

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0031.3
(547 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
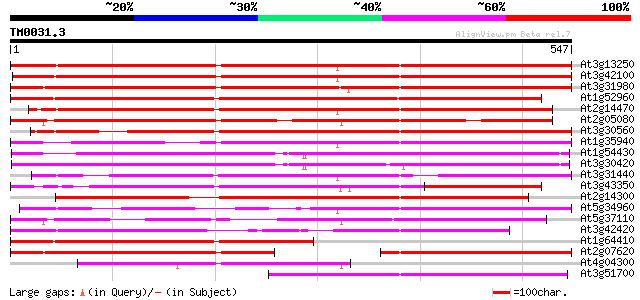
Score E
Sequences producing significant alignments: (bits) Value
At3g13250 hypothetical protein 505 e-143
At3g42100 putative protein 499 e-141
At3g31980 hypothetical protein 484 e-137
At1g52960 hypothetical protein 481 e-136
At2g14470 pseudogene 465 e-131
At2g05080 putative helicase 426 e-119
At3g30560 hypothetical protein 421 e-118
At1g35940 hypothetical protein 420 e-118
At1g54430 hypothetical protein 401 e-112
At3g30420 hypothetical protein 401 e-112
At3g31440 hypothetical protein 395 e-110
At3g43350 putative protein 387 e-108
At2g14300 pseudogene; similar to MURA transposase of maize Muta... 385 e-107
At5g34960 putative protein 327 8e-90
At5g37110 putative helicase 324 7e-89
At3g42420 putative protein 316 2e-86
At1g64410 unknown protein 279 3e-75
At2g07620 putative helicase 248 5e-66
At4g04300 hypothetical protein 229 3e-60
At3g51700 unknown protein 188 6e-48
>At3g13250 hypothetical protein
Length = 1419
Score = 505 bits (1301), Expect = e-143
Identities = 267/551 (48%), Positives = 366/551 (65%), Gaps = 10/551 (1%)
Query: 1 DEELQNLCMIEIEKILQGNERSLKEFPCLPYPKFSEIHNFEVIFVADELNYN-RAEMVKI 59
+ ++QN + EIEKI+ N +L++ P P I N + + DEL YN ++++ K
Sbjct: 874 EAQIQNYTLQEIEKIMLFNGATLEDIEHFPKPSREGIDNSNRLII-DELRYNNQSDLKKK 932
Query: 60 HDEFVSSLTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVL 119
H +++ LT EQ Y + + V +D FF+YGFGGTGKTF+W L AA+RS+G I L
Sbjct: 933 HSDWIQKLTPEQRGIYDQITNAVFNDLGGVFFVYGFGGTGKTFIWKTLAAAVRSKGQICL 992
Query: 120 NVASSGIASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAELLKKASLIIWDEAPML 179
NVASSGIASLLL G RTAHSRFSIP++ +E S C ++ S A+L+K+ASLIIWDEAPM+
Sbjct: 993 NVASSGIASLLLEGGRTAHSRFSIPLNPDEFSVCKIKPKSDLADLIKEASLIIWDEAPMM 1052
Query: 180 NKHCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGSAIN 239
+K C EALD+S +DI+K FGGKV+V G DF Q+LPVI+ R+EIV S++N
Sbjct: 1053 SKFCFEALDKSFSDIIKRVDNK----VFGGKVMVFGGDFRQVLPVINGAGRAEIVMSSLN 1108
Query: 240 SSYLWKHCKVMKLTVNMS-LQNATSTSSATEIKEFVDWLLQVGDGTVKTIDEEETLIEIP 298
+SYLW HCKV++LT NM L N S A EI+EF DWLL VGDG V ++ E +I+IP
Sbjct: 1109 ASYLWDHCKVLRLTKNMRLLNNDLSVDEAKEIQEFSDWLLAVGDGRVNEPNDGEVIIDIP 1168
Query: 299 PDLLIEQCKEPLLELVNFAY--PKLAHNLQKSSFFQERAILAPTLESLEEINNFMLAMIP 356
+LLI++ P+ + Y P H + FFQ RAILAP E + IN +ML +
Sbjct: 1169 EELLIQEADNPIEAISREIYGDPTKLHEISDPKFFQRRAILAPKNEDVNTINQYMLEHLD 1228
Query: 357 GDETEYLSCDTPCKSDEDSGDNAE*FTSEVLNDFKCSEIPNHAIKLKAGVPIMLIRNIDQ 416
+E YLS D+ SD DS N T + LN K S +P+H+++LK G P+ML+RN+D
Sbjct: 1229 SEERIYLSADSIDPSDSDSLKNPV-ITPDFLNSIKVSGMPHHSLRLKVGAPVMLLRNLDP 1287
Query: 417 AAGLCNGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFSVT 476
GLCNGTR+ + L +I+ V+ +++G+ +IP +++TPS++ +PFK +RRQF ++
Sbjct: 1288 KGGLCNGTRLQITQLCSHIVEAKVITGDRIGQIVYIPLINITPSDTKLPFKMRRRQFPLS 1347
Query: 477 LCFVMTINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSRKMLKMLIIDDE*VVSNTT 536
+ FVMTINKSQGQSL VGLYLP+PVF+HGQ YVAL RV S+ LK+LI+D E + T
Sbjct: 1348 VAFVMTINKSQGQSLEQVGLYLPKPVFSHGQLYVALSRVTSKTGLKILILDKEGKIQKQT 1407
Query: 537 RNVVYQEVLDN 547
NVV++EV N
Sbjct: 1408 TNVVFKEVFQN 1418
>At3g42100 putative protein
Length = 1752
Score = 499 bits (1285), Expect = e-141
Identities = 268/549 (48%), Positives = 359/549 (64%), Gaps = 10/549 (1%)
Query: 3 ELQNLCMIEIEKILQGNERSLKEFPCLPYPKFSEIHNFEVIFVADELNYN-RAEMVKIHD 61
E++N + EIEKI+ N +LKE P P I N + V DEL YN + + + HD
Sbjct: 1208 EIRNYTLQEIEKIMLRNGATLKEIQDFPQPSREGIDNSNRL-VVDELRYNIDSNLKEKHD 1266
Query: 62 EFVSSLTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLNV 121
E+ L +EQ Y + V +D FF+YGFGGTGKTF+W L AA+RSRG IVLNV
Sbjct: 1267 EWFQMLNTEQRGIYDEITGAVFNDLGGVFFIYGFGGTGKTFIWKTLAAAVRSRGQIVLNV 1326
Query: 122 ASSGIASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAELLKKASLIIWDEAPMLNK 181
ASSGIASLLL G RTAHSRF+IP++ +E S C + S A L+K+ASLIIWDEAPM++K
Sbjct: 1327 ASSGIASLLLEGGRTAHSRFAIPLNPDEFSVCKITPKSDLANLIKEASLIIWDEAPMMSK 1386
Query: 182 HCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGSAINSS 241
C E+LD+S DI+ + FGGKVVV G DF Q+LPVI+ R EIV S++N+S
Sbjct: 1387 FCFESLDKSFYDILNNKDNK----VFGGKVVVFGGDFRQVLPVINGAGRVEIVMSSLNAS 1442
Query: 242 YLWKHCKVMKLTVNMS-LQNATSTSSATEIKEFVDWLLQVGDGTVKTIDEEETLIEIPPD 300
YLW HCKV+KLT NM L S+ A EI++F DWLL VGDG + ++ E LI+IP +
Sbjct: 1443 YLWDHCKVLKLTKNMRLLSGGLSSEEAKEIQQFSDWLLAVGDGRINEPNDGEALIDIPEE 1502
Query: 301 LLIEQCKEPLLELVNFAY--PKLAHNLQKSSFFQERAILAPTLESLEEINNFMLAMIPGD 358
LLI++ P+ + Y P H + FFQ RAILAPT E + IN +ML + +
Sbjct: 1503 LLIKEAGNPIEAISKEIYGDPSELHMINDPKFFQRRAILAPTNEDVNTINQYMLEHLKSE 1562
Query: 359 ETEYLSCDTPCKSDEDSGDNAE*FTSEVLNDFKCSEIPNHAIKLKAGVPIMLIRNIDQAA 418
E YLS D+ +D DS N T + LN + + +P+HA++LK G P+ML+RN+D
Sbjct: 1563 ERIYLSADSIDPTDSDSLANPV-ITPDFLNSIQLTGMPHHALRLKVGAPVMLLRNLDPKG 1621
Query: 419 GLCNGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFSVTLC 478
GLCNGTR+ + L K ++ V+ +++G+ IP ++LTPS++ +PFK +RRQF +++
Sbjct: 1622 GLCNGTRLQITQLAKQVVQAKVITRDRIGDIVLIPLINLTPSDTKLPFKMRRRQFPLSVA 1681
Query: 479 FVMTINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSRKMLKMLIIDDE*VVSNTTRN 538
F MTINKSQGQSL VGLYLP+PVF+HGQ YVAL RV S+K LK+LI+D + + T N
Sbjct: 1682 FAMTINKSQGQSLEQVGLYLPKPVFSHGQLYVALSRVTSKKGLKILILDKDGNMQKQTTN 1741
Query: 539 VVYQEVLDN 547
VV++EV N
Sbjct: 1742 VVFKEVFQN 1750
>At3g31980 hypothetical protein
Length = 1099
Score = 484 bits (1245), Expect = e-137
Identities = 261/551 (47%), Positives = 361/551 (65%), Gaps = 11/551 (1%)
Query: 1 DEELQNLCMIEIEKILQGNERSLKEFPCLPYPKFSEIHNFEVIFVADELNYNRAEMVKIH 60
D E +N ++EIE +L N +L++F +P P E + F+ +E NYN ++ + H
Sbjct: 553 DAERRNYALLEIEDMLLCNGSTLQDFKDMPKPT-KEGTDHSNRFITEEKNYNIEKLREDH 611
Query: 61 DEFVSSLTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLN 120
D++ + +TSEQ+ Y ++ VL ++ FF+YGFGGT KTF+W L AA+R RGLI +N
Sbjct: 612 DDWFNKMTSEQKGIYDEIIKAVLENSGGIFFVYGFGGTSKTFMWKTLSAAVRMRGLISVN 671
Query: 121 VASSGIASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAELLKKASLIIWDEAPMLN 180
VASSGIASLLL G RTAHSRF IPI+ ++ +TC++ S A +LK+ASLIIWDEAPM++
Sbjct: 672 VASSGIASLLLQGGRTAHSRFGIPINPDDFTTCHIVPNSDLANMLKEASLIIWDEAPMMS 731
Query: 181 KHCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGSAINS 240
++C E+LDRSLND++ PFGGKVVV G DF Q+LPVI R+EIV +A+NS
Sbjct: 732 RYCFESLDRSLNDVIGNIDGK----PFGGKVVVFGGDFRQVLPVIHGAGRAEIVLAALNS 787
Query: 241 SYLWKHCKVMKLTVNMSL-QNATSTSSATEIKEFVDWLLQVGDGTVKTIDEEETLIEIPP 299
SYLW+HCKV+ LT NM L N A EIKEF +WLL VGDG V ++ E LI+IP
Sbjct: 788 SYLWEHCKVLTLTKNMRLMSNDLDKDEAEEIKEFSNWLLAVGDGRVSEPNDGEVLIDIPE 847
Query: 300 DLLIEQCKEPLLELVNFAYPKLAHNLQKSS---FFQERAILAPTLESLEEINNFMLAMIP 356
+LLI+ +P+ + Y L LQ ++ FFQ+RAIL P + IN+ ML +
Sbjct: 848 ELLIKDANDPIEAITKAVYGDL-DLLQPNNDPKFFQQRAILCPRNTDVNTINDIMLDKLN 906
Query: 357 GDETEYLSCDTPCKSDEDSGDNAE*FTSEVLNDFKCSEIPNHAIKLKAGVPIMLIRNIDQ 416
G+ YLS D+ D S +N T + LN K S +PNH + LK G P+ML+RNID
Sbjct: 907 GELVTYLSADSIDPQDAASLNNPV-LTPDFLNSIKLSGLPNHNLTLKIGTPVMLLRNIDP 965
Query: 417 AAGLCNGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFSVT 476
GLCNGTR+ V + +I+ V+ +++G+ I + ++PS++ +PF+ +RRQF +
Sbjct: 966 KGGLCNGTRLQVTQMGNHILEARVITGDRVGDKVIIIKSQISPSDTKLPFRMRRRQFPIA 1025
Query: 477 LCFVMTINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSRKMLKMLIIDDE*VVSNTT 536
+ F MTINKSQGQSL VG+YLP+PVF+HGQ YVAL RV S+K LK+LI+D E + T
Sbjct: 1026 VAFAMTINKSQGQSLKEVGIYLPKPVFSHGQLYVALSRVTSKKGLKVLIVDKEGNTQSQT 1085
Query: 537 RNVVYQEVLDN 547
NVV++E+ N
Sbjct: 1086 MNVVFKEIFQN 1096
>At1g52960 hypothetical protein
Length = 924
Score = 481 bits (1239), Expect = e-136
Identities = 246/518 (47%), Positives = 343/518 (65%), Gaps = 6/518 (1%)
Query: 1 DEELQNLCMIEIEKILQGNERSLKEFPCLPYPKFSEIHNFEVIFVADELNYNRAEMVKIH 60
DEE Q C+ EI ++L N SL ++ +P + F+ DE Y+RA +++ H
Sbjct: 407 DEERQQYCLQEIARLLTKNGVSLSKWKQMPQISDEHVEKCNH-FILDERKYDRAYLIEKH 465
Query: 61 DEFVSSLTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLN 120
+E+++ +TSEQ+ Y ++D VL D FF+YGFGGTGKTF+W +L AA+RS+G I LN
Sbjct: 466 EEWLTMVTSEQKKIYDEIMDAVLHDRGGVFFVYGFGGTGKTFLWKLLSAAIRSKGDISLN 525
Query: 121 VASSGIASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAELLKKASLIIWDEAPMLN 180
VASSGIA+LLL G RT HSRF IPI+ NE STCN+ +GS EL+K+A+LIIWDE PM++
Sbjct: 526 VASSGIAALLLDGGRTTHSRFGIPINPNESSTCNISRGSDLGELVKEANLIIWDETPMMS 585
Query: 181 KHCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGSAINS 240
KHC E+LDR+L DIM D PFGGK +V G DF Q+LPVI+ R EIV +A+NS
Sbjct: 586 KHCFESLDRTLRDIMNNPG----DKPFGGKGIVFGGDFRQVLPVINGAGREEIVFAALNS 641
Query: 241 SYLWKHCKVMKLTVNMSLQNATSTSSATEIKEFVDWLLQVGDGTVKTIDEEETLIEIPPD 300
SY+W+HCKV++LT NM L S +I+ F W+L VGDG + ++ LI+IP +
Sbjct: 642 SYIWEHCKVLELTKNMRLLANISEHEKRDIEYFSKWILDVGDGKISQPNDGIALIDIPEE 701
Query: 301 LLIEQCKEPLLELVNFAYPKLAHNLQKSSFFQERAILAPTLESLEEINNFMLAMIPGDET 360
LI +P+ ++ Y + FFQ RAIL PT E + IN M++M+ G+E
Sbjct: 702 FLINGDNDPVESIIEAVYGNTFMEEKDPKFFQGRAILCPTNEDVNSINEHMMSMLDGEER 761
Query: 361 EYLSCDTPCKSDEDSGDNAE*FTSEVLNDFKCSEIPNHAIKLKAGVPIMLIRNIDQAAGL 420
YLS D+ +D S +N + ++++ LN + S +PNH ++LK G P+ML+RN+D GL
Sbjct: 762 IYLSSDSIDPADTSSANN-DAYSADFLNSVRVSGLPNHCLRLKVGCPVMLLRNMDPNKGL 820
Query: 421 CNGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFSVTLCFV 480
CNGTR+ V + +I + N++G+ IPRM +TPS++ +PFK +RRQF +++ F
Sbjct: 821 CNGTRLQVTQMADTVIQARFITGNRVGKIVLIPRMLITPSDTRLPFKMRRRQFPLSVAFA 880
Query: 481 MTINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSR 518
MTINKSQGQ+L VGLYLPRPVF+HGQ YVA+ RV S+
Sbjct: 881 MTINKSQGQTLESVGLYLPRPVFSHGQLYVAISRVTSK 918
>At2g14470 pseudogene
Length = 1265
Score = 465 bits (1197), Expect = e-131
Identities = 249/515 (48%), Positives = 335/515 (64%), Gaps = 13/515 (2%)
Query: 19 NERSLKEFPCLPYPKFSEIHNFEVIFVADELNYNRAEMVKI-HDEFVSSLTSEQEIFYKN 77
N + EFP P I N + V +EL YNR +K H+E+ LT EQ Y
Sbjct: 757 NPEDIDEFP---KPTIDGIDNSNRLIV-EELRYNRESNLKEKHEEWKQMLTPEQRGVYNE 812
Query: 78 VLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLNVASSGIASLLLPGDRTA 137
+ + V ++ FF+YGFGGTGKTF+W L A +R R IVLNVASSGIASLLL G RTA
Sbjct: 813 ITEAVFNNLGGVFFVYGFGGTGKTFIWKTLSATIRYRDQIVLNVASSGIASLLLEGGRTA 872
Query: 138 HSRFSIPISINEISTCNLRQGSPKAELLKKASLIIWDEAPMLNKHCIEALDRSLNDIMKT 197
HSRF IP++ +E S C ++ S A L+KKASL+IWDEAPM+++ C EALD+S +DI+K
Sbjct: 873 HSRFGIPLNPDEFSVCKIKPKSDLANLVKKASLVIWDEAPMMSRFCFEALDKSFSDIIKN 932
Query: 198 QSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGSAINSSYLWKHCKVMKLTVNMS 257
+ FGGKVVV G DF Q+ PVI+ R+EIV S++N+SYLW +CKV+KLT N
Sbjct: 933 TD----NTVFGGKVVVFGGDFRQVFPVINGAGRAEIVMSSLNASYLWDNCKVLKLTKNTR 988
Query: 258 L-QNATSTSSATEIKEFVDWLLQVGDGTVKTIDEEETLIEIPPDLLIEQCKEPLLELVNF 316
L N S + A EI+EF DWLL VGDG + ++ +I+IP DLLI +P+ + N
Sbjct: 989 LLANNLSETEAKEIQEFSDWLLAVGDGRINESNDGVAIIDIPEDLLITNADKPIESITNE 1048
Query: 317 AY--PKLAHNLQKSSFFQERAILAPTLESLEEINNFMLAMIPGDETEYLSCDTPCKSDED 374
Y PK+ H + FFQ RAILA E + IN ++L + +E YLS D+ +D D
Sbjct: 1049 IYGDPKILHEITDPKFFQGRAILASKNEDVNTINEYLLDQLHAEERIYLSADSIDPTDSD 1108
Query: 375 SGDNAE*FTSEVLNDFKCSEIPNHAIKLKAGVPIMLIRNIDQAAGLCNGTRMIVNALTKY 434
S N T + LN K +PNH+++LK G P++L+RN+D GLCNGTR+ + L
Sbjct: 1109 SLSNPV-ITPDFLNSIKLPGLPNHSLRLKVGAPVLLLRNLDPKGGLCNGTRLQITQLCTQ 1167
Query: 435 IIVVTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFSVTLCFVMTINKSQGQSLSHV 494
I+ V+ +++G IP ++LTP+N+ +PFK +RRQF +++ FVMTINKS+GQSL HV
Sbjct: 1168 IVEAKVITGDRIGHIILIPTVNLTPTNTKLPFKMRRRQFPLSVAFVMTINKSEGQSLEHV 1227
Query: 495 GLYLPRPVFTHGQPYVALYRVKSRKMLKMLIIDDE 529
GLYLP+PVF+HGQ YVAL RV S+K LK+LI+D +
Sbjct: 1228 GLYLPKPVFSHGQLYVALSRVTSKKGLKILILDKD 1262
>At2g05080 putative helicase
Length = 1219
Score = 426 bits (1096), Expect = e-119
Identities = 238/535 (44%), Positives = 329/535 (61%), Gaps = 43/535 (8%)
Query: 1 DEELQNLCMIEIEKILQGNERSLKEFPCLPY----PKFSEIHNFEVIFVADELNYNRAEM 56
DEE + + EI+ IL+ N SL + +P P+F + + DE Y+R
Sbjct: 682 DEEKKVYALQEIDHILRRNGTSLTYYKTMPQVPRDPRFDTN-----VLILDEKGYDRESE 736
Query: 57 VKIHDEFVSSLTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGL 116
K H + + LT EQ+ Y N++ V + FF+YGFGGTGKTF+W L AALRS+G
Sbjct: 737 TKKHADSIKKLTLEQKSVYDNIIGAVNENVGGVFFVYGFGGTGKTFLWKTLSAALRSKGD 796
Query: 117 IVLNVASSGIASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAELLKKASLIIWDEA 176
IVLNVASSGIASLLL G RTAHSR IP++ NE +TCN++ GS +A L+K+ASLIIWDEA
Sbjct: 797 IVLNVASSGIASLLLEGGRTAHSRSGIPLNPNEFTTCNMKAGSDRANLVKEASLIIWDEA 856
Query: 177 PMLNKHCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGS 236
PM+++HC E+LDRSL+DI PFGGKVVV G DF Q+LPVI ++IV +
Sbjct: 857 PMMSRHCFESLDRSLSDICGNCDNK----PFGGKVVVFGGDFRQVLPVIPGADTADIVMA 912
Query: 237 AINSSYLWKHCKVMKLTVNMSLQNATSTSSATEIKEFVDWLLQVGDGTVKTIDEEETLIE 296
A+NSSYLW HCKV+ LT NM L + +W+L VGDG + ++ E LI+
Sbjct: 913 ALNSSYLWSHCKVLTLTKNMCLFSE-------------EWILAVGDGRIGEPNDGEALID 959
Query: 297 IPPDLLIEQCKEPLLELVNFAYPKLA--HNLQKSSFFQERAILAPTLESLEEINNFMLAM 354
IP + LI + K+P+ + Y + H + FFQERAIL PT E + +IN ML
Sbjct: 960 IPSEFLITKAKDPIQAICTEIYGDITKIHEQKDPVFFQERAILCPTNEDVNQINETMLDN 1019
Query: 355 IPGDETEYLSCDTPCKSDEDSGDNAE*FTSEVLNDFKCSEIPNHAIKLKAGVPIMLIRNI 414
+ G+E +LS D+ +D S +N T E LN+ K + NH ++LK G P+ML+RNI
Sbjct: 1020 LQGEELTFLSSDSLDTADIGSRNNPV-LTPEFLNNVKVLGLSNHKLRLKIGSPVMLLRNI 1078
Query: 415 DQAAGLCNGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFS 474
D GL NGTR+ + ++ +I+ +L ++ +++ +PF+ +R Q
Sbjct: 1079 DPIGGLMNGTRLQIMQMSPFILQAMILTGDR--------------ADTKLPFRMRRTQLP 1124
Query: 475 VTLCFVMTINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSRKMLKMLIIDDE 529
+ +CF MTINKSQGQSL VG++LPRP F+H Q YVA+ RV S+ LK+LI++DE
Sbjct: 1125 LAVCFAMTINKSQGQSLKRVGIFLPRPCFSHSQLYVAISRVTSKSGLKILIVNDE 1179
>At3g30560 hypothetical protein
Length = 1473
Score = 421 bits (1081), Expect = e-118
Identities = 234/527 (44%), Positives = 325/527 (61%), Gaps = 34/527 (6%)
Query: 21 RSLKEFPCLPYPKFSEIHNFEVIFVADELNYNRAEMVKIHDEFVSSLTSEQEIFYKNVLD 80
RSL+E LP K + F + + DE NYNR + IHD+++ LT+EQ+ Y ++D
Sbjct: 980 RSLEE-KMLPQLKPGDEPAFNQL-ILDERNYNRETLKTIHDDWLKMLTTEQKKVYDKIMD 1037
Query: 81 DVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLNVASSGIASLLLPGDRTAHSR 140
VL++ G I LNVASSGIASLLL G RTAHSR
Sbjct: 1038 AVLNNKG---------------------------GDICLNVASSGIASLLLEGGRTAHSR 1070
Query: 141 FSIPISINEISTCNLRQGSPKAELLKKASLIIWDEAPMLNKHCIEALDRSLNDIMKTQST 200
F IP++ +E STCN+ +GS AEL+ A LIIWDEAPM++K+C E+LD+SL DI+ T
Sbjct: 1071 FGIPLTPHETSTCNMERGSDLAELVTAAKLIIWDEAPMMSKYCFESLDKSLKDILSTPE- 1129
Query: 201 HGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGSAINSSYLWKHCKVMKLTVNMSLQN 260
D+PFGGK+++ G DF QILPVI R IV S++NSS+LW++CKV KLT NM L
Sbjct: 1130 ---DMPFGGKLIIFGGDFRQILPVILAAGRELIVKSSLNSSHLWQYCKVFKLTKNMRLLQ 1186
Query: 261 ATSTSSATEIKEFVDWLLQVGDGTVKTIDEEETLIEIPPDLLIEQCKEPLLELVNFAYPK 320
+ A EI++F W+L VG+G + ++ T I+I D+LI + P+ ++ Y
Sbjct: 1187 DIDINEAREIEDFSKWILAVGEGKLNQPNDGVTQIQIRDDILIPEGDNPIESIIKAVYGT 1246
Query: 321 LAHNLQKSSFFQERAILAPTLESLEEINNFMLAMIPGDETEYLSCDTPCKSDEDSGDNAE 380
+ FFQ+RAIL PT + + IN+ ML+ + G+E Y S D+ SD + N
Sbjct: 1247 SFDEERDPKFFQDRAILCPTNDDVNSINDHMLSKLTGEEKIYRSSDSIDPSDTRADKNPV 1306
Query: 381 *FTSEVLNDFKCSEIPNHAIKLKAGVPIMLIRNIDQAAGLCNGTRMIVNALTKYIIVVTV 440
+T + LN K S +PNH + LK G P+ML+RN+D GL NGTR+ + L ++ +
Sbjct: 1307 -YTPDFLNKIKISGLPNHLLWLKVGCPVMLLRNLDSHGGLMNGTRLQIVRLGDKLVQGRI 1365
Query: 441 LNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFSVTLCFVMTINKSQGQSLSHVGLYLPR 500
L ++G+ IPRM LTPS+ +PFK +RRQF +++ F MTINKSQGQSL +VG+YLP+
Sbjct: 1366 LTGTRVGKLVIIPRMPLTPSDRRLPFKMKRRQFPLSVAFAMTINKSQGQSLGNVGIYLPK 1425
Query: 501 PVFTHGQPYVALYRVKSRKMLKMLIIDDE*VVSNTTRNVVYQEVLDN 547
PVF+HGQ YVA+ RVKS+ LK+LI D + N T NVV++E+ N
Sbjct: 1426 PVFSHGQLYVAMSRVKSKGGLKVLITDSKGKQKNETTNVVFKEIFRN 1472
>At1g35940 hypothetical protein
Length = 1678
Score = 420 bits (1080), Expect = e-118
Identities = 238/550 (43%), Positives = 322/550 (58%), Gaps = 67/550 (12%)
Query: 1 DEELQNLCMIEIEKILQGNERSLKEFPCLPYPKFSEIHNFEVIFVADELNYNRAEMVKIH 60
+ E++N + EIEKI+ N +L++ P P
Sbjct: 1191 ETEIKNYTLQEIEKIMLSNGATLEDIDEFPKPT--------------------------R 1224
Query: 61 DEFVSSLTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLN 120
DE+ LT EQ Y + + V ++ FF+YGFGGTGKTF+W L AA+R RG IVLN
Sbjct: 1225 DEWKQMLTPEQRGVYNAITEAVFNNLGGVFFVYGFGGTGKTFIWKTLSAAIRCRGQIVLN 1284
Query: 121 VASSGIASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAELLKKASLIIWDEAPMLN 180
VASSGIASLLL G RTAHSRF IP++ +E S
Sbjct: 1285 VASSGIASLLLEGGRTAHSRFGIPLNHDEFSV---------------------------- 1316
Query: 181 KHCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGSAINS 240
+LD+S +DI+K + FGGKVVV G DF Q+LPVI+ R+EIV S++N+
Sbjct: 1317 -----SLDKSFSDIIKNTNNK----VFGGKVVVFGGDFRQVLPVINGAGRAEIVMSSLNA 1367
Query: 241 SYLWKHCKVMKLTVNMSL-QNATSTSSATEIKEFVDWLLQVGDGTVKTIDEEETLIEIPP 299
SYLW HCKV+KLT NM L N S + A EI+EF DWLL V DG + ++ I+IP
Sbjct: 1368 SYLWDHCKVLKLTKNMRLLANNLSATEAKEIQEFSDWLLAVSDGRINEPNDGVATIDIPE 1427
Query: 300 DLLIEQCKEPLLELVNFAY--PKLAHNLQKSSFFQERAILAPTLESLEEINNFMLAMIPG 357
DLLI +P+ + N Y PK+ H + FFQ RAILAP E + IN ++L +
Sbjct: 1428 DLLITNADKPIETITNEIYGDPKILHEITDPKFFQGRAILAPKNEDVNTINEYLLEQLDA 1487
Query: 358 DETEYLSCDTPCKSDEDSGDNAE*FTSEVLNDFKCSEIPNHAIKLKAGVPIMLIRNIDQA 417
+E YLS D+ +D DS +N T + LN K +PNH++ LK G P+ML+RN+D
Sbjct: 1488 EERIYLSADSIDPTDSDSLNNPV-ITPDFLNSIKLPGLPNHSLCLKVGAPVMLLRNLDPK 1546
Query: 418 AGLCNGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFSVTL 477
GLCNGTR+ + L I+ V+ +++G IP ++LTP+++ +PFK +RRQF +++
Sbjct: 1547 GGLCNGTRLQITQLCTQIVEAKVITGDRIGNIVLIPTVNLTPTDTKLPFKMRRRQFPLSV 1606
Query: 478 CFVMTINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSRKMLKMLIIDDE*VVSNTTR 537
F MTINKSQGQSL H+GLYLP+PVF+HGQ YVAL RV S+K LK+LI+D + + T
Sbjct: 1607 AFAMTINKSQGQSLEHIGLYLPKPVFSHGQLYVALSRVTSKKGLKILILDKDGKLQKQTT 1666
Query: 538 NVVYQEVLDN 547
NVV++EV N
Sbjct: 1667 NVVFKEVFQN 1676
>At1g54430 hypothetical protein
Length = 1639
Score = 401 bits (1031), Expect = e-112
Identities = 235/553 (42%), Positives = 328/553 (58%), Gaps = 48/553 (8%)
Query: 2 EELQNLCMIEIEKILQGNERSLKEFPCLPYPKFSEIHNFEVIFVADELNYNRAEMVKIHD 61
EEL+ +IEIE +L+ +E+SL ++P +P P+
Sbjct: 1117 EELEKYTLIEIETLLRQHEKSLSDYPEMPQPE---------------------------- 1148
Query: 62 EFVSSLTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLNV 121
+ L +Q I Y +VL V++ FFLYG GGTGKTF++ + +ALRS G V+ V
Sbjct: 1149 ---NKLNEQQRIIYDDVLKSVINKEGKLFFLYGAGGTGKTFLYKTIISALRSNGKNVMPV 1205
Query: 122 ASSGIASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAELLKKASLIIWDEAPMLNK 181
ASS IA+LLLPG RTAHSRF IPI+++E S C+++ GS A +L K LIIWDEAPM ++
Sbjct: 1206 ASSAIAALLLPGGRTAHSRFKIPINVHEDSICDIKIGSMLANVLSKVDLIIWDEAPMAHR 1265
Query: 182 HCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGSAINSS 241
H EA+DR+L DI+ FGGK V+LG DF QILPVI +G+R E V +AIN S
Sbjct: 1266 HTFEAVDRTLRDILSVGDEKALTKTFGGKTVLLGGDFRQILPVIPQGTRQETVSAAINRS 1325
Query: 242 YLWKHCKVMKLTVNMSLQNATSTSSATEIKEFVDWLLQVGDGTV--KT----IDEEETLI 295
YLW+ C L+ NM +Q EIK F +W+LQVGDG KT D+EE I
Sbjct: 1326 YLWESCHKYLLSQNMRVQ-------PEEIK-FAEWILQVGDGEAPRKTHGIDDDQEEDNI 1377
Query: 296 EIPPDLLIEQCKEPLLELVNFAYPKLAHNLQKSSFFQERAILAPTLESLEEINNFMLAMI 355
I +LL+ + + PL L +P + Q + A+L P E+++EIN+++L+ +
Sbjct: 1378 IIDKNLLLPETENPLEVLCRSVFPDFTNTFQDLENLKGTAVLTPRNETVDEINDYLLSKV 1437
Query: 356 PGDETEYLSCDTPCKSDEDSGDNAE-*FTSEVLNDFKCSEIPNHAIKLKAGVPIMLIRNI 414
PG EY S D+ + + + + E + E LN + +P H + LK GVPIML+RN+
Sbjct: 1438 PGLAKEYFSADSIDRDEALTEEGFEMSYPMEYLNSLEFPGLPAHRLCLKVGVPIMLLRNL 1497
Query: 415 DQAAGLCNGTRMIVNALTKYIIVVTVLNE-NKMGETTFIPRMSLTPSNSDIPFKFQRRQF 473
+Q GLCNGTR+IV L ++ +L++ K + IPR+ L+P +S PF +RRQF
Sbjct: 1498 NQKEGLCNGTRLIVTHLGDKVLKAEILSDTTKERKKVLIPRIILSPQDSKHPFTLRRRQF 1557
Query: 474 SVTLCFVMTINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSRKMLKMLIIDDE*VVS 533
V +C+ MTINKSQGQ+L+ V LYLP+PVF+HGQ YVAL RV S K L +L +
Sbjct: 1558 PVRMCYAMTINKSQGQTLNRVALYLPKPVFSHGQLYVALSRVTSPKGLTVLDTSKKKEGK 1617
Query: 534 NTTRNVVYQEVLD 546
T N+VY+EV +
Sbjct: 1618 YVT-NIVYREVFN 1629
>At3g30420 hypothetical protein
Length = 837
Score = 401 bits (1030), Expect = e-112
Identities = 235/556 (42%), Positives = 328/556 (58%), Gaps = 23/556 (4%)
Query: 2 EELQNLCMIEIEKILQGNERSLKEFPCLPYPKFSEIHNFEVIFVADELNYNRAEMVKIHD 61
+EL+ +IEIE +L+ +E+SL ++P +P P+ S + + E N + + H
Sbjct: 284 KELEKYTLIEIETLLRQHEKSLSDYPEMPQPEKSILEEVNNSLLRQEFQINIDKERETHA 343
Query: 62 EFVSSLTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLNV 121
S L +Q I Y +VL V + FFLYG GGTGKTF++ + +ALRS G V+ V
Sbjct: 344 NLFSKLNEQQRIIYDDVLKSVTNKEGKLFFLYGDGGTGKTFLYKTIISALRSNGKNVMPV 403
Query: 122 ASSGIASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAELLKKASLIIWDEAPMLNK 181
ASS IA+LLLPG RTAHS F IPI+++E C+++ GS A +L K LIIWDEAPM ++
Sbjct: 404 ASSAIAALLLPGGRTAHSWFKIPINVHEDFICDIKIGSMLANVLSKVDLIIWDEAPMAHR 463
Query: 182 HCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGSAINSS 241
H EA+DR+L DI+ GGK V+LG DF QILPVI + +R E V +AIN S
Sbjct: 464 HTFEAVDRTLRDILSVGDEKALTKTLGGKTVLLGGDFRQILPVIPQRTRQETVSAAINRS 523
Query: 242 YLWKHCKVMKLTVNMSLQNATSTSSATEIKEFVDWLLQVGDGTV--KT----IDEEETLI 295
YLW+ C L+ NM +Q EIK F +W+LQ+GDG KT D+EE I
Sbjct: 524 YLWESCHKYLLSQNMRVQ-------PEEIK-FAEWILQIGDGEAPRKTHGIDDDQEEDNI 575
Query: 296 EIPPDLLIEQCKEPLLELVNFAYPKLAHNLQKSSFFQERAILAPTLESLEEINNFMLAMI 355
I +LL+ + + PL L P + Q + A+L P E+++EIN+++L+ +
Sbjct: 576 IIDKNLLLPETENPLEVLCQSVSPDFTNTFQDLENLKGTAVLTPRNETVDEINDYLLSKV 635
Query: 356 PGDETEYLSCDTPCKSDEDSGDNAE*F----TSEVLNDFKCSEIPNHAIKLKAGVPIMLI 411
PG EY S D+ D+D E F E LN + +P H + LK GVPIML+
Sbjct: 636 PGLAKEYFSADS---IDQDEALTEEGFEMSYPMEYLNSLEFPGLPAHRLCLKVGVPIMLL 692
Query: 412 RNIDQAAGLCNGTRMIVNALTKYIIVVTVLNE-NKMGETTFIPRMSLTPSNSDIPFKFQR 470
RN++Q GLCNGTR+ V L ++ +L++ K + IPR+ L+P +S PF +R
Sbjct: 693 RNLNQKEGLCNGTRLTVTHLGDKVLKAEILSDTTKKRKKVLIPRIILSPQDSKHPFTLRR 752
Query: 471 RQFSVTLCFVMTINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSRKMLKMLIIDDE* 530
RQF V +C+ MT+NKSQGQ+L+ V LYLP+PVF+HGQ YVAL RV S K L +L +
Sbjct: 753 RQFPVRMCYAMTVNKSQGQTLNRVALYLPKPVFSHGQLYVALSRVTSPKGLTVLDTSKKK 812
Query: 531 VVSNTTRNVVYQEVLD 546
T N+VY+EV +
Sbjct: 813 EGKYVT-NIVYREVFN 827
>At3g31440 hypothetical protein
Length = 536
Score = 395 bits (1016), Expect = e-110
Identities = 231/530 (43%), Positives = 313/530 (58%), Gaps = 58/530 (10%)
Query: 22 SLKEFPCLPYPKFSEIHNFEVIFVADELNYNRAEMVKI-HDEFVSSLTSEQEIFYKNVLD 80
+L++ P P I N + V +EL YNR +K H+E+ LTSEQ
Sbjct: 59 TLEDIDEFPKPTRDGIDNSNRLIV-EELRYNRESNLKEKHEEWKQMLTSEQR-------- 109
Query: 81 DVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLNVASSGIASLLLPGDRTAHSR 140
GT KT +W L+AA+R R IVLN+ASSGIASLLL G RTAHSR
Sbjct: 110 ----------------GTWKTIIWKTLFAAIRRRDQIVLNMASSGIASLLLEGGRTAHSR 153
Query: 141 FSIPISINEISTCNLRQGSPKAELLKKASLIIWDEAPMLNKHCIEALDRSLNDIMKTQST 200
F I ++ +E S C ++ S A L+K+ASL+I D+APM+++ C EALD+S +DI+K
Sbjct: 154 FGIRLNPDEFSVCKIKPKSDLANLVKEASLVICDKAPMMSRFCFEALDKSFSDIIKNT-- 211
Query: 201 HGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGSAINSSYLWKHCKVMKLTVNMSL-Q 259
Y+ FGGKVVV DF Q+LPVI+ R+EIV S++N+SYLW HCKV+KLT NM L
Sbjct: 212 --YNKVFGGKVVVFSGDFRQVLPVINGAGRAEIVMSSLNASYLWDHCKVLKLTKNMRLLA 269
Query: 260 NATSTSSATEIKEFVDWLLQVGDGTVKTIDEEETLIEIPPDLLIEQCKEPLLELVNFAY- 318
N S + A EI EF DWLL VGDG + +++ +I+IP DLLI +P+ + N Y
Sbjct: 270 NNLSETEAKEIHEFSDWLLAVGDGRINEPNDDVAIIDIPKDLLITNADKPIEWITNEIYG 329
Query: 319 -PKLAHNLQKSSFFQERAILAPTLESLEEINNFMLAMIPGDETEYLSCDTPCKSDEDSGD 377
PK+ H + FFQ RAILAP E + IN ++L + +E YLS D+ +D DS +
Sbjct: 330 DPKILHEITDPKFFQGRAILAPKNEDVNTINEYLLEQLHAEERIYLSADSIDPTDSDSLN 389
Query: 378 NAE*FTSEVLNDFKCSEIPNHAIKLKAGVPIMLIRNIDQAAGLCNGTRMIVNALTKYIIV 437
N T + LN K GLCNG R+ + L I+
Sbjct: 390 NPV-ITPDFLNSIKLP------------------------GGLCNGARLQITQLFTEIVE 424
Query: 438 VTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFSVTLCFVMTINKSQGQSLSHVGLY 497
V+ +++G IP ++LTP+++ +PFK +RRQF +++ F MTINKSQGQSL HVGLY
Sbjct: 425 AKVITGDRIGHIVLIPTVNLTPTDTKLPFKMRRRQFPLSVAFAMTINKSQGQSLEHVGLY 484
Query: 498 LPRPVFTHGQPYVALYRVKSRKMLKMLIIDDE*VVSNTTRNVVYQEVLDN 547
LP+PVF+HGQ YVAL RV S+K LK+LI+D + T N+V++EV N
Sbjct: 485 LPKPVFSHGQLYVALSRVTSKKGLKILILDKNGKLQKQTTNIVFKEVFQN 534
>At3g43350 putative protein
Length = 830
Score = 387 bits (994), Expect = e-108
Identities = 226/523 (43%), Positives = 304/523 (57%), Gaps = 71/523 (13%)
Query: 1 DEELQNLCMIEIEKILQGNERSLKEFPCLPYPKFSEIHNFEVIFVADELNYNRAEMVKIH 60
DEE Q C+ EI ++L N SL K++ H F++ + D Y KI+
Sbjct: 136 DEERQQYCLQEIARLLTKNGVSLS--------KWNRCHKFQMNTMVD---YGDFRAKKIY 184
Query: 61 DEFVSSLTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLN 120
DE ++D VL D FF+YGFGGTGKTF+W +L AA+RS+G I LN
Sbjct: 185 DE---------------IMDVVLHDRGGVFFVYGFGGTGKTFLWKLLSAAVRSKGDISLN 229
Query: 121 VASSGIASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAELLKKASLIIWDEAPMLN 180
VASSGIA+L L G RTAHSRF IPI+ NE STCN+ +GS EL+K+A LIIWDEAPM++
Sbjct: 230 VASSGIAALRLDGGRTAHSRFDIPINPNESSTCNISRGSDLGELVKEAKLIIWDEAPMMS 289
Query: 181 KHCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGSAINS 240
KHC E+LDR+L DI+ D P GGKV+V G DF Q+LPVI+ R EIV +A+NS
Sbjct: 290 KHCFESLDRTLKDIVNNPG----DKPLGGKVIVFGGDFRQVLPVINGAGREEIVFAALNS 345
Query: 241 SYLWKHCKVMKLTVNMSLQNATSTSSATEIKEFVDWLLQVGDGTVKTIDEEETLIEIPPD 300
SY+W+H KV++LT NM L S +I++F W+L VGDG + ++ LI+IP +
Sbjct: 346 SYIWEHSKVLELTKNMRLLADISEHEKRDIEDFSKWILDVGDGKISQPNDGIALIDIPEE 405
Query: 301 LLIEQCKEPLLELVNFAYPKL---AHNLQKSSF--FQERAILAPTLESLEEINNFMLAMI 355
LI +P+ ++ Y + +K+ + +Q RAIL PT E + IN M+ M+
Sbjct: 406 FLINGDNDPVESIIEAVYGNTFMEEKDPKKTDYPQYQGRAILCPTNEDVNSINEHMMRML 465
Query: 356 PGDETEYLSCDTPCKSDEDSGDNAE*FTSEVLNDFKCSEIPNHAIKLKAGVPIMLIRNID 415
G+E YLS D+ +D S +NA + ++ LN+ + +PNH ++LK G P+ML+RN+D
Sbjct: 466 DGEERIYLSSDSIDPADISSANNAA-YLADFLNNVRVYGLPNHCLRLKVGCPVMLLRNMD 524
Query: 416 QAAGLCNGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFSV 475
GLCNGTR+ V +T II Q
Sbjct: 525 PNKGLCNGTRLQVTQMTDTII-----------------------------------QARF 549
Query: 476 TLCFVMTINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSR 518
F MTINKSQGQ+L VGLYLPRPVF+HGQ YVA+ RV S+
Sbjct: 550 ITAFAMTINKSQGQTLESVGLYLPRPVFSHGQLYVAISRVTSK 592
Score = 133 bits (335), Expect = 2e-31
Identities = 61/114 (53%), Positives = 85/114 (74%)
Query: 405 GVPIMLIRNIDQAAGLCNGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDI 464
G P+ML+RN+D GLCNGTR+ V + +I + N++G+ IPRM +TPS++ +
Sbjct: 595 GCPVMLLRNMDPNKGLCNGTRLQVTQMADTVIQARFITGNRVGKIVLIPRMLITPSDTRL 654
Query: 465 PFKFQRRQFSVTLCFVMTINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSR 518
PFK +R+QF++++ F MTINKSQGQ+L VGLYLPRPVF+HGQ YVA+ RV S+
Sbjct: 655 PFKMRRKQFALSVAFAMTINKSQGQTLESVGLYLPRPVFSHGQLYVAISRVTSK 708
Score = 131 bits (329), Expect = 1e-30
Identities = 60/114 (52%), Positives = 84/114 (73%)
Query: 405 GVPIMLIRNIDQAAGLCNGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDI 464
G P+ML+RN+D GLCNGTR+ V + +I + N++G+ IPRM +TP ++ +
Sbjct: 711 GCPVMLLRNMDPNKGLCNGTRLQVTQMADTVIQARFITGNRVGKIVLIPRMLITPLDTRL 770
Query: 465 PFKFQRRQFSVTLCFVMTINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSR 518
PFK +R+QF++++ F MTINKSQGQ+L VGLYLPRPVF+HGQ YVA+ RV S+
Sbjct: 771 PFKMRRKQFALSVAFAMTINKSQGQTLESVGLYLPRPVFSHGQLYVAISRVTSK 824
>At2g14300 pseudogene; similar to MURA transposase of maize Mutator
transposon
Length = 1230
Score = 385 bits (989), Expect = e-107
Identities = 205/463 (44%), Positives = 285/463 (61%), Gaps = 30/463 (6%)
Query: 45 VADELNYNRAEMVKIHDEFVSSLTSE-QEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFV 103
+ DE NYNR + K HD ++ +LT E +++++ ++DDVL+D FFLY FGGTGKTF+
Sbjct: 797 IIDERNYNRESLKKKHDNWLKTLTPEHKKVYHDEIMDDVLNDKGGVFFLYAFGGTGKTFL 856
Query: 104 WNILYAALRSRGLIVLNVASSGIASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAE 163
W +L AA+R +G LNVASS IASLLL G RTAHSRF IP++ +E STCN+ +GS AE
Sbjct: 857 WKVLSAAIRCKGDTCLNVASSSIASLLLEGGRTAHSRFGIPLTPHETSTCNMERGSDLAE 916
Query: 164 LLKKASLIIWDEAPMLNKHCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILP 223
L+ A LIIWDE D+PFG KV++ G DF QIL
Sbjct: 917 LVTAAKLIIWDE----------------------------DMPFGRKVILFGGDFRQILH 948
Query: 224 VISKGSRSEIVGSAINSSYLWKHCKVMKLTVNMSLQNATSTSSATEIKEFVDWLLQVGDG 283
VI R IV S++NSSYLW+HCKV+KLT NM L + A EI++F+ W+L VG+G
Sbjct: 949 VIPAAGRELIVKSSLNSSYLWQHCKVLKLTKNMRLLQDIDINEAREIEDFLKWILTVGEG 1008
Query: 284 TVKTIDEEETLIEIPPDLLIEQCKEPLLELVNFAYPKLAHNLQKSSFFQERAILAPTLES 343
+ + T I+IP D+LI + P+ ++ Y + FFQ +AIL PT +
Sbjct: 1009 KLNEPSDGVTHIQIPDDILIPEGDNPIESIIKAVYGTTFAQKRDPKFFQHKAILCPTNDD 1068
Query: 344 LEEINNFMLAMIPGDETEYLSCDTPCKSDEDSGDNAE*FTSEVLNDFKCSEIPNHAIKLK 403
+ IN+ ML+ + G+E Y S ++ SD + N +T + LN K S + NH ++LK
Sbjct: 1069 VNSINDHMLSKLTGEERIYRSSNSIDPSDTRADKNPI-YTPDFLNKIKISGLANHLLRLK 1127
Query: 404 AGVPIMLIRNIDQAAGLCNGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSD 463
G P+ML+RN GL NGTR+ + L ++ +L ++G+ IPRMSLTPS+
Sbjct: 1128 VGCPVMLLRNFYPHGGLMNGTRLQIVRLGDKLVQGRILTGTRVGKLVIIPRMSLTPSDRR 1187
Query: 464 IPFKFQRRQFSVTLCFVMTINKSQGQSLSHVGLYLPRPVFTHG 506
+PFK +RR F +++ F MTINKSQGQSL +VG+YLP+ VF+HG
Sbjct: 1188 LPFKMKRRHFPLSVAFAMTINKSQGQSLGNVGMYLPKAVFSHG 1230
>At5g34960 putative protein
Length = 1033
Score = 327 bits (839), Expect = 8e-90
Identities = 202/541 (37%), Positives = 284/541 (52%), Gaps = 104/541 (19%)
Query: 10 IEIEKILQGNERSLKEFPCLPYPKFSEIHNFEVIFVADELNYNRAEMVKI-HDEFVSSLT 68
+ EKI+ N +L++ P P I N + V +EL YNR +K H+E+ LT
Sbjct: 592 VSFEKIMLSNGATLEDIDEFPKPTRDGIDNSNRLIV-EELRYNRESNLKEKHEEWKQMLT 650
Query: 69 SEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLNVASSGIAS 128
EQ Y + + A + IVLNVASSGIAS
Sbjct: 651 PEQRGVYNEITE----------------------------AVFNNLDQIVLNVASSGIAS 682
Query: 129 LLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAELLKKASLIIWDEAPMLNKHCIEALD 188
LLL G RTAHSRF I ++ +E S C ++ S A L+K+ASL+IWDEAPM+++
Sbjct: 683 LLLEGGRTAHSRFGISLNPDEFSVCKIKPKSDLANLVKEASLVIWDEAPMMSR------- 735
Query: 189 RSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGSAINSSYLWKHCK 248
Q+L VI+ R+EIV S++N+SYLW HCK
Sbjct: 736 -------------------------------QVLLVINGAGRAEIVMSSLNASYLWDHCK 764
Query: 249 VMKLTVNMSLQNATSTSSATEIKEFVDWLLQVGDGTVKTIDEEETLIEIPPDLLIEQCKE 308
V+K+ + DG +I+IP DLLI +
Sbjct: 765 VLKIN-------------------------EPNDGV--------AIIDIPEDLLITNTDK 791
Query: 309 PLLELVNFAY--PKLAHNLQKSSFFQERAILAPTLESLEEINNFMLAMIPGDETEYLSCD 366
P+ + N Y PK+ H + FFQ RAILAPT E + IN ++L + +E YLS D
Sbjct: 792 PIESITNEIYGDPKILHEITDPKFFQGRAILAPTNEDVNTINEYLLEQLHAEERIYLSAD 851
Query: 367 TPCKSDEDSGDNAE*FTSEVLNDFKCSEIPNHAIKLKAGVPIMLIRNIDQAAGLCNGTRM 426
+ +D +S +N T + LN K + +PNH+++LK P+ML+RN+D GLCNGTR+
Sbjct: 852 SIDPTDSNSLNNPV-ITPDFLNSIKLAGLPNHSLRLKVSAPVMLLRNLDPKGGLCNGTRL 910
Query: 427 IVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFSVTLCFVMTINKS 486
+ L I+ V+ + +G IP ++LTP+++ +PFK +RRQF +++ F MTIN S
Sbjct: 911 QITQLCTQIVEAKVITGDIIGHIVLIPTVNLTPTDTKLPFKMRRRQFPLSVAFAMTINTS 970
Query: 487 QGQSLSHVGLYLPRPVFTHGQPYVALYRVKSRKMLKMLIIDDE*VVSNTTRNVVYQEVLD 546
QGQSL HVGLYLP+ VF+HGQ YVAL RV S+K LK LI+D + + T NVV++EV
Sbjct: 971 QGQSLEHVGLYLPKAVFSHGQLYVALSRVTSKKGLKFLILDKDGKLQKQTTNVVFKEVFQ 1030
Query: 547 N 547
N
Sbjct: 1031 N 1031
>At5g37110 putative helicase
Length = 1307
Score = 324 bits (831), Expect = 7e-89
Identities = 194/529 (36%), Positives = 279/529 (52%), Gaps = 94/529 (17%)
Query: 1 DEELQNLCMIEIEKILQGNERSLKEFPCLPY----PKFSEIHNFEVIFVADELNYNRAEM 56
DEE + + EI+ IL+ N SL + +P P+F + + DE Y+R +
Sbjct: 852 DEEKKLYALQEIDHILRRNGTSLTYYKTMPQVPRDPRFDTN-----VLILDEKGYDRDNL 906
Query: 57 VKIHDEFVSSLTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGL 116
+ H +++ LT EQ+ Y +++ V + F+YGFGGT
Sbjct: 907 TEKHAKWIKMLTPEQKSIYDDIIGAVNENVGVVVFVYGFGGT------------------ 948
Query: 117 IVLNVASSGIASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAELLKKASLIIWDEA 176
G RTAHSRF IP++ NE +TCN++ GS +A L+K+ASLIIWDEA
Sbjct: 949 ---------------EGGRTAHSRFGIPLNPNEFTTCNMKVGSDRANLVKEASLIIWDEA 993
Query: 177 PMLNKHCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGS 236
PM++++C E+LDRSL+DI +G + PFGGKVVV G
Sbjct: 994 PMMSRYCFESLDRSLSDICG----NGDNKPFGGKVVVFG--------------------- 1028
Query: 237 AINSSYLWKHCKVMKLTVNMSLQNATSTSSATEIKEFVDWLLQVGDGTVKTIDEEETLIE 296
S S A +IKEF +W+L VGDG + ++ E LI
Sbjct: 1029 ------------------------GLSVSEAKDIKEFSEWILAVGDGRIVEPNDGEALIV 1064
Query: 297 IPPDLLIEQCKEPLLELVNFAYPKLA--HNLQKSSFFQERAILAPTLESLEEINNFMLAM 354
IP + LI + K+P+ + Y + H FFQE+AIL PT E + +IN ML
Sbjct: 1065 IPSEFLITKAKDPIEAICTEIYGDITKIHEQNDPIFFQEKAILCPTNEDVNQINETMLDN 1124
Query: 355 IPGDETEYLSCDTPCKSDEDSGDNAE*FTSEVLNDFKCSEIPNHAIKLKAGVPIMLIRNI 414
+ G+E +LS D+ +D G N T + LN K S +PNH ++LK G P+ML+RNI
Sbjct: 1125 LQGEEFTFLSSDSLDPADI-GGKNNPALTPDFLNSVKVSRLPNHKLRLKIGCPVMLLRNI 1183
Query: 415 DQAAGLCNGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFS 474
D GL NGTR+ + + +I+ +L ++ G IPR+ L PS++ +PF+ +R Q
Sbjct: 1184 DPIGGLMNGTRLRITQMGPFILQAMILTGDRAGHLVLIPRLKLAPSDTKLPFRMRRTQLP 1243
Query: 475 VTLCFVMTINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSRKMLKM 523
+ +CF MTINKSQGQSL VG++L RP F+HGQ YVA+ RV S+ LK+
Sbjct: 1244 LAVCFAMTINKSQGQSLKRVGIFLLRPCFSHGQLYVAISRVTSKTRLKI 1292
>At3g42420 putative protein
Length = 1018
Score = 316 bits (809), Expect = 2e-86
Identities = 190/488 (38%), Positives = 268/488 (53%), Gaps = 70/488 (14%)
Query: 1 DEELQNLCMIEIEKILQGNERSLKEFPCLPYPKFSEIHNFEVIFVADELNYNRAEMVKIH 60
+ E++N + EIEKI+ N ++ E P P I N + V DE+ Y+R + H
Sbjct: 600 EAEIKNYTLQEIEKIMLFNGGTITEIENFPQPTRECIDNSNRLIV-DEMRYDRQYLTGKH 658
Query: 61 DEFVSSLTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLN 120
E++ LT E+ Y + V D FF+YGFGGT KTF+W L AA+RSRG IVLN
Sbjct: 659 AEWIDMLTPEKRGVYDEITGAVFKDLGGVFFVYGFGGTRKTFIWKTLSAAIRSRGDIVLN 718
Query: 121 VASSGIASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAELLKKASLIIWDEAPMLN 180
VAS+GIASLLL G RTAHSRFSIP++ +E ++C ++ S A L++KASLIIWDEAP+++
Sbjct: 719 VASNGIASLLLEGGRTAHSRFSIPLTPDEYTSCRIKPKSDLANLIRKASLIIWDEAPVMS 778
Query: 181 KHCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGSAINS 240
K C E+LD+S +D I+G+ N
Sbjct: 779 KWCFESLDKSFSD---------------------------------------IIGNKDNK 799
Query: 241 SYLWKHCKVMKLTVNMS-LQNATSTSSATEIKEFVDWLLQVGDGTVKTIDEEETLIEIPP 299
HCKV+KLT NM L S A +IKEF DWLL VG+ ++ I E
Sbjct: 800 D----HCKVLKLTKNMRLLSKDLSEEEAKDIKEFSDWLLAVGN-PIEAISHE-------- 846
Query: 300 DLLIEQCKEPLLELVNFAYPKLAHNLQKSSFFQERAILAPTLESLEEINNFMLAMIPGDE 359
+ P + + FF +R IL+PT + + IN +ML + G+E
Sbjct: 847 ---------------IYGDPAMLKVNEDQKFFLKRVILSPTNDDVHTINQYMLNNMEGEE 891
Query: 360 TEYLSCDTPCKSDEDSGDNAE*FTSEVLNDFKCSEIPNHAIKLKAGVPIMLIRNIDQAAG 419
+LS D+ SD DS N T ++LN K S +P+H ++LK G PI+L+RN+D G
Sbjct: 892 RIFLSADSIDPSDSDSLKNPV-ITPDLLNSIKLSGLPHHELRLKIGAPIILLRNLDPKGG 950
Query: 420 LCNGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFSVTLCF 479
LCNGTR+ + +T ++ V+ ++ G+ IP +++TPSN +PF+ +RRQF V+L F
Sbjct: 951 LCNGTRLQITQMTIQVLQAKVIIGDRSGDIVLIPLINITPSNMKLPFRMRRRQFPVSLAF 1010
Query: 480 VMTINKSQ 487
MTINKSQ
Sbjct: 1011 AMTINKSQ 1018
>At1g64410 unknown protein
Length = 753
Score = 279 bits (713), Expect = 3e-75
Identities = 146/296 (49%), Positives = 197/296 (66%), Gaps = 5/296 (1%)
Query: 1 DEELQNLCMIEIEKILQGNERSLKEFPCLPYPKFSEIHNFEVIFVADELNYNRAEMVKIH 60
DEE Q C+ EI ++L N SL ++ +P + F+ DE Y+RA + + H
Sbjct: 457 DEERQQYCLQEIARLLTKNGVSLSKWKQMPQISDEHVEKCNH-FILDERKYDRAYLTEKH 515
Query: 61 DEFVSSLTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLN 120
+E+++ +T EQ+ Y ++D VL D FF+YGFGGTGKTF+W +L AA+RS+G I LN
Sbjct: 516 EEWLTMVTLEQKKIYDEIMDVVLHDRGGVFFVYGFGGTGKTFLWKLLSAAIRSKGDISLN 575
Query: 121 VASSGIASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAELLKKASLIIWDEAPMLN 180
VASSGIA+LLL G RTAHSRF IPI+ NE STCN+ +G EL+K+A+LIIWDEA M++
Sbjct: 576 VASSGIAALLLDGGRTAHSRFGIPINPNESSTCNISRGLDFGELVKEANLIIWDEAHMMS 635
Query: 181 KHCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGSAINS 240
KHC E+LDR+L DIM D PFGGKV+V G DF Q+L VI+ R EIV +A+NS
Sbjct: 636 KHCFESLDRTLRDIMNNPG----DKPFGGKVIVFGGDFRQVLSVINGAGREEIVFAALNS 691
Query: 241 SYLWKHCKVMKLTVNMSLQNATSTSSATEIKEFVDWLLQVGDGTVKTIDEEETLIE 296
SY+W+HCKV++LT NM L S +I+ F W+L VGDG + ++ LI+
Sbjct: 692 SYIWEHCKVLELTKNMRLLANISEHEKRDIEYFSKWILDVGDGKISQPNDGIALID 747
>At2g07620 putative helicase
Length = 1241
Score = 248 bits (634), Expect = 5e-66
Identities = 130/258 (50%), Positives = 178/258 (68%), Gaps = 5/258 (1%)
Query: 1 DEELQNLCMIEIEKILQGNERSLKEFPCLPYPKFSEIHNFEVIFVADELNYNRAEMVKIH 60
D E N ++EIE +L N +L++F +P P E + F+ +E NYN ++ + H
Sbjct: 782 DAEKINYALLEIEDMLLCNGSTLEDFKHMPKPT-KEGTDHSNRFITEEKNYNVEKLKEDH 840
Query: 61 DEFVSSLTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLN 120
D++ + +TSEQ+ Y ++ VL ++ FF+YGFGGTGKTF+W L AA+R +GLI +N
Sbjct: 841 DDWFNKMTSEQKEIYDEIMKAVLENSGGIFFVYGFGGTGKTFMWKTLSAAVRMKGLISVN 900
Query: 121 VASSGIASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAELLKKASLIIWDEAPMLN 180
VASSGIA LLL G RTAHSRF IPI+ ++ +TC++ S A +LK+ASLIIWDEAPM++
Sbjct: 901 VASSGIAFLLLQGGRTAHSRFGIPINPDDFTTCHIVPNSDLANMLKEASLIIWDEAPMMS 960
Query: 181 KHCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGSAINS 240
++C E+LDRSLND++ PFGGKVVV G DF Q+L VI R+EIV +A+NS
Sbjct: 961 RYCFESLDRSLNDVIGNVDGK----PFGGKVVVFGGDFRQVLHVIHGAGRAEIVLAALNS 1016
Query: 241 SYLWKHCKVMKLTVNMSL 258
SYLW+HC V+ LT NMSL
Sbjct: 1017 SYLWEHCNVLTLTKNMSL 1034
Score = 163 bits (413), Expect = 2e-40
Identities = 84/186 (45%), Positives = 119/186 (63%), Gaps = 1/186 (0%)
Query: 362 YLSCDTPCKSDEDSGDNAE*FTSEVLNDFKCSEIPNHAIKLKAGVPIMLIRNIDQAAGLC 421
YLS D+ D S +N FT LN K S + NH + LK G P+ML++NID GLC
Sbjct: 1054 YLSADSIDPQDPASLNNPV-FTPYFLNSIKLSGLSNHNLTLKIGTPVMLLKNIDPKGGLC 1112
Query: 422 NGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFSVTLCFVM 481
NGTR+ V + +I+ V+ +++ + I + ++PS++ +PF+ +RRQF + + F M
Sbjct: 1113 NGTRLQVTQMGNHILEARVITGDRVRDKVIIIKAQISPSDTKLPFRMRRRQFPIAVAFAM 1172
Query: 482 TINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSRKMLKMLIIDDE*VVSNTTRNVVY 541
I KSQGQSL V +YLPRPVF+HGQ YVAL RV S+K LK+LI+D E + T NVV+
Sbjct: 1173 RIKKSQGQSLKEVEIYLPRPVFSHGQLYVALSRVTSKKGLKVLIVDKEGNTQSQTMNVVF 1232
Query: 542 QEVLDN 547
+E+ N
Sbjct: 1233 KEIFQN 1238
>At4g04300 hypothetical protein
Length = 286
Score = 229 bits (584), Expect = 3e-60
Identities = 132/285 (46%), Positives = 171/285 (59%), Gaps = 23/285 (8%)
Query: 67 LTSEQEIFYKNVLDDVLSD-NS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLNVASSG 125
LT EQ Y + + V +D FFF+YG GG GKTF+W L A RS+G LN+ASSG
Sbjct: 2 LTPEQRGIYDQITNAVFNDMGGVFFFVYGSGGIGKTFIWKTLAAVGRSKGQTCLNIASSG 61
Query: 126 IASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKA---------------ELLKKASL 170
IASLLL G R AH RFSIP++ +E S C ++ S A +L+KKASL
Sbjct: 62 IASLLLEGGRIAHYRFSIPLNPDEFSVCKIKPKSDLADLIKEASLIIWDKLVDLIKKASL 121
Query: 171 IIWDEAPMLNKHCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSR 230
IIWD+APM +K C EALD+S +DI+K F GKV+V G DF Q+LPVI+ R
Sbjct: 122 IIWDKAPMKSKFCFEALDKSFSDIIKRVDNK----VFCGKVMVFGGDFRQVLPVINGAGR 177
Query: 231 SEIVGSAINSSYLWKHCKVMKLTVNMS-LQNATSTSSATEIKEFVDWLLQVGDGTVKTID 289
+E V S++N+ Y+W HCKV+KLT NM L N S A EI+EF DWLL VGDG V +
Sbjct: 178 AETVMSSLNAVYIWDHCKVLKLTKNMRLLNNDLSVDEAKEIQEFFDWLLVVGDGRVNEPN 237
Query: 290 EEETLIEIPPDLLIEQCKEPLLELVNFAYPKLA--HNLQKSSFFQ 332
+ E LI+IP +LLI++ P+ + Y H + FF+
Sbjct: 238 DGEALIDIPEELLIQEADIPIEAISREIYGDATKLHEINDPKFFR 282
>At3g51700 unknown protein
Length = 344
Score = 188 bits (478), Expect = 6e-48
Identities = 107/294 (36%), Positives = 166/294 (56%), Gaps = 3/294 (1%)
Query: 253 TVNMSLQNATSTSSATEIKE-FVDWLLQVGDGTVKTIDEEETLIEIPPDLLIEQCKEPLL 311
+++M L +++ T T I E F W+ +G + ++ ET I+I DLLI +CK+P+
Sbjct: 39 SIDMVLLDSSGTKIHTTIDEAFTKWITNIGGENINKPNDGETKIDIHEDLLITECKDPIK 98
Query: 312 ELVNFAYPKLAHNLQKSSFFQERAILAPTLESLEEINNFMLAMIPGDETEYLSCDTPCKS 371
+V+ Y + F+QERAIL T + +EIN++ML+ + G+ET+ DT +
Sbjct: 99 TIVDEVYGESFTESYNPDFYQERAILCHTNDVADEINDYMLSQLQGEETKCYGADTIYPT 158
Query: 372 DEDSGDNAE*FTSEVLNDFKCSEIPNHAIKLKAGVPIMLIRNIDQAAGLCNGTRMIVNAL 431
D + E LN K P+ ++LK G P+ML+R++ L GTR+ + +
Sbjct: 159 HASPNDKML-YPLEFLNSIKIPGFPDFKLRLKVGAPVMLLRDLAPYGWLRKGTRLQITRV 217
Query: 432 TKYIIVVTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFSVTLCFVMTINKSQGQSL 491
+++ ++ N GE IPR+ + P K +RRQF V L F MTI++SQ Q+L
Sbjct: 218 ETFVLEAMIITGNNHGEKVLIPRIPSDLREAKFPIKMRRRQFPVKLAFAMTIDESQRQTL 277
Query: 492 SHVGLYLPRPVFTHGQPYVALYRVKSRKMLKMLIID-DE*VVSNTTRNVVYQEV 544
S VG+YLPR + HGQ YVA+ +VKSR LK+LI D D T+NVV++E+
Sbjct: 278 SKVGIYLPRQLLFHGQRYVAISKVKSRAGLKVLITDKDGKPDQEETKNVVFKEL 331
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.139 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,764,313
Number of Sequences: 26719
Number of extensions: 494458
Number of successful extensions: 1320
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 33
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1187
Number of HSP's gapped (non-prelim): 59
length of query: 547
length of database: 11,318,596
effective HSP length: 104
effective length of query: 443
effective length of database: 8,539,820
effective search space: 3783140260
effective search space used: 3783140260
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0031.3


