

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0031.15
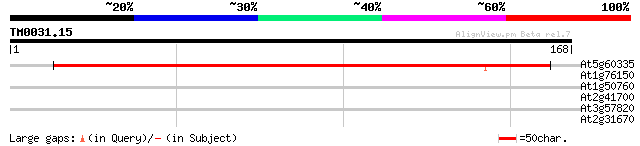
(168 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g60335 putative protein 182 1e-46
At1g76150 unknown protein 34 0.042
At1g50760 hypothetical protein 27 4.0
At2g41700 ATP-binding cassette transporter ABCA1 27 5.2
At3g57820 putative protein 27 6.8
At2g31670 unknown protein 26 8.9
>At5g60335 putative protein
Length = 187
Score = 182 bits (461), Expect = 1e-46
Identities = 90/150 (60%), Positives = 116/150 (77%), Gaps = 1/150 (0%)
Query: 14 LSLRCFSSAATPQVLKPGDVLRKARAFTEEDVLHYSKVTHDLNPLHTDSAAAQNVGFEGP 73
L LR FSS A+ +LK GDVLR+ R F+ ED+ Y++V+HD NPLH D +A+ GFE
Sbjct: 33 LVLRSFSSVASKTLLKVGDVLRETRVFSSEDIKAYAEVSHDWNPLHFDPESARKAGFENR 92
Query: 74 LVHGMLVASLFPHIISSHFPGAVYVSQSLNFKFPVYIGDQVIGEVQATNLRANKNRYLVK 133
LVHGMLV+S+FP IIS+HFPGAVYVSQSL+F+ PVYIGD+++G VQA LR KN+Y+VK
Sbjct: 93 LVHGMLVSSMFPRIISAHFPGAVYVSQSLHFRSPVYIGDEILGLVQAIALRETKNKYIVK 152
Query: 134 FKTRCIKS-DELVAIEGEAVAMLPTLTVEQ 162
F T+C K+ +ELV I+GEA A+LP L + Q
Sbjct: 153 FSTKCFKNHNELVVIDGEATAILPNLDMLQ 182
>At1g76150 unknown protein
Length = 309
Score = 33.9 bits (76), Expect = 0.042
Identities = 26/91 (28%), Positives = 43/91 (46%), Gaps = 6/91 (6%)
Query: 50 KVTHDLNPLHTDSAAAQNVGFEGPLVHGMLVASL-FPHIISSHFPGAVYVSQSLNFKF-- 106
+++ D NPLH+D A+ GF P++HG+ II G ++++ +F
Sbjct: 204 RLSGDYNPLHSDPEFAKLAGFPRPILHGLCTLGFAIKAIIKCVCKGDPTAVKTISGRFLT 263
Query: 107 PVYIGDQVIGEVQATNLRANKNRYLVKFKTR 137
V+ G+ +I E+ LR Y K K R
Sbjct: 264 TVFPGETLITEMWLEGLRV---IYQTKVKER 291
>At1g50760 hypothetical protein
Length = 649
Score = 27.3 bits (59), Expect = 4.0
Identities = 16/52 (30%), Positives = 26/52 (49%), Gaps = 5/52 (9%)
Query: 97 YVSQSLNFKFPVYIGDQVIGEVQATNLRANKNRYLVKFKTRCIKSDELVAIE 148
Y N+KFP + ++ + ++ + + L+ F RCIK ELV IE
Sbjct: 312 YTRSVTNWKFPQFYPEKAM----CVHVSPSLDEELISF-ARCIKDSELVGIE 358
>At2g41700 ATP-binding cassette transporter ABCA1
Length = 1884
Score = 26.9 bits (58), Expect = 5.2
Identities = 15/44 (34%), Positives = 22/44 (49%), Gaps = 3/44 (6%)
Query: 68 VGFEGPLVHGMLVASLFPHIISSHFPGAVYVSQ---SLNFKFPV 108
VG+ LV S+ HI+ H P A VS+ ++FK P+
Sbjct: 739 VGYTLTLVKTSPTVSVAAHIVHRHIPSATCVSEVGNEISFKLPL 782
>At3g57820 putative protein
Length = 89
Score = 26.6 bits (57), Expect = 6.8
Identities = 17/51 (33%), Positives = 27/51 (52%), Gaps = 1/51 (1%)
Query: 97 YVSQSLNFKFPVYIGDQVIGE-VQATNLRANKNRYLVKFKTRCIKSDELVA 146
Y S + K+ IGD++I + + ++R V+FK R K+DEL A
Sbjct: 26 YPSFTFGLKWVYQIGDKIIRKRIHVLVEHVQQSRCAVEFKLRKKKNDELKA 76
>At2g31670 unknown protein
Length = 263
Score = 26.2 bits (56), Expect = 8.9
Identities = 21/84 (25%), Positives = 39/84 (46%), Gaps = 6/84 (7%)
Query: 11 LQSLSLRCFS--SAATPQVLKPGDVLRKARAFTEEDVLHYSKVTHDLNPLHTDSAAAQNV 68
L+ L R FS S++TPQ ++ F +D +K+T +N L+ + Q +
Sbjct: 28 LRLLPRRSFSVMSSSTPQ----SQIIEHIVLFKTKDDADSTKITSMINNLNALAYLDQVL 83
Query: 69 GFEGPLVHGMLVASLFPHIISSHF 92
+H + A+ F H++ S +
Sbjct: 84 HISTSPLHRISSATAFTHVLHSRY 107
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.134 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,314,897
Number of Sequences: 26719
Number of extensions: 124055
Number of successful extensions: 287
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 284
Number of HSP's gapped (non-prelim): 6
length of query: 168
length of database: 11,318,596
effective HSP length: 92
effective length of query: 76
effective length of database: 8,860,448
effective search space: 673394048
effective search space used: 673394048
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0031.15


