++K L NRLK LG I+ NQ +F+PGR ++N IIA EV+++++ KR+ ++A+
Sbjct: 699 YKIISKILVNRLKKHLGGAITENQAAFVPGRLITNNAIIAHEVYYALKARKRQANSYMAL 758
Query: 430 KIDLEKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCVSLSSFQVLFNGS 478
K D+ KAYDRLEW FL++T+ ++G + + + I+ CV++ F VL NGS
Sbjct: 759 KTDITKAYDRLEWDFLEETMRQMGFNTKWIERIMICVTMVRFSVLINGS 807
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 271 bits (692), Expect = 2e-72
Identities = 185/569 (32%), Positives = 276/569 (47%), Gaps = 56/569 (9%)
Query: 1 FLVSFTYGSPNLSIRNFLWQELRRIAANTGEPWVVMGDFNTYLNAPDKWGGDPPNLLAMG 60
F +S YG P S R+ LWQ L I+ N W+++GDFN L+ +K GG
Sbjct: 480 FYLSCVYGHPTQSERHQLWQTLEHISDNRNAEWLLVGDFNEILSNAEKIGGPMREEWTFR 539
Query: 61 KFRDCLDDCYLSDLGFKGPPFTWEGRG----VKERLDWALGNDQWLRSFPEASIFHLPQL 116
FR+ + C + D+ KG F+W G VK LD N W +FP A I L
Sbjct: 540 NFRNMVSHCDIEDMRSKGDRFSWVGERHTHTVKCCLDRVFINSAWTATFPYAEIEFLDFT 599
Query: 117 KSDHKPLLLQLSP---------------IDI-------DHSQRPNRQV----FGEIGRRK 150
SDHKP+L+ + IDI S R NR E
Sbjct: 600 GSDHKPVLVHFNESFPRRSKLFRFDNRLIDIPTFKRIVQTSWRTNRNSRSTPITERISSC 659
Query: 151 RHLMRRLEGINSRLRMQHIPYLE-----------KLQKRLWKEYQTTLIQ----EELLWR 195
R M RL+ ++ Q I L+ ++ ++L + Q +L + EE+ W+
Sbjct: 660 RQAMARLKHASNLNSEQRIKKLQSSLNRAMESTRRVDRQLIPQLQESLAKAFSDEEIYWK 719
Query: 196 QKSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVTDPITLRSMAIDFFQHLY 255
QKSR W+ GD+NT +FH T R +N++ + D+ G T + + A DFF +++
Sbjct: 720 QKSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQGRMFTGDKEIGNHAQDFFTNIF 779
Query: 256 TSPGTDVLYHVRNAFPTLPRDTL-LEISNPLEADEIKAAVFSMRALKAPGPDGLNTLFFQ 314
++ G V F + +T+ L+++ EI A+ + KAPGPDGL F++
Sbjct: 780 STNGIKVSPIDFADFKSTVTNTVNLDLTKEFSDTEIYDAICQIGDDKAPGPDGLTARFYK 839
Query: 315 SQWDTIGSSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVIYKTL 374
+ WD +G V+ ++ + +N T I +IPKI P LS YR I +CNV+YK +
Sbjct: 840 NCWDIVGYDVILEVKKFFETSFMKPSINHTNICMIPKITNPTTLSDYRPIALCNVLYKVI 899
Query: 375 TKALANRLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYLKR-KKGWIAIKIDL 433
+K L NRLK L ++S +Q +FIPGR +DN++IA EV HS++ KR K ++A+K D+
Sbjct: 900 SKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVMIAHEVMHSLKVRKRVSKTYMAVKTDV 959
Query: 434 EKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCVSLSSFQVLFNGSK----TEFPSFQGHP 489
KAYDR+EW FL+ T+ G + I+ V + VL NGS T +
Sbjct: 960 SKAYDRVEWDFLETTMRLFGFCNKWIGWIMAAVKSVHYSVLINGSPHGYITPTRGIRQGD 1019
Query: 490 PR*PII-----PIFVRSLHGAASSSDSKG 513
P P + I ++G ASS D +G
Sbjct: 1020 PLSPYLFILCGDILSHLINGRASSGDLRG 1048
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 269 bits (687), Expect = 8e-72
Identities = 184/569 (32%), Positives = 275/569 (47%), Gaps = 56/569 (9%)
Query: 1 FLVSFTYGSPNLSIRNFLWQELRRIAANTGEPWVVMGDFNTYLNAPDKWGGDPPNLLAMG 60
F +S YG P S R+ LWQ L I+ N W+++GDFN L+ +K GG
Sbjct: 254 FYLSCVYGHPTQSERHQLWQTLEHISDNRNAEWLLVGDFNEILSNAEKIGGPMREEWTFR 313
Query: 61 KFRDCLDDCYLSDLGFKGPPFTWEGRG----VKERLDWALGNDQWLRSFPEASIFHLPQL 116
FR+ + C + D+ KG F+W G VK LD N W +FP A L
Sbjct: 314 NFRNMVSHCDIEDMRSKGDRFSWVGERHTHTVKCCLDRVFINSAWTATFPYAETEFLDFT 373
Query: 117 KSDHKPLLLQLSP---------------IDI-------DHSQRPNRQV----FGEIGRRK 150
SDHKP+L+ + IDI S R NR E
Sbjct: 374 GSDHKPVLVHFNESFPRRSKLFRFDNRLIDIPTFKRIVQTSWRTNRNSRSTPITERISSC 433
Query: 151 RHLMRRLEGINSRLRMQHIPYLE-----------KLQKRLWKEYQTTLIQ----EELLWR 195
R M RL+ ++ Q I L+ ++ ++L + Q +L + EE+ W+
Sbjct: 434 RQAMARLKHASNLNSEQRIKKLQSSLNRAMESTRRVDRQLIPQLQESLAKAFSDEEIYWK 493
Query: 196 QKSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVTDPITLRSMAIDFFQHLY 255
QKSR W+ GD+NT +FH T R +N++ + D+ G T + + A DFF +++
Sbjct: 494 QKSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQGRMFTGDKEIGNHAQDFFTNIF 553
Query: 256 TSPGTDVLYHVRNAFPTLPRDTL-LEISNPLEADEIKAAVFSMRALKAPGPDGLNTLFFQ 314
++ G V F + +T+ L+++ EI A+ + KAPGPDGL F++
Sbjct: 554 STNGIKVSPIDFADFKSTVTNTVNLDLTKEFSDTEIYDAICQIGDDKAPGPDGLTARFYK 613
Query: 315 SQWDTIGSSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVIYKTL 374
+ WD +G V+ ++ + +N T I +IPKI P LS YR I +CNV+YK +
Sbjct: 614 NCWDIVGYDVILEVKKFFETSFMKPSINHTNICMIPKITNPTTLSDYRPIALCNVLYKVI 673
Query: 375 TKALANRLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYLKR-KKGWIAIKIDL 433
+K L NRLK L ++S +Q +FIPGR +DN++IA EV HS++ KR K ++A+K D+
Sbjct: 674 SKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVMIAHEVMHSLKVRKRVSKTYMAVKTDV 733
Query: 434 EKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCVSLSSFQVLFNGSK----TEFPSFQGHP 489
KAYDR+EW FL+ T+ G + I+ V + VL NGS T +
Sbjct: 734 SKAYDRVEWDFLETTMRLFGFCNKWIGWIMAAVKSVHYSVLINGSPHGYITPTRGIRQGD 793
Query: 490 PR*PII-----PIFVRSLHGAASSSDSKG 513
P P + I ++G ASS D +G
Sbjct: 794 PLSPYLFILCGDILSHLINGRASSGDLRG 822
>At4g10830 putative protein
Length = 1294
Score = 257 bits (656), Expect = 3e-68
Identities = 167/524 (31%), Positives = 256/524 (47%), Gaps = 47/524 (8%)
Query: 1 FLVSFTYGSPNLSIRNFLWQELRRIAANTGEPWVVMGDFNTYLNAPDKWGGDPPNLLAMG 60
F +S YG P S R+ LW ++ +PW+++GDFN L+ +K GG +
Sbjct: 460 FYLSRVYGHPCQSERHSLWTHFENLSKTRNDPWILIGDFNEILSNNEKIGGPQRDEWTFR 519
Query: 61 KFRDCLDDCYLSDLGFKGPPFTWEGRG----VKERLDWALGNDQWLRSFPEASIFHLPQL 116
FR+ + C L D+ G F+W G VK LD A N + FP A + L
Sbjct: 520 GFRNMVSTCDLKDIRSIGDRFSWVGERHSHTVKCCLDRAFINSEGAFLFPFAELEFLEFT 579
Query: 117 KSDHKPLLLQLS--------PIDIDHS--QRPNRQVFGEIGRRK------RHL------- 153
SDHKPL L L P D + P+ + + + G K +HL
Sbjct: 580 GSDHKPLFLSLEKTETRKMRPFRFDKRLLEVPHFKTYVKAGWNKAINGQRKHLPDQVRTC 639
Query: 154 ------MRRLEGINSRLRMQHIPY--------LEKLQKRLWKEYQTTLI----QEELLWR 195
++ +NSR+R+ + + + ++R Q L EE W+
Sbjct: 640 RQAMAKLKHKSNLNSRIRINQLQAALDKAMSSVNRTERRTISHIQRELTVAYRDEERYWQ 699
Query: 196 QKSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVTDPITLRSMAIDFFQHLY 255
QKSR W+ GD+NT FFH T R N++ + DE G+ + A +FF +Y
Sbjct: 700 QKSRNQWMKEGDRNTEFFHACTKTRFSVNRLVTIKDEEGMIYRGDKEIGVHAQEFFTKVY 759
Query: 256 TSPGTDV-LYHVRNAFPTLPRDTLLEISNPLEADEIKAAVFSMRALKAPGPDGLNTLFFQ 314
S G V + P + +++ L EI A+ + KAPGPDGL F++
Sbjct: 760 ESNGRPVSIIDFAGFKPIVTEQINDDLTKDLSDLEIYNAICHIGDDKAPGPDGLTARFYK 819
Query: 315 SQWDTIGSSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVIYKTL 374
S W+ +G V++ ++ + + +N T I +IPKI P LS YR I +CNV+YK +
Sbjct: 820 SCWEIVGPDVIKEVKIFFRTSYMKQSINHTNICMIPKITNPETLSDYRPIALCNVLYKII 879
Query: 375 TKALANRLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYLKR-KKGWIAIKIDL 433
+K L RLKG L ++S +Q +FIPGR +DN++IA E+ HS++ KR + ++A+K D+
Sbjct: 880 SKCLVERLKGHLDAIVSDSQAAFIPGRLVNDNVMIAHEMMHSLKTRKRVSQSYMAVKTDV 939
Query: 434 EKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCVSLSSFQVLFNG 477
KAYDR+EW+FL+ T+ G + I+ V ++ VL NG
Sbjct: 940 SKAYDRVEWNFLETTMRLFGFSETWIKWIMGAVKSVNYSVLVNG 983
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 253 bits (647), Expect = 4e-67
Identities = 172/530 (32%), Positives = 257/530 (48%), Gaps = 60/530 (11%)
Query: 1 FLVSFTYGSPNLSIRNFLWQELRRIAANTGEPWVVMGDFNTYLNAPDKWGGDPPNLLAMG 60
+ +S YG P +R LW+ L I E W ++GDFN + +K GG + +
Sbjct: 77 WFISCVYGLPVTHMRPKLWEHLNSIGLKRAEAWCLIGDFNDIRSNDEKLGGPRRSPSSFQ 136
Query: 61 KFRDCLDDCYLSDLGFKGPPFTWEG----RGVKERLDWALGNDQWLRSFPEASIFHLPQL 116
F L +C + +LG G FTW G + V+ +LD GN W FP A + L +
Sbjct: 137 CFEHMLLNCSMHELGSTGNSFTWGGNRNDQWVQCKLDRCFGNPAWFSIFPNAHQWFLEKF 196
Query: 117 KSDHKPLLLQLSP---------------------IDIDH-------SQRPNRQVFGEIG- 147
SDH+P+L++ + I++ H SQ + F I
Sbjct: 197 GSDHRPVLVKFTNDNELFRGQFRYDKRLDDDPYCIEVIHRSWNSAMSQGTHSSFFSLIEC 256
Query: 148 RRKRHLMRRLEGINSRLRMQHI---------------PYLEKLQKRLWKEYQTTLIQEEL 192
RR + + N++ R++ + P +E ++ +L Y EEL
Sbjct: 257 RRAISVWKHSSDTNAQSRIKRLRKDLDAEKSIQIPCWPRIEYIKDQLSLAYG----DEEL 312
Query: 193 LWRQKSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVTDPITLRSMAIDFFQ 252
WRQKSR WL GDKNT FFH + R +N++ L DE+ T +A FF+
Sbjct: 313 FWRQKSRQKWLAGGDKNTGFFHATVHSERLKNELSFLLDENDQEFTRNSDKGKIASSFFE 372
Query: 253 HLYTSPGTDVLYHVRNAFPTLPRDTLLEISNPLEAD----EIKAAVFSMRALKAPGPDGL 308
+L+TS T +L H N L E+++ L + E+ AVFS+ APGPDG
Sbjct: 373 NLFTS--TYILTH-NNHLEGLQAKVTSEMNHNLIQEVTELEVYNAVFSINKESAPGPDGF 429
Query: 309 NTLFFQSQWDTIGSSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICN 368
LFFQ WD + ++ I + ++ N T I LIPKI P +S R I +C+
Sbjct: 430 TALFFQQHWDLVKHQILTEIFGFFETGVLPQDWNHTHICLIPKITSPQRMSDLRPISLCS 489
Query: 369 VIYKTLTKALANRLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYLKR-KKGWI 427
V+YK ++K L RLK L ++S Q +F+P R SDNI++A E+ HS+R R K +
Sbjct: 490 VLYKIISKILTQRLKKHLPAIVSTTQSAFVPQRLISDNILVAHEMIHSLRTNDRISKEHM 549
Query: 428 AIKIDLEKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCVSLSSFQVLFNG 477
A K D+ KAYDR+EW FL+ + +G + + I++CV+ S+ VL NG
Sbjct: 550 AFKTDMSKAYDRVEWPFLETMMTALGFNNKWISWIMNCVTSVSYSVLING 599
>At1g24640 hypothetical protein
Length = 1270
Score = 249 bits (635), Expect = 9e-66
Identities = 159/522 (30%), Positives = 255/522 (48%), Gaps = 46/522 (8%)
Query: 1 FLVSFTYGSPNLSIRNFLWQELRRIAANTGEPWVVMGDFNTYLNAPDKWGGDPPNLLAMG 60
F VS YG P+ S R+ W+ + RI + W + GDFN L+ +K GG + L
Sbjct: 62 FCVSCVYGDPDRSKRSQAWERISRIGVGRRDKWCMFGDFNDILHNGEKNGGPRRSDLDCK 121
Query: 61 KFRDCLDDCYLSDLGFKGPPFTWEGRG----VKERLDWALGNDQWLRSFPEASIFHLPQL 116
F + + C L ++ G FTW GR ++ RLD A GN +W FP ++ L
Sbjct: 122 AFNEMIKGCDLVEMPAHGNGFTWAGRRGDHWIQCRLDRAFGNKEWFCFFPVSNQTFLDFR 181
Query: 117 KSDHKPLLLQLSPIDIDHSQR---PNRQVFGE--------------------------IG 147
SDH+P+L++L + + R +F E
Sbjct: 182 GSDHRPVLIKLMSSQDSYRGQFRFDKRFLFKEDVKEAIIRTWSRGKHGTNISVADRLRAC 241
Query: 148 RRKRHLMRRLEGINSRLRMQHIPY-LEKLQKRLWKEYQTTLI----------QEELLWRQ 196
R+ ++ +NS ++ + LEK Q +W +Q + +EE W+Q
Sbjct: 242 RKSLSSWKKQNNLNSLDKINQLEAALEKEQSLVWPIFQRVSVLKKDLAKAYREEEAYWKQ 301
Query: 197 KSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVTDPITLRSMAIDFFQHLYT 256
KSR WL G++N+++FH + R+R +IE L D +G T +A +F +L+
Sbjct: 302 KSRQKWLRSGNRNSKYFHAAVKQNRQRKRIEKLKDVNGNMQTSEAAKGEVAAAYFGNLFK 361
Query: 257 SPGTDVLYHVRNAFPTLPRDTLLE-ISNPLEADEIKAAVFSMRALKAPGPDGLNTLFFQS 315
S + + + E + + A EIK AVFS++ APGPDG++ LFFQ
Sbjct: 362 SSNPSGFTDWFSGLVPRVSEVMNESLVGEVSAQEIKEAVFSIKPASAPGPDGMSALFFQH 421
Query: 316 QWDTIGSSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVIYKTLT 375
W T+G+ V ++ E N T + LIPK P + R I +C+V+YK ++
Sbjct: 422 YWSTVGNQVTSEVKKFFADGIMPAEWNYTHLCLIPKTQHPTEMVDLRPISLCSVLYKIIS 481
Query: 376 KALANRLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYLKR-KKGWIAIKIDLE 434
K +A RL+ L +++S Q +F+ R +DNI++A E+ HS++ R ++A+K D+
Sbjct: 482 KIMAKRLQPWLPEIVSDTQSAFVSERLITDNILVAHELVHSLKVHPRISSEFMAVKSDMS 541
Query: 435 KAYDRLEWSFLKDTLLEVGLDRNFCDLILDCVSLSSFQVLFN 476
KAYDR+EWS+L+ LL +G + + I+ CVS ++ VL N
Sbjct: 542 KAYDRVEWSYLRSLLLSLGFHLKWVNWIMVCVSSVTYSVLIN 583
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 247 bits (630), Expect = 3e-65
Identities = 166/528 (31%), Positives = 252/528 (47%), Gaps = 60/528 (11%)
Query: 3 VSFTYGSPN-LSIRNFLWQELRRIAANTGEPWVVMGDFNTYLNAPDKWGGDPPNLLAMGK 61
V +PN + R+ W ++ + A W++ GDFN L+ +K GG
Sbjct: 44 VEILEAAPNFIDNRSVFWDKISSLGAQRSSAWLLTGDFNDILDNSEKQGGPLRWEGFFLA 103
Query: 62 FRDCLDDCYLSDLGFKGPPFTWEGRG----VKERLDWALGNDQWLRSFPEASIFHLPQLK 117
FR + L D+ G +W G +K RLD ALGN W FP + +L
Sbjct: 104 FRSFVSQNGLWDINHTGNSLSWRGTRYSHFIKSRLDRALGNCSWSELFPMSKCEYLRFEG 163
Query: 118 SDHKPL--------LLQLSPIDIDHSQRPNRQ----------------VFGEIGRRKRHL 153
SDH+PL L + P D R + V +I R ++ +
Sbjct: 164 SDHRPLVTYFGAPPLKRSKPFRFDRRLREKEEIRALVKEVWELARQDSVLYKISRCRQSI 223
Query: 154 MRRLEGINSRLRMQHIPYLEKLQKRL-------------WKEYQTTLIQEELLWRQKSRV 200
++ + NS + L+ L +E + QEEL W+Q SRV
Sbjct: 224 IKWTKEQNSNSAKAIKKAQQALESALSADIPDPSLIGSITQELEAAYRQEELFWKQWSRV 283
Query: 201 AWLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVTDPITLRSMAIDFFQHLYTSPGT 260
WLN GD+N +FH +T RR N + + D G + + S +FQ+++T+
Sbjct: 284 QWLNSGDRNKGYFHATTRTRRMLNNLSVIEDGSGQEFHEEEQIASTISSYFQNIFTTSNN 343
Query: 261 DVLYHVRNAFPTLP----RDTLLEISNPLEADEIKAAVFSMRALKAPGPDGLNTLFFQSQ 316
L V+ A + + L++IS+ LE IK A+FS+ A KAPGPDG + FF +
Sbjct: 344 SDLQVVQEALSPIISSHCNEELIKISSLLE---IKEALFSISADKAPGPDGFSASFFHAY 400
Query: 317 WDTIGSSVVQ-----FIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVIY 371
WD I + V + F+ C+ +N+T + LIPKI P +S YR I +CNV Y
Sbjct: 401 WDIIEADVSRDIRSFFVDSCLSP-----RLNETHVTLIPKISAPRKVSDYRPIALCNVQY 455
Query: 372 KTLTKALANRLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYLKRKKGW-IAIK 430
K + K L RL+ L +LIS +Q +F+PGR +DN++I E+ H +R KK +AIK
Sbjct: 456 KIVAKILTRRLQPWLSELISLHQSAFVPGRAIADNVLITHEILHFLRVSGAKKYCSMAIK 515
Query: 431 IDLEKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCVSLSSFQVLFNGS 478
D+ KAYDR++W+FL++ L+ +G + ++ CV S+ L NGS
Sbjct: 516 TDMSKAYDRIKWNFLQEVLMRLGFHDKWIRWVMQCVCTVSYSFLINGS 563
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 238 bits (607), Expect = 2e-62
Identities = 161/491 (32%), Positives = 242/491 (48%), Gaps = 61/491 (12%)
Query: 40 NTYLNAPDKWGGDPPNLLAMGKFRDCLDDCYLSDLGFKGPPFTWEGRG----VKERLDWA 95
N L+ +K GG P + + FR + L DL + G PF+W G V++RLD A
Sbjct: 36 NEILDNSEKRGGPPRDQGSFIDFRSFISKNGLWDLKYSGNPFSWRGMRYDWFVRQRLDRA 95
Query: 96 LGNDQWLRSFPEASIFHLPQLKSDHKPLLLQLSPIDI--------DHSQRPN-------- 139
+ N+ WL SFP +L SDH+PL++ + + D+ R N
Sbjct: 96 MSNNSWLESFPSGRSEYLRFEGSDHRPLVVFVDEARVKRRGQFRFDNRLRDNDVVNALIQ 155
Query: 140 --------RQVFGEIGRRKRHLMR--RLEGINSRLRMQHIPYLEKLQKRLWKEYQ----- 184
V ++ + +R ++ RL+ +NS +EK QK L +
Sbjct: 156 ETWTNAGDASVLTKMNQCRREIINWTRLQNLNSA------ELIEKTQKALEEALTADPPN 209
Query: 185 -------TTLIQ-----EELLWRQKSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTDE 232
T ++ EE W+Q+SRV WL+ GD+NT +FH T RR +N++ + D
Sbjct: 210 PTTIGALTATLEHAYKLEEQFWKQRSRVLWLHSGDRNTGYFHAVTRNRRTQNRLTVMEDI 269
Query: 233 HGVSVTDPITLRSMAIDFFQHLYTSPGTDVLYHVRNAFPTL----PRDTLLEISNPLEAD 288
+GV+ + + + +FQ ++TS V A + D L I N +
Sbjct: 270 NGVAQHEEHQISQIISGYFQQIFTSESDGDFSVVDEAIEPMVSQGDNDFLTRIPND---E 326
Query: 289 EIKAAVFSMRALKAPGPDGLNTLFFQSQWDTIGSSVVQFIQDCIQRPEKIREVNDTLIVL 348
E+K AVFS+ A KAPGPDG F+ S W I + V + I+ R +N+T I L
Sbjct: 327 EVKDAVFSINASKAPGPDGFTAGFYHSYWHIISTDVGREIRLFFTSKNFPRRMNETHIRL 386
Query: 349 IPKIDRPNLLSQYRLIGICNVIYKTLTKALANRLKGVLGDLISPNQCSFIPGRQSSDNII 408
IPK P ++ YR I +CN+ YK + K + R++ +L LIS NQ +F+PGR SDN++
Sbjct: 387 IPKDLGPRKVADYRPIALCNIFYKIVAKIMTKRMQLILPKLISENQSAFVPGRVISDNVL 446
Query: 409 IAQEVFHSMRYLKRKKGW-IAIKIDLEKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCVS 467
I EV H +R KK +A+K D+ KAYDR+EW FLK L G + D +L+CV+
Sbjct: 447 ITHEVLHFLRTSSAKKHCSMAVKTDMSKAYDRVEWDFLKKVLQRFGFHSIWIDWVLECVT 506
Query: 468 LSSFQVLFNGS 478
S+ L NG+
Sbjct: 507 SVSYSFLINGT 517
>At2g15540 putative non-LTR retroelement reverse transcriptase
Length = 1225
Score = 234 bits (598), Expect = 2e-61
Identities = 153/501 (30%), Positives = 255/501 (50%), Gaps = 49/501 (9%)
Query: 3 VSFTYGSPNLSIRNFLWQELRRIAANTGEPWVVMGDFNTYLNAPDKWGGDPPNLLAMGKF 62
++F YG P + R +W+ L RI EPW ++GDFN + +K GG + + F
Sbjct: 101 ITFVYGDPVVQYRELVWKRLTRIGIVRSEPWFMIGDFNEIIGNHEKRGGKKRSESSFLPF 160
Query: 63 RDCLDDCYLSDLGFKGPPFTWEGRG------------VKERLDWALGNDQWLRSFPEASI 110
+++C + D G F+W G+ +K RLD A+GN++W + ++
Sbjct: 161 CCMIENCGMIDFPSTGSLFSWVGKRSCGVAGRKRRDLIKCRLDRAMGNEEWHSIYSHTNV 220
Query: 111 FHLPQLKSDHKPLLLQLSPIDIDHSQRPNRQ-VFGEIGRRKRHLMRRL-EGINSRLRMQH 168
+L SDHKPLL + + RP + +F + K + EG R +
Sbjct: 221 EYLQHRGSDHKPLLASIQ----NKPYRPYKHFIFDKRWINKPGFKESVQEGWAFPSRGEG 276
Query: 169 IPYLEKLQK-----RLWKEYQTTLIQEELLWRQKSRVAWLNHGDKNTRFFHTSTMVRRKR 223
+P+ +K++ +WK+ T E+L+ S++ L + D+N F T ++ K
Sbjct: 277 VPFFQKIKNCRQTISIWKKSNKTNT-EKLILELHSQLD-LAYEDEN---FSTEDLLALKW 331
Query: 224 NKIEALTDEHGVSVTDPITLRSMAIDFFQHLYTSPGTDVLYHVRNAFPTLPRDTLLE-IS 282
+A DE + + V P L + + + ++
Sbjct: 332 KLCQAYRDE-------------------EIFWKLKNPQVFDSALQEVPVLITEEMNKSLT 372
Query: 283 NPLEADEIKAAVFSMRALKAPGPDGLNTLFFQSQWDTIGSSVVQFIQDCIQRPEKIREVN 342
+ +E+K A+FS+ KAPGPDG+ F+Q WD G +++ +Q+ +N
Sbjct: 373 KVISPEEVKRALFSLNPDKAPGPDGMTAFFYQHYWDLTGPDLIKLVQNFHSTGFFDERLN 432
Query: 343 DTLIVLIPKIDRPNLLSQYRLIGICNVIYKTLTKALANRLKGVLGDLISPNQCSFIPGRQ 402
+T I LIPK +RP ++++R I +CNV YK ++K L++RLK +L +LIS Q +F+ R
Sbjct: 433 ETNICLIPKTERPRKMAEFRPISLCNVSYKVISKVLSSRLKRLLPELISETQSAFVAERL 492
Query: 403 SSDNIIIAQEVFHSMRYLKR-KKGWIAIKIDLEKAYDRLEWSFLKDTLLEVGLDRNFCDL 461
+DNI+IAQE FH++R KK ++AIK D+ KAYDR+EWSFL+ +L++G + + D
Sbjct: 493 ITDNILIAQENFHALRTNPACKKKYMAIKTDMSKAYDRVEWSFLRALMLKMGFAQKWVDW 552
Query: 462 ILDCVSLSSFQVLFNGSKTEF 482
I+ C+S S+++L NGS F
Sbjct: 553 IIFCISSVSYKILLNGSPKGF 573
>At3g30570 putative reverse transcriptase
Length = 1099
Score = 226 bits (576), Expect = 6e-59
Identities = 154/501 (30%), Positives = 230/501 (45%), Gaps = 95/501 (18%)
Query: 7 YGSPNLSIRNFLWQELRRIAANTGEPWVVMGDFNTYLNAPDKWGGDPPNLLAMGKFRDCL 66
Y P S R+ LW L + P ++ DFN + ++ GG P F + +
Sbjct: 155 YAPPTPSRRSKLWDTLSAVIRGVKGPVIIGEDFNMIVRVDERSGGSGPLSPDSIAFGEWI 214
Query: 67 DDCYLSDLGFKGPPFTWEGRG--------------VKERLDWALGNDQWLRSFPEASIFH 112
+D L TW R V +RLD L + EA++ H
Sbjct: 215 NDSSL---------LTWVSRVTNSVGSGSVRRDFFVAKRLDRVLCCSHARLKWQEATLTH 265
Query: 113 LPQLKSDHKPLLLQLS-PIDIDHSQRP--------------------------------- 138
LP L SDH PL LQL+ + + +RP
Sbjct: 266 LPFLASDHAPLYLQLALGVGKNRFRRPFRFEAAWLSHPGFKELLLASWKGNIPTPEALKG 325
Query: 139 --------NRQVFGEIGRRKRHLMRRLEGINSRLRMQHIPYLEKLQKRLWKEYQTTLIQE 190
N++VFG+I +K LM ++ + L L + + L E+ L QE
Sbjct: 326 IQTTLRKWNKEVFGDIQEKKEKLMVEIKTVQDSLYSNQTDDLLRRESELIIEFDVVLEQE 385
Query: 191 ELLWRQKSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVTDPITLRSMAIDF 250
E LW KSR W GD+NT FFHTST++RR+ N+IE L +E G +++ L +AID+
Sbjct: 386 ETLWLHKSREKWFVQGDRNTSFFHTSTIIRRRHNRIEMLKNEKGGWISNTNELEKLAIDY 445
Query: 251 FQHLYTSPGTDVLYHVRNA--FPTLPRDTLLEISNPLEADEIKAAVFSMRALKAPGPDGL 308
++ LY+ D + A F L ++ + ++ P A E+++A+
Sbjct: 446 YKRLYSLDDVDEVVERLPAGRFLGLNQEEQIRLNKPFSAMEVESAL-------------- 491
Query: 309 NTLFFQSQWDTIGSSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICN 368
+ +F+ D +E ND ++VLI K+ +P ++Q+R I +CN
Sbjct: 492 --------------KITRFVLDFFLSGNLPQETNDAMVVLIAKVGKPEKITQFRPISLCN 537
Query: 369 VIYKTLTKALANRLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYLKRKKGWIA 428
V+ + +T + RLK V+ LI P+Q SFIPGR S+DNI I +E HSMR K KG +
Sbjct: 538 VLLEIITNTMVERLKSVITKLIGPSQTSFIPGRLSTDNIEIVEEDVHSMRCKKGHKGCML 597
Query: 429 IKIDLEKAYDRLEWSFLKDTL 449
+K DLEKAYDR+ W FL+DTL
Sbjct: 598 LKFDLEKAYDRVRWDFLEDTL 618
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 216 bits (550), Expect = 7e-56
Identities = 120/311 (38%), Positives = 182/311 (57%), Gaps = 12/311 (3%)
Query: 172 LEKLQKRLWKEYQTTLIQEELLWRQKSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTD 231
+ +L RL + Y+ EE W QKSR W+ GD N++FFH T RR RN+I L D
Sbjct: 102 ITELTLRLKEAYR----DEEQYWYQKSRSLWMKLGDNNSKFFHALTKQRRARNRITGLHD 157
Query: 232 EHGVSVTDPITLRSMAIDFFQHLYTSPGTDVLYHVRNAFPTLPRDTLLEISNPLEAD--- 288
E+G+ + ++++A+ +FQ+L+T+ V L D I++ L AD
Sbjct: 158 ENGIWSIEDDDIQNIAVSYFQNLFTTANPQVFDEALGEVQVLITD---RINDLLTADATE 214
Query: 289 -EIKAAVFSMRALKAPGPDGLNTLFFQSQWDTIGSSVVQFIQDCIQRPEKIREVNDTLIV 347
E++AA+F + KAPGPDG+ LFFQ W I S ++ + +Q + +N T I
Sbjct: 215 CEVRAALFMIHPEKAPGPDGMTALFFQKSWAIIKSDLLSLVNSFLQEGVFDKRLNTTNIC 274
Query: 348 LIPKIDRPNLLSQYRLIGICNVIYKTLTKALANRLKGVLGDLISPNQCSFIPGRQSSDNI 407
LIPK +RP +++ R I +CNV YK ++K L RLK VL +LIS Q +F+ GR SDNI
Sbjct: 275 LIPKTERPTRMTELRPISLCNVGYKVISKILCQRLKTVLPNLISETQSAFVDGRLISDNI 334
Query: 408 IIAQEVFHSMRYLKR-KKGWIAIKIDLEKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCV 466
+IAQE+FH +R K ++AIK D+ KAYD++EW+F++ L ++G + I+ C+
Sbjct: 335 LIAQEMFHGLRTNSSCKDKFMAIKTDMSKAYDQVEWNFIEALLRKMGFCEKWISWIMWCI 394
Query: 467 SLSSFQVLFNG 477
+ ++VL NG
Sbjct: 395 TTVQYKVLING 405
>At4g08830 putative protein
Length = 947
Score = 215 bits (547), Expect = 1e-55
Identities = 115/315 (36%), Positives = 184/315 (57%), Gaps = 20/315 (6%)
Query: 132 IDHSQRPNRQVFGEIGRRKRHLMRRLEGINSRLRMQHIPYLEKLQKRLWKEYQTTLIQEE 191
+D ++ NR VFG++ ++K LMR ++ + L + + + ++ L K +
Sbjct: 43 LDRLKKWNRDVFGDVKKKKEELMREIQVVQDALDVHQSDDMLRKEEDLIKAFD------- 95
Query: 192 LLWRQKSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVTDPITLRSMAIDFF 251
KSR W+ GD+NT +FHT+T++RR++N++E L +E G + D L ++A+ ++
Sbjct: 96 -----KSREKWIALGDRNTNYFHTTTIIRRRQNRVERLHNEEGTWIFDAKELEALAVQYY 150
Query: 252 QHLYTSPGTDVLYHV----RNAFPTLPRDTLLEISNPLEADEIKAAVFSMRALKAPGPDG 307
Q LY+ DVL V ++ L R+ +L ++ P A E++ A+ SM KAPGPDG
Sbjct: 151 QRLYSMD--DVLQVVNPLPQDGLERLTREEVLSLNKPFLATEVEVAIRSMGKYKAPGPDG 208
Query: 308 LNTLFFQSQWDTIGSSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGIC 367
+F+QS W+ +G+SV++F D + ND L+VLIPK+ +P ++Q+R I +C
Sbjct: 209 YQPIFYQSCWEVVGNSVIRFALDFFSSGILPPQTNDALVVLIPKVPKPERMNQFRPISLC 268
Query: 368 NVIYKTLTKALANRLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYLKRKKGWI 427
N I+K +TK + RLK V+G LI P+Q SFIPGR S DNI++ QE HS+ K +K +
Sbjct: 269 NAIFKMITKMMVLRLKKVIGKLIGPSQSSFIPGRLSLDNIVVVQEAVHSLNRKKGRKERL 328
Query: 428 AIKIDLEKAYDRLEW 442
ID +A EW
Sbjct: 329 CHMID--RAVAVKEW 341
>At2g11240 pseudogene
Length = 1044
Score = 213 bits (542), Expect = 6e-55
Identities = 149/458 (32%), Positives = 222/458 (47%), Gaps = 59/458 (12%)
Query: 68 DCYLSDLGFKGPPFTWEGRG----VKERLDWALGNDQWLRSFPEASIFHLPQLKSDHKPL 123
+C L DL G +W G+ V RLD AL N W +P + +L SDH+PL
Sbjct: 3 ECDLYDLRHSGNFLSWRGKRHDHVVHCRLDRALSNGAWAEDYPASRCIYLCFEGSDHRPL 62
Query: 124 LLQLSPIDIDHSQRPNRQVFGEIGRRKRH----------------------LMR-RLEGI 160
L D S++ + VF R K + + R RLE +
Sbjct: 63 LTHF-----DLSKKKKKGVFRYDRRLKNNDEVTALVQEAWNLYDTDIVEEKISRCRLEIV 117
Query: 161 N-SRLRMQHIPYL-----EKLQK---------RLWKEYQTTLI----QEELLWRQKSRVA 201
SR + Q L +KL++ L T L+ EE W+Q+SR
Sbjct: 118 KWSRAKQQSSQKLIEENRQKLEEAMSSQDHNQELLSTINTNLLLAYKAEEEYWKQRSRQL 177
Query: 202 WLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVTDPITLRSMAIDFFQHLYTSPGTD 261
WL GDKN+ +FH T R NK + E GV + + ++ ++FQ L+++
Sbjct: 178 WLALGDKNSGYFHAITRGRTVINKFSVIEKEDGVPEYEEAGILNVISEYFQKLFSANEGA 237
Query: 262 VLYHVRNAFPTLPRDTLLEISNPLEADEIKAAVFSMRALKAPGPDGLNTLFFQSQWDTIG 321
++ A IS +EIK+A FS+ A KAPGPDG + FFQS W T+G
Sbjct: 238 RAATIKEAIKPF-------ISPEQNPEEIKSACFSIHADKAPGPDGFSASFFQSNWMTVG 290
Query: 322 SSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVIYKTLTKALANR 381
++V IQ +N T I LIPKI + YR I +C V YK ++K L+ R
Sbjct: 291 PNIVLEIQSFFSSSTLQPTINKTHITLIPKIQSLKRMVDYRPIALCTVFYKIISKLLSRR 350
Query: 382 LKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYL-KRKKGWIAIKIDLEKAYDRL 440
L+ +L ++IS NQ +F+P R S+DN++I E H ++ L K+ ++A+K ++ KAYDR+
Sbjct: 351 LQPILQEIISENQSAFVPKRASNDNVLITHEALHYLKSLGAEKRCFMAVKTNMSKAYDRI 410
Query: 441 EWSFLKDTLLEVGLDRNFCDLILDCVSLSSFQVLFNGS 478
EW F+K + E+G + + IL C++ S+ L NGS
Sbjct: 411 EWDFIKLVMQEMGFHQTWISWILQCITTVSYSFLLNGS 448
>At4g04000 putative reverse transcriptase
Length = 1077
Score = 200 bits (508), Expect = 5e-51
Identities = 137/439 (31%), Positives = 206/439 (46%), Gaps = 63/439 (14%)
Query: 91 RLDWALGNDQWLRSFPEASIFHLPQLKSDHKPLLLQLSPIDIDHSQRPNRQVFG------ 144
RLD A+ N W+ ++P +L SDH+PL+ DH Q+ + +F
Sbjct: 172 RLDRAMANSDWIIAYPSGRSEYLRFEGSDHRPLVTSF-----DHVQKKTKNIFRYDRTLK 226
Query: 145 ---EIGR-----------------------------RKRHLMRRLEGINSRLRMQHI--- 169
EI + ++ HL + E ++ R++++
Sbjct: 227 DNPEISKLMNETWKSNPSAKVDYRLHLCRAALLNWSKEHHLNSQKEILSLRVQLEEALTD 286
Query: 170 -----PYLEKLQKRLWKEYQTTLIQEELLWRQKSRVAWLNHGDKNTRFFHTSTMVRRKRN 224
++ L + L Y+ +EE W+Q+SR WL GDKN+ FFH +T RR N
Sbjct: 287 DAATQDSVDLLNQNLLLAYR----KEETFWKQRSRNLWLALGDKNSCFFHAATNSRRAIN 342
Query: 225 KIEALTDEHGVSVTDPITLRSMAIDFFQHLYTSPGTDVLYHVRNAF-PTLPRDTLLEISN 283
I + + G SV + + D+FQ ++TS D V A P + D + +
Sbjct: 343 TISVIENSAGTSVYEDSEIIETISDYFQDIFTSQEGDRTSVVAEALSPCITTDMNNTLIS 402
Query: 284 PLEADEIKAAVFSMRALKAPGPDGLNTLFFQSQWDTIGSSVVQFIQDCIQRPEKIREVND 343
EIK A S+ KAPG DG + FFQS W T+ + +Q +++N
Sbjct: 403 LPTVAEIKQACLSIHPDKAPGRDGFSASFFQSNWSTVSEEITAEVQAFFISEILPQKINH 462
Query: 344 TLIVLIPKIDRPNLLSQYRLIGICNVIYKTLTKALANRLKGVLGDLISPNQCSFIPGRQS 403
T + LIPKI P +S YR I +C+V YK + K LA RL+ +L IS NQ +F+P R
Sbjct: 463 THVRLIPKIPAPQKVSDYRPIALCSVYYKIIAKLLAKRLQPILHSCISENQSAFVPQRAI 522
Query: 404 SDNIIIAQEVFHSMRYLKRKKG----WIAIKIDLEKAYDRLEWSFLKDTLLEVGLDRNFC 459
SDN++I E H YLK ++A+K D+ KAYDRLEW F+K L E+G +F
Sbjct: 523 SDNVLITHEALH---YLKNSGALVNCYMAVKTDMSKAYDRLEWDFVKAALEEMGFHHHFV 579
Query: 460 DLILDCVSLSSFQVLFNGS 478
I++ +S + NGS
Sbjct: 580 GWIIEVLSRMCLKAQENGS 598
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 196 bits (497), Expect = 9e-50
Identities = 108/291 (37%), Positives = 164/291 (56%), Gaps = 2/291 (0%)
Query: 189 QEELLWRQKSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVTDPITLRSMAI 248
+EE WR KSR W+ GDKN++FF + R N + L DE+G T +A+
Sbjct: 60 EEEDFWRLKSRDKWMVGGDKNSKFFQATVKANRVSNSLRFLVDENGNEQTVNREKGKIAV 119
Query: 249 DFFQHLYTSPGTDVLYHVRNAF-PTLPRDTLLEISNPLEADEIKAAVFSMRALKAPGPDG 307
FF+ L++S + V F + D +++ + EI AVFS+ A APGPDG
Sbjct: 120 TFFEDLFSSSYPSSMDSVLEGFNKRVTEDMNQDLTKKVNEQEIYKAVFSINAESAPGPDG 179
Query: 308 LNTLFFQSQWDTIGSSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGIC 367
LFFQ QW + + ++ I+ Q + N T + LIPKI +P ++ R I +C
Sbjct: 180 FTALFFQRQWPLVKNQIISDIELFFQTGILPEDWNHTHLCLIPKITKPARMADIRPISLC 239
Query: 368 NVIYKTLTKALANRLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYLKR-KKGW 426
+V+YK ++K L+ RLK L ++SP Q +F+ R SDNII+A E+ H++R ++ K +
Sbjct: 240 SVMYKIISKILSARLKKYLPVIVSPTQSAFVAERLVSDNIILAHEIVHNLRTNEKISKDF 299
Query: 427 IAIKIDLEKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCVSLSSFQVLFNG 477
+ K D+ KAYDR+EW FLK LL +G + + + ++ CVS S+ VL NG
Sbjct: 300 MVFKTDMSKAYDRVEWPFLKGILLALGFNSTWINWMMACVSSVSYSVLING 350
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.343 0.152 0.539
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,787,739
Number of Sequences: 26719
Number of extensions: 1182374
Number of successful extensions: 3273
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 62
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 3004
Number of HSP's gapped (non-prelim): 124
length of query: 1248
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1137
effective length of database: 8,352,787
effective search space: 9497118819
effective search space used: 9497118819
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0037a.3


