

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
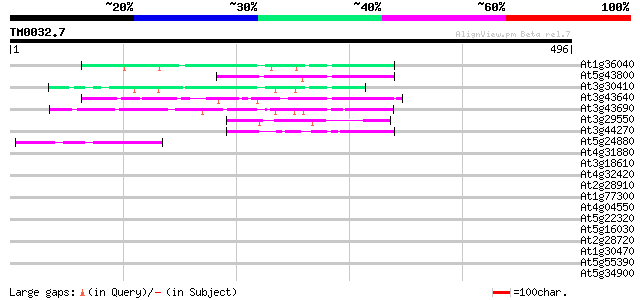
Query= TM0032.7
(496 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g36040 putative envelope-like protein gb|AAD20656 82 5e-16
At5g43800 putative protein 74 2e-13
At3g30410 hypothetical protein 61 1e-09
At3g43640 putative protein 60 4e-09
At3g43690 unknown protein 58 1e-08
At3g29550 hypothetical protein 48 1e-05
At3g44270 putative protein 42 6e-04
At5g24880 glutamic acid-rich protein 42 8e-04
At4g31880 unknown protein 42 0.001
At3g18610 unknown protein 42 0.001
At4g32420 cyclophilin (AtCYP95) 37 0.033
At2g28910 unknown protein 36 0.043
At1g77300 hypothetical protein 35 0.073
At4g04550 putative protein 35 0.095
At5g22320 unknown protein 35 0.12
At5g16030 unknown protein 35 0.12
At2g28720 putative histone H2B 35 0.12
At1g30470 unknown protein 35 0.12
At5g55390 putative protein 34 0.16
At5g34900 putative protein 34 0.16
>At1g36040 putative envelope-like protein gb|AAD20656
Length = 444
Score = 82.4 bits (202), Expect = 5e-16
Identities = 83/312 (26%), Positives = 124/312 (39%), Gaps = 51/312 (16%)
Query: 64 STPEVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVK------TPSEKKKRKVPVDVEDS 117
+TP P PP P T + K K K + T + K R V DS
Sbjct: 57 ATPSDPPPPPVAPMFIPKTEPGICMSKKTKSKVSTLTQRKRQCTAASAKFRSKAQLVRDS 116
Query: 118 ESDAEADVQDIRPS---------------EKKKYSGKRIPQNVPVVPIDRVSFHLEESVQ 162
+ DV +I P +KY + P P F EE +
Sbjct: 117 SDLQDGDVTEIAPPLFLSCYRENRRLLAVRDRKYPELKFPSETPYSSC----FLTEEGLG 172
Query: 163 KWKFVCKIRMAKVREVGPDVLECKDVIVLIEKAGLMKTIQNVGRCYEKLVREFLVNLSVD 222
++K + +R + D L+ GL+ T+ + ++V EF NL
Sbjct: 173 RYKVIGNRCFNDMRFLPLDGNNTASTQQLLFNGGLLPTVTEIDSYVHEVVMEFYANL--- 229
Query: 223 VGLPESDE----FRKVFVRVECVVFSPAVINQAL----------GRSAIEFADKEVSMDD 268
P+ +E VFVR FS A+INQ G S I+ + SMD+
Sbjct: 230 ---PDGEEGDNLAYSVFVRRNMYEFSLAIINQMFQLPNPSYPLDGMSEIQVPE---SMDE 283
Query: 269 VAKELTAGHVKKWPSKKLLSTGNLSVKYAILNRIDAVNWVPTQHTSGVSATLAKLIYMIG 328
VA L+ G W K+L++ LS + A+LN+I NW PT + S + L+YM+
Sbjct: 284 VAIALSNGKANSW---KMLTSRLLSPELALLNKICCHNWSPTVNRSVLKPERMTLLYMVA 340
Query: 329 KSIAFDFGAFVF 340
K++ F+FG +F
Sbjct: 341 KALPFNFGKLIF 352
>At5g43800 putative protein
Length = 515
Score = 73.9 bits (180), Expect = 2e-13
Identities = 50/163 (30%), Positives = 83/163 (50%), Gaps = 12/163 (7%)
Query: 184 ECKDVIVLIEKAGLMKTIQNVGRCYEKLVREFLVNLSVDVGLPESDEFRKVFVRVECVVF 243
E +D++ +IE L+ T+ + ++++ EF N + E + KVFVR + F
Sbjct: 201 EMRDMMKVIEDNHLLDTVAEIDPFVKEVIWEFYANFHI---FNEETKKSKVFVRGKMYEF 257
Query: 244 SPAVINQALGRSAI------EFADKEVSMDDVAKELTAGHVKKWPSKKLLSTGNLSVKYA 297
SP +IN+ AI EF +S +AK L+ G V W K L T + A
Sbjct: 258 SPQMINKTFRSPAIKWHAKAEFERVTMSKHALAKYLSDGKVVTW---KDLRTPVMPTHLA 314
Query: 298 ILNRIDAVNWVPTQHTSGVSATLAKLIYMIGKSIAFDFGAFVF 340
L I +NW+P+++ S V+ AK++Y++ + I F+FG V+
Sbjct: 315 GLYLICCINWIPSKNNSNVTVDRAKMLYLLNERIPFNFGKLVY 357
>At3g30410 hypothetical protein
Length = 421
Score = 61.2 bits (147), Expect = 1e-09
Identities = 76/307 (24%), Positives = 121/307 (38%), Gaps = 55/307 (17%)
Query: 35 PVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTYKSRQATVKGKG 94
P S P +P E + ++ KVS P + + C AT K +
Sbjct: 27 PPSSVPMFIPKTEPGIYMS---KKTKSKVSAPS------QSKRQCTA------ATSKTRS 71
Query: 95 KQKVVKTPSEKKKRK-------VPVDVEDSESDAEADVQDIRP-------SEKKKYSGKR 140
K ++ S KK + V +S + DV I P E ++ R
Sbjct: 72 KAPALRRLSSKKSTRSASAAGQTEEHVFESSDLQDGDVTQIAPPLFLSGYQENRRLLSAR 131
Query: 141 IPQNVPVVPIDRVSFHLEESVQKWKFVCKIRMAKVREVGPDVLECKDVIVLIEKAGLMKT 200
PQ + F E + ++K + +R + D + L+ AGL+ T
Sbjct: 132 DPQVSSETSYNNC-FLTAEGLGRYKLIGGRYFNDMRFLSLDGNNTASTLQLLSTAGLLLT 190
Query: 201 IQNVGRCYEKLVREFLVNLSVDVGLPESDEFRK----VFVRVECVVFSPAVINQA----- 251
+ + +++V EF NL P+ +E + VFVR FSPA+INQ
Sbjct: 191 VTEIDSYVQEVVMEFYANL------PDGEEGDQLAYFVFVRGNMYEFSPAIINQIFQLPN 244
Query: 252 ----LGRSAIEFADKEVSMDDVAKELTAGHVKKWPSKKLLSTGNLSVKYAILNRIDAVNW 307
L S I+ + SMD+V + W K+L++ LS + A+LN+I NW
Sbjct: 245 PPYPLNMSKIKVPE---SMDEVDVARSNSKANSW---KMLTSQLLSPEIALLNKIYCHNW 298
Query: 308 VPTQHTS 314
+PT + S
Sbjct: 299 MPTVNRS 305
>At3g43640 putative protein
Length = 428
Score = 59.7 bits (143), Expect = 4e-09
Identities = 73/293 (24%), Positives = 120/293 (40%), Gaps = 43/293 (14%)
Query: 64 STPEVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDVEDSESDAEA 123
+TP P PP P T + K K K V P+++K++ + S S A A
Sbjct: 57 ATPSDPPPPPVAPMFIPKTEPGICMSKKTKSK---VSAPTQRKRQCTAASAK-SRSKAPA 112
Query: 124 DVQDIRPSEKKKYSGKRIPQNVPVVPIDRVSFHLEESVQKWKFVCKIRMAKVREVGPDVL 183
S K S Q +V R S L++ V E+ P +
Sbjct: 113 PALRRLSSRKSNRSASAAGQTEDLV---RDSSDLQDG-------------DVTEIAPPLF 156
Query: 184 -----ECKDVIVLIEKAGLMKTIQNVGRCYEKLVREFLV----NLSVDVGLPESDEFRKV 234
E + ++ + ++ G K I N RC+ + FL N + L +
Sbjct: 157 LSRYRENRRLLAVRDRLGRYKVIGN--RCFNDM--RFLPLDGNNTASTQQLLFNAGLLPT 212
Query: 235 FVRVECVVFSPAVINQALGRSAIEFADKEVSMDDVAKELTAGHVKKWPSKKLLSTGNLSV 294
F ++ + +++ + D SMD+VA L+ G W K+L++ LS
Sbjct: 213 FTEID------SYVHEVVMEFYANLPDVPESMDEVAIALSNGKANSW---KMLTSRLLSP 263
Query: 295 KYAILNRIDAVNWVPTQHTSGVSATLAKLIYMIGKSIAFDFGAFVF*ADIEAC 347
+ A+LN+I NW PT + S + L+YM+ K++ F+FG +F +I AC
Sbjct: 264 ELALLNKICCHNWSPTVNHSVLKPERMTLLYMVAKALPFNFGKLIF-DEIWAC 315
>At3g43690 unknown protein
Length = 539
Score = 58.2 bits (139), Expect = 1e-08
Identities = 76/317 (23%), Positives = 130/317 (40%), Gaps = 37/317 (11%)
Query: 36 VKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTYKSRQATVKGKGK 95
V PP P + S S++ P ++ P P+P PV +
Sbjct: 135 VSIPPPSAPPLVLSDSKDAE----PAGLTNPSAPPSPLAPKNITPVASPVADVPMPDP-- 188
Query: 96 QKVVKTPSEKKKRKVPVDVEDSESDAEADVQDIRPSEKKKYSGKRIPQNVPVVPIDRVSF 155
+ T + VP + A A Q ++ + +R P+VP F
Sbjct: 189 -LISPTAETAEGASVPDAAVSYAARAAAHRQVFAERDELDRTLRR-----PLVPPHTKRF 242
Query: 156 HLEESVQKWKFVCK--IRMAKVREVGPDVLECKDVIVLIEKAGLMKTIQNVGRCYEKLVR 213
+ +++K + K K + P+VL +E +G+ +T+ V + ++VR
Sbjct: 243 LSAAAAERYKHIAKRDFIFQKTLPLDPEVLTATKYF--LEHSGMAQTVVAVEQFVPEVVR 300
Query: 214 EFLVNLSVDVGLPESDEFRK-----VFVRVECVVFSPAVINQ--ALGRSAIE----FADK 262
EF NL PE E+R+ V+VR + FSPA+IN ++ SA++
Sbjct: 301 EFYANL------PEM-EYRECGLDLVYVRGKMYEFSPALINHMFSIDDSALDPEAPVTLS 353
Query: 263 EVSMDDVAKELTAGHVKKWPSKKLLSTGNLSVKYAILNRIDAVNWVPTQHTSGVSATLAK 322
S DD+A +T G ++W +L +L +L+++ NW PT +TS + +
Sbjct: 354 TASRDDLALMMTGGTTRRW--LRLQPADHLDTM-KMLHKVCCGNWFPTTNTSTLRVDRLR 410
Query: 323 LIYMIGKSIAFDFGAFV 339
LI M +F+ G V
Sbjct: 411 LIDMGTHGKSFNLGKLV 427
>At3g29550 hypothetical protein
Length = 223
Score = 48.1 bits (113), Expect = 1e-05
Identities = 47/156 (30%), Positives = 66/156 (42%), Gaps = 52/156 (33%)
Query: 192 IEKAGLMKTIQNVGRCYEKLVREFLVNL------SVDVGLPESDEFRKVFVRVECVVFSP 245
IEKAGL++T+ ++ E+LV EF NL + +VG V VR FSP
Sbjct: 38 IEKAGLIRTVTDLKPYKEELVFEFWANLPTVKVDTTNVG---------VLVRNWEYEFSP 88
Query: 246 AVINQALGRSAIEFADKEVSM-----DDVAKELTAGHVKKWPSKKLLSTGNLSVKYAILN 300
IN+ G ++++ + + M D+VAK LT G VK
Sbjct: 89 ENINELFGLASVDVRQQRMDMVGLMEDEVAKFLTDGKVKS-------------------- 128
Query: 301 RIDAVNWVPTQHTSGVSATLAKLIYMIGKSIAFDFG 336
G+ AKL+YMI +I FDFG
Sbjct: 129 ------------LKGLLPDRAKLVYMIMHNIPFDFG 152
>At3g44270 putative protein
Length = 324
Score = 42.4 bits (98), Expect = 6e-04
Identities = 45/149 (30%), Positives = 66/149 (44%), Gaps = 27/149 (18%)
Query: 192 IEKAGLMKTIQNVGRCYEKLVREFLVNLSVDVGLPESDEFRKVFVRVECVVFSPAVINQA 251
I AGL++T+ + E+LV EF NL V+V+ S +V+ Q
Sbjct: 147 IYDAGLIRTVTELKPFEEELVFEFWANLPT--------------VKVDST--SVSVMQQM 190
Query: 252 LGRSAIEFADKEVSMDDVAKELTAGHVKKWPSKKLLSTGNLSVKYAILNRIDAVNWVPTQ 311
+E D+VA+ LT G VK K+LL + S+ L ++ NW PT
Sbjct: 191 DMAGLLE--------DEVAQFLTDGKVKLL--KRLLLSA-FSLPGRELFKLCCTNWSPTS 239
Query: 312 HTSGVSATLAKLIYMIGKSIAFDFGAFVF 340
+ S AKL+YMI + F+F F
Sbjct: 240 NNSYAQPDRAKLVYMIMHKMPFNFVRLAF 268
>At5g24880 glutamic acid-rich protein
Length = 443
Score = 42.0 bits (97), Expect = 8e-04
Identities = 34/130 (26%), Positives = 58/130 (44%), Gaps = 12/130 (9%)
Query: 6 MSKVQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVST 65
+++++ K N E+ T+E + D + KS E V ++I + E P KV T
Sbjct: 267 VAELEEKLIKNEDDIEEKTEEMKEQDNNQANKSEE------EEDVKKKIDENETPEKVDT 320
Query: 66 PEVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDVEDSESDAEADV 125
K +S T + ++ VK +GK++V + EK+K K E E + + V
Sbjct: 321 ES------KEVESVEETTQEKEEEVKEEGKERVEEEEKEKEKVKEDDQKEKVEEEEKEKV 374
Query: 126 QDIRPSEKKK 135
+ EK K
Sbjct: 375 KGDEEKEKVK 384
>At4g31880 unknown protein
Length = 873
Score = 41.6 bits (96), Expect = 0.001
Identities = 38/123 (30%), Positives = 56/123 (44%), Gaps = 15/123 (12%)
Query: 25 DEEEINDEDSPVKSPPDVVPGVETSVSQ-EISDQENPGKVSTPEVTPTPPKR------SK 77
+EE ++E+S + PP V +S S+ +IS GK P + SK
Sbjct: 744 EEEASSEEESEEEEPPKTVGKSGSSRSKKDISSVSKSGKSKASSKKKEEPSKATTSSKSK 803
Query: 78 SCPV-TYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDVEDSESDAEADVQDIRPSEKKKY 136
S PV + ++ T KGK K TP+ K K SES++E ++ P+ K K
Sbjct: 804 SGPVKSVPAKSKTGKGKAKSGSASTPASKAKESA------SESESEETPKEPEPATKAK- 856
Query: 137 SGK 139
SGK
Sbjct: 857 SGK 859
>At3g18610 unknown protein
Length = 636
Score = 41.6 bits (96), Expect = 0.001
Identities = 35/143 (24%), Positives = 60/143 (41%), Gaps = 3/143 (2%)
Query: 12 KEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPT 71
KEP V +D +DE ++E VK P V + S ++ + TP PT
Sbjct: 228 KEPIVVK--KDSSDESSSDEETPVVKKKPTTVVKDAKAESSSSEEESSSDDEPTPAKKPT 285
Query: 72 PPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDVEDSESDAEADVQDIRPS 131
K +K S + + + K P++K K ++S SD +D D S
Sbjct: 286 VVKNAKPAAKDSSSSEED-SDEEESDDEKPPTKKAKVSSKTSKQESSSDESSDESDKEES 344
Query: 132 EKKKYSGKRIPQNVPVVPIDRVS 154
+ +K + K+ +V +V ++ S
Sbjct: 345 KDEKVTPKKKDSDVEMVDAEQKS 367
Score = 30.4 bits (67), Expect = 2.3
Identities = 24/120 (20%), Positives = 50/120 (41%), Gaps = 4/120 (3%)
Query: 10 QNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVT 69
+ E T E + + +DE+ K P + + S S+E D + +++ +
Sbjct: 52 EKAEKTVPKKVESSSSDASDSDEEEKTKETPSKLK--DESSSEEEDDSSSDEEIAPAKKR 109
Query: 70 PTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDVEDSESDAEADVQDIR 129
P P K++K + S + + V K P+ +K KV D +S ++ + ++
Sbjct: 110 PEPIKKAK--VESSSSDDDSTSDEETAPVKKQPAVLEKAKVESSSSDDDSSSDEETVPVK 167
>At4g32420 cyclophilin (AtCYP95)
Length = 837
Score = 36.6 bits (83), Expect = 0.033
Identities = 30/121 (24%), Positives = 53/121 (43%), Gaps = 13/121 (10%)
Query: 47 ETSVSQEISDQENPGKVSTPEVTPTPP-------KRSKSCPVTYKSRQATVKGKGKQKVV 99
ET S+ SD E+ +S+P + KRS +S+Q + + K+ +
Sbjct: 216 ETDSSE--SDSESDSDLSSPSFLSSSSHERQKKRKRSSKKDKHRRSKQRDKRHEKKRSMR 273
Query: 100 KTPSEKKKRKVPVDVEDSESDAEADVQDIR----PSEKKKYSGKRIPQNVPVVPIDRVSF 155
++K R+ P +EDS S +EA + D+ ++K +R + P V + S
Sbjct: 274 DKRPKRKSRRSPDSLEDSNSGSEASLSDVNVEIGAKKRKHRVSRRTGNSAPAVEKEAESL 333
Query: 156 H 156
H
Sbjct: 334 H 334
>At2g28910 unknown protein
Length = 332
Score = 36.2 bits (82), Expect = 0.043
Identities = 30/86 (34%), Positives = 40/86 (45%), Gaps = 7/86 (8%)
Query: 61 GKVSTPEVTPTPPKRSKSCPVTYKSRQATV------KGKGKQKVVKTPSEKKKRKVPVDV 114
GK EV+ + S+S S + K KG V KT S +KK+K D
Sbjct: 129 GKGEVEEVSSEEEEESESSDSDVDSEMERIIAERFGKKKGGSSVKKTSSVRKKKKRVSDE 188
Query: 115 EDSESDAEADVQDIRPSEKKKYSGKR 140
DS+SD+ D + R S KK+ S KR
Sbjct: 189 SDSDSDS-GDRKRRRRSMKKRSSHKR 213
>At1g77300 hypothetical protein
Length = 408
Score = 35.4 bits (80), Expect = 0.073
Identities = 22/86 (25%), Positives = 37/86 (42%), Gaps = 1/86 (1%)
Query: 35 PVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTYKSRQATVKGKG 94
P+ P +V+ + + +S QE P + T +PT S+ P S + T G G
Sbjct: 276 PLLQPTEVLKELSSGISITAVQQEVPAEKKTKSTSPTSSSLSRMSPGGTNSDKTTKHGSG 335
Query: 95 KQKVVKTPSEKKKRKVPVDVEDSESD 120
+ K + P + + K E S+ D
Sbjct: 336 EDKKI-LPRPRPRMKTSRSSESSKRD 360
>At4g04550 putative protein
Length = 307
Score = 35.0 bits (79), Expect = 0.095
Identities = 28/94 (29%), Positives = 37/94 (38%), Gaps = 8/94 (8%)
Query: 248 INQALGRSAIEFADKEVSM-----DDVAKELTAGHVKKWPSKKLLSTGNLSVKYAILNRI 302
IN + G ++ +V + D+V LT G VK S L S L +
Sbjct: 123 INASFGLQLVDARALQVQIASLIDDEVTSYLTNGQVKVLQS---LPMSTFSENCRKLFKF 179
Query: 303 DAVNWVPTQHTSGVSATLAKLIYMIGKSIAFDFG 336
NW PT S L+Y I +AFDFG
Sbjct: 180 SCRNWSPTTSEGYASIHRVLLMYRIAHKLAFDFG 213
>At5g22320 unknown protein
Length = 452
Score = 34.7 bits (78), Expect = 0.12
Identities = 29/102 (28%), Positives = 47/102 (45%), Gaps = 16/102 (15%)
Query: 8 KVQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPE 67
K ++KE T +P D D+ E + + K D + ETS + EI +EN K S
Sbjct: 339 KEEHKEKT-IPSNNDDDDDAEKKQKRATPKEELDAIDDAETSFA-EIFSRENVPKGSEDG 396
Query: 68 VTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRK 109
+ K ++++V+ G KV+ T + KKK+K
Sbjct: 397 IE--------------KKKKSSVQETGLVKVIDTKANKKKKK 424
>At5g16030 unknown protein
Length = 339
Score = 34.7 bits (78), Expect = 0.12
Identities = 32/139 (23%), Positives = 62/139 (44%), Gaps = 15/139 (10%)
Query: 6 MSKVQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVP-----GVETSVSQEISDQENP 60
M+ + +P N P+ + ++ D + + VV G++++ S ++S +P
Sbjct: 158 MNNIAQNKPNNNPNVRVIHEDLYAPDPELLALAEKKVVGMKRDVGIQSTTSVDLSSG-SP 216
Query: 61 GKVSTPEVTPTPPKR---SKSCPVTYKSRQATVKGKGKQKVVKTPS-EKKKRKVPVDVED 116
TP + KR + PV +K KG+Q+ VK EK++ K + E+
Sbjct: 217 SPAKTPPIMERSLKRHVEADDWPV-----DINLKVKGQQQDVKLEEKEKEEEKQDMSNEE 271
Query: 117 SESDAEADVQDIRPSEKKK 135
E + E + QD+ + K+
Sbjct: 272 DEEEEEEEKQDMSEEDDKE 290
>At2g28720 putative histone H2B
Length = 151
Score = 34.7 bits (78), Expect = 0.12
Identities = 29/120 (24%), Positives = 59/120 (49%), Gaps = 22/120 (18%)
Query: 97 KVVKTPSEKKK-RKVPVDVEDSESDAEADVQDIRPSEKKKYSGKRIPQNVPVVPIDRVSF 155
K K P+EKK K P + E V + P+EKK +GK++P+ +++
Sbjct: 4 KAGKKPAEKKPAEKAPAE--------EEKVAEKAPAEKKPKAGKKLPKEAVTGGVEKKKK 55
Query: 156 HLEESVQKWK-FVCKIRMAKVREVGPDVLECKDVIVLIEKAGLMKTIQNVGRCYEKLVRE 214
+++S + +K ++ K+ +++V P D+ + + G+M + + +EKL +E
Sbjct: 56 RVKKSTETYKIYIFKV----LKQVHP------DIGISSKAMGIMNSF--INDIFEKLAQE 103
>At1g30470 unknown protein
Length = 843
Score = 34.7 bits (78), Expect = 0.12
Identities = 37/123 (30%), Positives = 52/123 (42%), Gaps = 16/123 (13%)
Query: 11 NKEPTNVPHCEDVTDEEEINDED------SPVKSPPDVVPGVETSV----SQEISDQENP 60
N E T + DV E+E ND+D S VK+P VPG ET+ + EN
Sbjct: 486 NPEETTILSNGDVQIEKEDNDDDDDTDNKSAVKTPG--VPGDETTEKLPDESGVEPTENS 543
Query: 61 GKVSTPEVTPTPPKRSKSCPVTYKSRQ---ATVKGKGKQKVVKTPSEKKKRKVPVDVEDS 117
K S E T + PK S VT R A K ++P K+ +V + +
Sbjct: 544 PKASGIEPTESSPKAS-GAEVTGNLRDSDPAESHADAKSSEPESPHGTKETEVAAEADTK 602
Query: 118 ESD 120
E++
Sbjct: 603 ETE 605
>At5g55390 putative protein
Length = 1332
Score = 34.3 bits (77), Expect = 0.16
Identities = 31/120 (25%), Positives = 48/120 (39%), Gaps = 17/120 (14%)
Query: 13 EPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPG-KVSTPEVTPT 71
E + P V E +D SPV+S PD + V TS+S + + + S P +T T
Sbjct: 1050 ELSRAPESIKVKIPEMTSDWQSPVRSSPDDIYAVCTSISTTTPQRSHEAVEASLPAITRT 1109
Query: 72 PPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDVEDSESDAEADVQDIRPS 131
K+ + V+G GK +V + D S + D+ +RPS
Sbjct: 1110 KSNLGKN----IREHGCKVQGTGKPEVSR------------DRPSSVRTSREDIYTVRPS 1153
>At5g34900 putative protein
Length = 767
Score = 34.3 bits (77), Expect = 0.16
Identities = 27/119 (22%), Positives = 52/119 (43%), Gaps = 6/119 (5%)
Query: 26 EEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEV-TPTPPKRSKSCPVTYK 84
E+ I + ++ S P + V E++++ + K TP TP S+ K
Sbjct: 374 EDSIREPNTQYGSYPGDDENTQCDVGDEVAEESSKDKSPTPRSSTPNFNILSEESLDVQK 433
Query: 85 SRQATVKGKGKQKVVKTPSE-----KKKRKVPVDVEDSESDAEADVQDIRPSEKKKYSG 138
++ + +G+ + K VK K +++VP + A+ DV + S+K+K G
Sbjct: 434 DKKRSSRGRNENKRVKPSVHAEDNLKTRKQVPRKRQKQVDSADVDVPTRKESKKRKIIG 492
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.333 0.142 0.464
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,712,092
Number of Sequences: 26719
Number of extensions: 463569
Number of successful extensions: 3741
Number of sequences better than 10.0: 130
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 112
Number of HSP's that attempted gapping in prelim test: 3528
Number of HSP's gapped (non-prelim): 275
length of query: 496
length of database: 11,318,596
effective HSP length: 103
effective length of query: 393
effective length of database: 8,566,539
effective search space: 3366649827
effective search space used: 3366649827
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0032.7


