

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0031.13
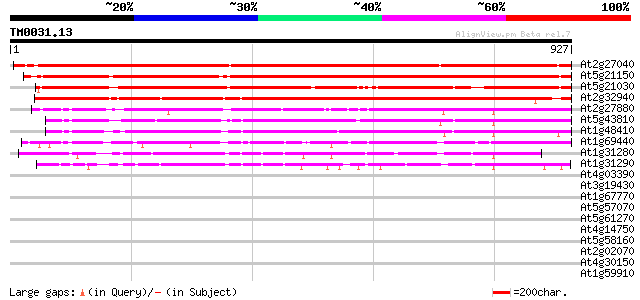
(927 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g27040 Argonaute (AGO1)-like protein 1357 0.0
At5g21150 zwille/pinhead-like protein 1236 0.0
At5g21030 zwille/pinhead-like protein 1062 0.0
At2g32940 Argonaute (AGO1)-like protein 941 0.0
At2g27880 Argonaute (AGO1)-like protein 525 e-149
At5g43810 PINHEAD (gb|AAD40098.1); translation initiation factor 493 e-139
At1g48410 Argonaute protein (AGO1) 479 e-135
At1g69440 hypothetical protein 386 e-107
At1g31280 unknown protein 326 4e-89
At1g31290 hypothetical protein 320 2e-87
At4g03390 SRF3 (LRR receptor-like protein kinase like protein) 40 0.005
At3g19430 putative late embryogenesis abundant protein 39 0.018
At1g67770 terminal ear1-like protein 35 0.15
At5g57070 putative protein 34 0.44
At5g61270 bHLH transcription factor like protein 33 0.57
At4g14750 unknown protein 33 0.57
At5g58160 strong similarity to unknown protein (gb|AAD23008.1) 33 0.97
At2g02070 putative C2H2-type zinc finger protein 33 0.97
At4g30150 hypothetical protein 32 1.3
At1g59910 hypothetical protein 32 1.3
>At2g27040 Argonaute (AGO1)-like protein
Length = 924
Score = 1357 bits (3511), Expect = 0.0
Identities = 669/926 (72%), Positives = 792/926 (85%), Gaps = 18/926 (1%)
Query: 7 ENGNG-NGNGNEDLMPPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLPIARKGLGS 65
E+ NG NG+G + +PPPPP ++P +VEPV+V E +KK P R+P+ARKG G+
Sbjct: 12 ESANGANGSGVTEALPPPPP--VIPPNVEPVRVKT-----ELAEKKGPVRVPMARKGFGT 64
Query: 66 KGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQETYGSELN 125
+G K+PLLTNHFKV VAN GHFF YSVAL Y+DGRPVE KGVGRK++DKV +TY S+L+
Sbjct: 65 RGQKIPLLTNHFKVDVANLQGHFFHYSVALFYDDGRPVEQKGVGRKILDKVHQTYHSDLD 124
Query: 126 GKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDG-ASTNDSDKKRMRRP 184
GK+FAYDGEKTLFT G+L NK++F+VVLE+V + R NGN SP+G S +D D+KR+RRP
Sbjct: 125 GKEFAYDGEKTLFTYGALPSNKMDFSVVLEEVSATRANGNGSPNGNESPSDGDRKRLRRP 184
Query: 185 YHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCLLVRQSF 244
SK F+VEIS+AAKIPLQA+ NA+RGQESEN QEAIRVLDIILRQHAA+QGCLLVRQSF
Sbjct: 185 NRSKNFRVEISYAAKIPLQALANAMRGQESENSQEAIRVLDIILRQHAARQGCLLVRQSF 244
Query: 245 FHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFLIANQNVR 304
FHNDP N VGG +LGCRGFHSSFRTTQ G+SLN+DV+TTMII+PGPVVDFLIANQN R
Sbjct: 245 FHNDPTNCEPVGGNILGCRGFHSSFRTTQGGMSLNMDVTTTMIIKPGPVVDFLIANQNAR 304
Query: 305 DPFSLDWAKAKRTLKNLRIKASPSNQEYKITGLSELPCKEQTFTMKKKGGN-NGEEDATE 363
DP+S+DW+KAKRTLKNLR+K SPS QE+KITGLS+ PC+EQTF +KK+ N NGE + TE
Sbjct: 305 DPYSIDWSKAKRTLKNLRVKVSPSGQEFKITGLSDKPCREQTFELKKRNPNENGEFETTE 364
Query: 364 EEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRS 423
+TV +YF + R IDL+YSADLPCINVGKPKRPTY+P+ELC+LV LQRYTKALTT QRS
Sbjct: 365 --VTVADYFRDTRHIDLQYSADLPCINVGKPKRPTYIPLELCALVPLQRYTKALTTFQRS 422
Query: 424 SLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKFGN 483
+LVEKSRQKP ERM VL++ALK SNY EP+L++CGI+I+S FTQVEGRVL AP+LK G
Sbjct: 423 ALVEKSRQKPQERMTVLSKALKVSNYDAEPLLRSCGISISSNFTQVEGRVLPAPKLKMGC 482
Query: 484 GEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARCDVRGLVRDLIKCARLKGIPIDEPY 543
G + PRNGRWN NNK+ V P KI+ W VVNFSARC+VR +V DLIK KGI I P+
Sbjct: 483 GSETFPRNGRWNFNNKEFVEPTKIQRWVVVNFSARCNVRQVVDDLIKIGGSKGIEIASPF 542
Query: 544 EEIFEENGQFRRAPPLVRVEKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAE 603
+ +FEE QFRRAPP++RVE MF+ IQ +LPG P F+LC+LP++KNSDLYGPWKKKNL E
Sbjct: 543 Q-VFEEGNQFRRAPPMIRVENMFKDIQSKLPGVPQFILCVLPDKKNSDLYGPWKKKNLTE 601
Query: 604 YGIVTQCISPTRV-NDQYLTNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDV 662
+GIVTQC++PTR NDQYLTN+L+KINAKLGGLNS+L VE P+ ++SKVPTIILGMDV
Sbjct: 602 FGIVTQCMAPTRQPNDQYLTNLLLKINAKLGGLNSMLSVERTPAFTVISKVPTIILGMDV 661
Query: 663 SHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRE 722
SHGSPGQSD+PSIAAVVSSREWPLISKYRA VRTQ K EMI++L K+ + ED+GII+E
Sbjct: 662 SHGSPGQSDVPSIAAVVSSREWPLISKYRASVRTQPSKAEMIESLVKK-NGTEDDGIIKE 720
Query: 723 LLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVA 782
LL+DFY+SS KRKP++IIIFRDGVSESQFNQVLNIEL+QIIEACK LD WNPKFL++VA
Sbjct: 721 LLVDFYTSSNKRKPEHIIIFRDGVSESQFNQVLNIELDQIIEACKLLDANWNPKFLLLVA 780
Query: 783 QKNHHTKFFQPGSPDNVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIG 842
QKNHHTKFFQP SP+NVPPGT+IDNKICHP+NNDFY+CAHAGMIGT+RPTHYHVL D+IG
Sbjct: 781 QKNHHTKFFQPTSPENVPPGTIIDNKICHPKNNDFYLCAHAGMIGTTRPTHYHVLYDEIG 840
Query: 843 FSPDELQELVHSLSYVYQRSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLT 902
FS DELQELVHSLSYVYQRST+AISVVAPICYAHLAA Q+G FMKFED+S+TSSSHGG+T
Sbjct: 841 FSADELQELVHSLSYVYQRSTSAISVVAPICYAHLAAAQLGTFMKFEDQSETSSSHGGIT 900
Query: 903 AAGVAPV-VPQLPKLQDSVSSSMFFC 927
A G P+ V QLP+L+D+V++SMFFC
Sbjct: 901 APG--PISVAQLPRLKDNVANSMFFC 924
>At5g21150 zwille/pinhead-like protein
Length = 892
Score = 1236 bits (3197), Expect = 0.0
Identities = 609/906 (67%), Positives = 737/906 (81%), Gaps = 26/906 (2%)
Query: 23 PPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLPIAR-KGLGSKGTKLPLLTNHFKVTV 81
PPPPP VPA++ P EPVKK + LP+AR +G GSKG K+PLLTNHF V
Sbjct: 12 PPPPPFVPANLVP--------EVEPVKKNI--LLPMARPRGSGSKGQKIPLLTNHFGVKF 61
Query: 82 ANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQETYGSELNGKDFAYDGEKTLFTIG 141
G+FF YSVA++YEDGRPVE KG+GRK++DKVQETY S+L K FAYDGEKTLFT+G
Sbjct: 62 NKPSGYFFHYSVAINYEDGRPVEAKGIGRKILDKVQETYQSDLGAKYFAYDGEKTLFTVG 121
Query: 142 SLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAAKIP 201
+L NKL+F+VVLE++ S+RN+ G TND+D+KR RRP +K F VEIS+AAKIP
Sbjct: 122 ALPSNKLDFSVVLEEIPSSRNHA-----GNDTNDADRKRSRRPNQTKKFMVEISYAAKIP 176
Query: 202 LQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLG 261
+QAI +AL+G+E+EN Q+A+RVLDIILRQ AA+QGCLLVRQSFFHND KN+ +GGGV G
Sbjct: 177 MQAIASALQGKETENLQDALRVLDIILRQSAARQGCLLVRQSFFHNDVKNFVPIGGGVSG 236
Query: 262 CRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFLIANQNVRDPFSLDWAKAKRTLKNL 321
CRGFHSSFRTTQ GLSLNID STTMI+QPGPVVDFL+ANQN +DP+ +DW KA+R LKNL
Sbjct: 237 CRGFHSSFRTTQGGLSLNIDTSTTMIVQPGPVVDFLLANQNKKDPYGMDWNKARRVLKNL 296
Query: 322 RIKASPSNQEYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLR 381
R++ + SN+EYKI+GLSE CK+Q K + GE + E EITV Y+ R I++R
Sbjct: 297 RVQITLSNREYKISGLSEHSCKDQL-----KPNDKGEFE--EVEITVLNYYKE-RNIEVR 348
Query: 382 YSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLN 441
YS D PCINVGKPKRPTY P+E C+LVSLQRYTK+LT QR++LVEKSRQKP ERM L
Sbjct: 349 YSGDFPCINVGKPKRPTYFPIEFCNLVSLQRYTKSLTNFQRAALVEKSRQKPPERMASLT 408
Query: 442 QALKTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKFGNGEDFNPRNGRWNLNNKKV 501
+ LK SNY +P+L++ G++I + FTQVEGR+L P LK G GE+ +P G+WN K +
Sbjct: 409 KGLKDSNYNADPVLQDSGVSIITNFTQVEGRILPTPMLKVGKGENLSPIKGKWNFMRKTL 468
Query: 502 VRPAKIEHWAVVNFSARCDVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVR 561
P + WAVVNFSARCD L+RDLIKC R KGI ++ P++++ EN QFR AP VR
Sbjct: 469 AEPTTVTRWAVVNFSARCDTNTLIRDLIKCGREKGINVEPPFKDVINENPQFRNAPATVR 528
Query: 562 VEKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRVNDQYL 621
VE MFE+I+ +LP P FLLC+L ERKNSD+YGPWKKKNL + GIVTQCI+PTR+NDQYL
Sbjct: 529 VENMFEQIKSKLPKPPLFLLCILAERKNSDVYGPWKKKNLVDLGIVTQCIAPTRLNDQYL 588
Query: 622 TNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSS 681
TNVL+KINAKLGGLNS+L +E +P++P V++VPTII+GMDVSHGSPGQSDIPSIAAVVSS
Sbjct: 589 TNVLLKINAKLGGLNSLLAMERSPAMPKVTQVPTIIVGMDVSHGSPGQSDIPSIAAVVSS 648
Query: 682 REWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIII 741
R+WPLISKY+ACVRTQS K+EMIDNLFK V+ K DEG+ RELL+DFY SS RKP++III
Sbjct: 649 RQWPLISKYKACVRTQSRKMEMIDNLFKPVNGK-DEGMFRELLLDFYYSSENRKPEHIII 707
Query: 742 FRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPDNVPP 801
FRDGVSESQFNQVLNIEL+Q+++ACKFLD+TW+PKF VIVAQKNHHTKFFQ PDNVPP
Sbjct: 708 FRDGVSESQFNQVLNIELDQMMQACKFLDDTWHPKFTVIVAQKNHHTKFFQSRGPDNVPP 767
Query: 802 GTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQR 861
GT+ID++ICHPRN DFY+CAHAGMIGT+RPTHYHVL D+IGF+ D+LQELVHSLSYVYQR
Sbjct: 768 GTIIDSQICHPRNFDFYLCAHAGMIGTTRPTHYHVLYDEIGFATDDLQELVHSLSYVYQR 827
Query: 862 STTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGVAPVVPQLPKLQDSVS 921
STTAISVVAP+CYAHLAA Q+G MK+E+ S+TSSSHGG+T G P VP +P+L ++VS
Sbjct: 828 STTAISVVAPVCYAHLAAAQMGTVMKYEELSETSSSHGGITTPGAVP-VPPMPQLHNNVS 886
Query: 922 SSMFFC 927
+SMFFC
Sbjct: 887 TSMFFC 892
>At5g21030 zwille/pinhead-like protein
Length = 850
Score = 1062 bits (2747), Expect = 0.0
Identities = 556/898 (61%), Positives = 681/898 (74%), Gaps = 65/898 (7%)
Query: 43 LPP------EPVKKKLPTRLPIARKGLGSKGTKLPLLTNHFKVTVANSDGH-FFQYSVAL 95
LPP EP+K K + LP+ R+G GSKG K+ LLTNHF+V + H FF YSV +
Sbjct: 5 LPPPQHMEREPLKSK-SSLLPMTRRGNGSKGQKILLLTNHFRVNFRKPNSHNFFHYSVTI 63
Query: 96 SYEDGRPVEGKGVGRKVIDKVQETYGSELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLE 155
+YEDG P+ KG GRK+++KVQ+T ++L K FAYDG+K L+T+G L R+ L+F+VVLE
Sbjct: 64 TYEDGSPLLAKGFGRKILEKVQQTCQADLGCKHFAYDGDKNLYTVGPLPRSSLDFSVVLE 123
Query: 156 DVISNRNNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAA-KIPLQAIVNALRGQES 214
S RN KR++ P+ SK F V I FA +IP++AI NAL+G+++
Sbjct: 124 TAPSRRNAD--------------KRLKLPHQSKKFNVAILFAPPEIPMEAIANALQGKKT 169
Query: 215 ENYQEAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQS 274
++ +AIRV+D IL Q+AA+QGCLLVRQSFFHND K +A++G GV C+GFHSSFRTTQ
Sbjct: 170 KHLLDAIRVMDCILSQNAARQGCLLVRQSFFHNDAKYFANIGEGVDCCKGFHSSFRTTQG 229
Query: 275 GLSLNIDVSTTMIIQPGPVVDFLIANQNVRDPFSLDWAKAKRTLKNLRIKASPSNQEYKI 334
GLSLNIDVST MI++PGPVVDFLIANQ V DPFS++W KAK TLKNLR+K PSNQEYKI
Sbjct: 230 GLSLNIDVSTAMIVKPGPVVDFLIANQGVNDPFSINWKKAKNTLKNLRVKVLPSNQEYKI 289
Query: 335 TGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKP 394
TGLS L CK+QTFT KK+ N E E EITV +YF R+I+LRYS LPCINVGKP
Sbjct: 290 TGLSGLHCKDQTFTWKKRNQNREFE---EVEITVSDYFTRIREIELRYSGGLPCINVGKP 346
Query: 395 KRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPM 454
RPTY P+ELC LVSLQRYTKALT QRS+L+++SRQ P +R+ VL +ALKTSNY ++PM
Sbjct: 347 NRPTYFPIELCELVSLQRYTKALTKFQRSNLIKESRQNPQQRIGVLTRALKTSNYNDDPM 406
Query: 455 LKNCGITIASGFTQVEGRVLQAPRLKFGNGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVN 514
L+ CG+ I S FTQVEGRVL P+LK G +D P NG WN NK PA + WAVVN
Sbjct: 407 LQECGVRIGSDFTQVEGRVLPTPKLKAGKEQDIYPINGSWNFKNK----PATVTRWAVVN 462
Query: 515 FSARCDVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVRVEKMFERIQK--- 571
FSARCD + ++ DL +C ++KGI +D PY +FEEN QF+ A VRV+KMF+ +Q
Sbjct: 463 FSARCDPQKIIDDLTRCGKMKGINVDSPYHVVFEENPQFKDATGSVRVDKMFQHLQSILG 522
Query: 572 ELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCI-SPTRVNDQYLTNVLMKINA 630
E+P P FLLC+L E+KNSD+Y +K+ + + +CI P +NDQYLTN+L+KINA
Sbjct: 523 EVP--PKFLLCIL-EKKNSDVY----EKSCSMWN--CECIVPPQNLNDQYLTNLLLKINA 573
Query: 631 KLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSD-IPSIAAVVSSREWPLISK 689
KLGGLNSVL +E++ ++P+V +VPTII+GMDVSHGSPGQSD IPSIAAVVSSREWPLISK
Sbjct: 574 KLGGLNSVLDMELSGTMPLVMRVPTIIIGMDVSHGSPGQSDHIPSIAAVVSSREWPLISK 633
Query: 690 YRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSES 749
YRACVRTQSPKVEMID+LFK VS+K+D+GI+RELL+DF+SSSGK KP++IIIFRDGVSES
Sbjct: 634 YRACVRTQSPKVEMIDSLFKPVSDKDDQGIMRELLLDFHSSSGK-KPNHIIIFRDGVSES 692
Query: 750 QFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPDNVPPGTVIDNKI 809
QFNQVLNIEL+Q++ Q NHHTKFFQ SP+NV PGT+ID+ I
Sbjct: 693 QFNQVLNIELDQMM-------------------QINHHTKFFQTESPNNVLPGTIIDSNI 733
Query: 810 CHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISVV 869
CH NNDFY+CAHAG IGT+RPTHYHVL D+IGF D+LQELVHSLSYVYQRSTTAIS+V
Sbjct: 734 CHQHNNDFYLCAHAGKIGTTRPTHYHVLYDEIGFDTDQLQELVHSLSYVYQRSTTAISLV 793
Query: 870 APICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGVAPVVPQLPKLQDSVSSSMFFC 927
APICYAHLAA Q+ MKFED S+TSSSHGG+T AG P VP +PKL +V+SSMFFC
Sbjct: 794 APICYAHLAAAQMATAMKFEDMSETSSSHGGITTAGAVP-VPPMPKLNTNVASSMFFC 850
>At2g32940 Argonaute (AGO1)-like protein
Length = 887
Score = 941 bits (2431), Expect = 0.0
Identities = 485/903 (53%), Positives = 641/903 (70%), Gaps = 38/903 (4%)
Query: 41 LDLPPEPVKKKLPTRLPI---ARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSY 97
L L P ++ + P+ R+G+G+ G + L TNHF V+V D F+QY+V+++
Sbjct: 7 LPLSPISIEPEQPSHRDYDITTRRGVGTTGNPIELCTNHFNVSVRQPDVVFYQYTVSITT 66
Query: 98 EDGRPVEGKGVGRKVIDKVQETYGSELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDV 157
E+G V+G G+ RK++D++ +TY S+L+GK AYDGEKTL+T+G L +N+ +F V++E
Sbjct: 67 ENGDAVDGTGISRKLMDQLFKTYSSDLDGKRLAYDGEKTLYTVGPLPQNEFDFLVIVEGS 126
Query: 158 ISNRNNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQES--E 215
S R+ G DG S++ + K R +R + +++KV+I +AA+IPL+ ++ RG + +
Sbjct: 127 FSKRDCG--VSDGGSSSGTCK-RSKRSFLPRSYKVQIHYAAEIPLKTVLGTQRGAYTPDK 183
Query: 216 NYQEAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSG 275
+ Q+A+RVLDI+LRQ AA++GCLLVRQ+FFH+D + VGGGV+G RG HSSFR T G
Sbjct: 184 SAQDALRVLDIVLRQQAAERGCLLVRQAFFHSDG-HPMKVGGGVIGIRGLHSSFRPTHGG 242
Query: 276 LSLNIDVSTTMIIQPGPVVDFLIANQNVRDPFSLDWAK-AKRTLKNLRIKASPSNQEYKI 334
LSLNIDVSTTMI++PGPV++FL ANQ+V P +DW K A + LK++R+KA+ N E+KI
Sbjct: 243 LSLNIDVSTTMILEPGPVIEFLKANQSVETPRQIDWIKVAAKMLKHMRVKATHRNMEFKI 302
Query: 335 TGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKP 394
GLS PC +Q F+MK K +GE + EITVY+YF + SA PC++VGKP
Sbjct: 303 IGLSSKPCNQQLFSMKIK---DGEREVPIREITVYDYFKQTYTEPIS-SAYFPCLDVGKP 358
Query: 395 KRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPM 454
RP Y+P+E C+LVSLQRYTK L+ QR LVE SRQKPLER+ LN A+ T Y +P
Sbjct: 359 DRPNYLPLEFCNLVSLQRYTKPLSGRQRVLLVESSRQKPLERIKTLNDAMHTYCYDKDPF 418
Query: 455 LKNCGITIASGFTQVEGRVLQAPRLKFGNGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVN 514
L CGI+I TQVEGRVL+ P LKFG EDF P NGRWN NNK ++ P I+ WA+VN
Sbjct: 419 LAGCGISIEKEMTQVEGRVLKPPMLKFGKNEDFQPCNGRWNFNNKMLLEPRAIKSWAIVN 478
Query: 515 FSARCDVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVRVEKMFERIQKELP 574
FS CD + R+LI C KGI ID P+ + EE+ Q+++A P+ RVEKM ++ + P
Sbjct: 479 FSFPCDSSHISRELISCGMRKGIEIDRPFA-LVEEDPQYKKAGPVERVEKMIATMKLKFP 537
Query: 575 GAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRVNDQYLTNVLMKINAKLGG 634
P F+LC+LPERK SD+YGPWKK L E GI TQCI P +++DQYLTNVL+KIN+KLGG
Sbjct: 538 DPPHFILCILPERKTSDIYGPWKKICLTEEGIHTQCICPIKISDQYLTNVLLKINSKLGG 597
Query: 635 LNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACV 694
+NS+LG+E + +IP+++K+PT+ILGMDVSHG PG++D+PS+AAVV S+ WPLIS+YRA V
Sbjct: 598 INSLLGIEYSYNIPLINKIPTLILGMDVSHGPPGRADVPSVAAVVGSKCWPLISRYRAAV 657
Query: 695 RTQSPKVEMIDNLFKQV--SEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFN 752
RTQSP++EMID+LF+ + +EK D GI+ EL ++FY +S RKP IIIFRDGVSESQF
Sbjct: 658 RTQSPRLEMIDSLFQPIENTEKGDNGIMNELFVEFYRTSRARKPKQIIIFRDGVSESQFE 717
Query: 753 QVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPDNVPPGTVIDNKICHP 812
QVL IE++QII+A + L E+ PKF VIVAQKNHHTK FQ P+NVP GTV+D KI HP
Sbjct: 718 QVLKIEVDQIIKAYQRLGESDVPKFTVIVAQKNHHTKLFQAKGPENVPAGTVVDTKIVHP 777
Query: 813 RNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAIS----- 867
N DFYMCAHAG IGTSRP HYHVLLD+IGFSPD+LQ L+HSLSY S +S
Sbjct: 778 TNYDFYMCAHAGKIGTSRPAHYHVLLDEIGFSPDDLQNLIHSLSYKLLNSIFNVSSLLCV 837
Query: 868 ---VVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGVAPVVPQLPKLQDSVSSSM 924
VAP+ YAHLAA Q+ QF KFE S+ VP+LP+L ++V +M
Sbjct: 838 FVLSVAPVRYAHLAAAQVAQFTKFEGISEDGK-------------VPELPRLHENVEGNM 884
Query: 925 FFC 927
FFC
Sbjct: 885 FFC 887
>At2g27880 Argonaute (AGO1)-like protein
Length = 997
Score = 525 bits (1351), Expect = e-149
Identities = 350/921 (38%), Positives = 499/921 (54%), Gaps = 84/921 (9%)
Query: 37 KVDLLDLPPEPVKKKLPTRLPIARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALS 96
K ++ LPP K P+ R G G+ G K+ + NHF V VA+ D + Y V+++
Sbjct: 131 KPEMTSLPPASSKA---VTFPV-RPGRGTLGKKVMVRANHFLVQVADRD--LYHYDVSIN 184
Query: 97 YEDGRPVEGKGVGRKVIDKVQETY-GSELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLE 155
E V K V R V+ + + Y S L GK AYDG K+L+T G L + EF V L
Sbjct: 185 PE----VISKTVNRNVMKLLVKNYKDSHLGGKSPAYDGRKSLYTAGPLPFDSKEFVVNLA 240
Query: 156 DVISNRNNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESE 215
+ + DG+S D + FKV + L + L ++ E
Sbjct: 241 EKRA---------DGSSGKD------------RPFKVAVKNVTSTDLYQLQQFLDRKQRE 279
Query: 216 NYQEAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLG-----CRGFHSSFR 270
+ I+VLD++LR + + V +SFFH A G G LG RG+ S R
Sbjct: 280 APYDTIQVLDVVLRDKPSND-YVSVGRSFFHTSLGKDARDGRGELGDGIEYWRGYFQSLR 338
Query: 271 TTQSGLSLNIDVSTTMIIQPGPVVDFLIANQNVRD---PF-SLDWAKAKRTLKNLRIKAS 326
TQ GLSLNIDVS +P V DF+ N+RD P D K K+ L+ L++K
Sbjct: 339 LTQMGLSLNIDVSARSFYEPIVVTDFISKFLNIRDLNRPLRDSDRLKVKKVLRTLKVKLL 398
Query: 327 PSN--QEYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSA 384
N + KI+G+S LP +E FT++ K E TV +YF ++Y A
Sbjct: 399 HWNCTKSAKISGISSLPIRELRFTLEDKS-----------EKTVVQYFAEKYNYRVKYQA 447
Query: 385 DLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQAL 444
LP I G RP Y+P+ELC + QRYTK L Q ++L++ + Q+P +R N + +
Sbjct: 448 -LPAIQTGSDTRPVYLPMELCQIDEGQRYTKRLNEKQVTALLKATCQRPPDRENSIKNLV 506
Query: 445 KTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKF---GNGEDFNPRNGRWNLNNKKV 501
+NY N+ + K G+++ + +E RVL P LK+ G + NPR G+WN+ +KK+
Sbjct: 507 VKNNY-NDDLSKEFGMSVTTQLASIEARVLPPPMLKYHDSGKEKMVNPRLGQWNMIDKKM 565
Query: 502 VRPAKIEHWAVVNFSARCDVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVR 561
V AK+ W V+FS R D RGL ++ C +L G+ + + E + F PP
Sbjct: 566 VNGAKVTSWTCVSFSTRID-RGLPQEF--CKQLIGMCVSKGMEFKPQPAIPFISCPP-EH 621
Query: 562 VEKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRVND--- 618
+E+ I K PG L+ +LP+ S YG K+ E GIV+QC P +VN
Sbjct: 622 IEEALLDIHKRAPGL-QLLIVILPDVTGS--YGKIKRICETELGIVSQCCQPRQVNKLNK 678
Query: 619 QYLTNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAV 678
QY+ NV +KIN K GG N+VL + +IP+++ PTII+G DV+H PG+ PSIAAV
Sbjct: 679 QYMENVALKINVKTGGRNTVLNDAIRRNIPLITDRPTIIMGADVTHPQPGEDSSPSIAAV 738
Query: 679 VSSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKE----DEGIIRELLIDFYSSSGKR 734
V+S +WP I+KYR V Q+ + E+I +L+K V + + G+IRE I F ++G+
Sbjct: 739 VASMDWPEINKYRGLVSAQAHREEIIQDLYKLVQDPQRGLVHSGLIREHFIAFRRATGQI 798
Query: 735 KPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFF--Q 792
P II +RDGVSE QF+QVL E+ I +AC L E + P+ ++ QK HHT+ F Q
Sbjct: 799 -PQRIIFYRDGVSEGQFSQVLLHEMTAIRKACNSLQENYVPRVTFVIVQKRHHTRLFPEQ 857
Query: 793 PGSPD------NVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPD 846
G+ D N+ PGTV+D KICHP DFY+ +HAG+ GTSRP HYHVLLD+ GF+ D
Sbjct: 858 HGNRDMTDKSGNIQPGTVVDTKICHPNEFDFYLNSHAGIQGTSRPAHYHVLLDENGFTAD 917
Query: 847 ELQELVHSLSYVYQRSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGV 906
+LQ L ++L Y Y R T ++S+V P YAHLAA + +M+ E SD SS + GV
Sbjct: 918 QLQMLTNNLCYTYARCTKSVSIVPPAYYAHLAAFRARYYMESE-MSDGGSSRSRSSTTGV 976
Query: 907 APVVPQLPKLQDSVSSSMFFC 927
V+ QLP ++D+V MF+C
Sbjct: 977 GQVISQLPAIKDNVKEVMFYC 997
>At5g43810 PINHEAD (gb|AAD40098.1); translation initiation factor
Length = 988
Score = 493 bits (1270), Expect = e-139
Identities = 322/902 (35%), Positives = 487/902 (53%), Gaps = 78/902 (8%)
Query: 60 RKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQET 119
R G G+ GTK + NHF + D + QY V ++ E V K V R +I ++
Sbjct: 131 RPGFGTLGTKCIVKANHFLADLPTKDLN--QYDVTITPE----VSSKSVNRAIIAELVRL 184
Query: 120 YG-SELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDK 178
Y S+L + AYDG K+L+T G L EF+V + D NG
Sbjct: 185 YKESDLGRRLPAYDGRKSLYTAGELPFTWKEFSVKIVDEDDGIING-------------- 230
Query: 179 KRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCL 238
P +++KV I F A+ + + L G+ ++ QEA+++LDI+LR+ + K+ C
Sbjct: 231 -----PKRERSYKVAIKFVARANMHHLGEFLAGKRADCPQEAVQILDIVLRELSVKRFCP 285
Query: 239 LVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDF-- 296
+ R SFF D K +G G+ GF+ S R TQ GLSLNID+++ I+P PV++F
Sbjct: 286 VGR-SFFSPDIKTPQRLGEGLESWCGFYQSIRPTQMGLSLNIDMASAAFIEPLPVIEFVA 344
Query: 297 -LIANQNVRDPFS-LDWAKAKRTLKNLRIKASPS---NQEYKITGLSELPCKEQTFTMKK 351
L+ + P S D K K+ L+ ++++ + ++Y++ GL+ P +E F +
Sbjct: 345 QLLGKDVLSKPLSDSDRVKIKKGLRGVKVEVTHRANVRRKYRVAGLTTQPTRELMFPV-- 402
Query: 352 KGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQ 411
+E+ T + +V EYF ++++ LPC+ VG K+ +Y+P+E C +V Q
Sbjct: 403 ------DENCTMK--SVIEYFQEMYGFTIQHT-HLPCLQVGNQKKASYLPMEACKIVEGQ 453
Query: 412 RYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEG 471
RYTK L Q ++L++ + Q+P +R N + + ++ + Y +P K G+ I+ VE
Sbjct: 454 RYTKRLNEKQITALLKVTCQRPRDRENDILRTVQHNAYDQDPYAKEFGMNISEKLASVEA 513
Query: 472 RVLQAPRLKF---GNGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARCD---VRGLV 525
R+L AP LK+ G +D P+ G+WN+ NKK++ + WA VNFS RG
Sbjct: 514 RILPAPWLKYHENGKEKDCLPQVGQWNMMNKKMINGMTVSRWACVNFSRSVQENVARGFC 573
Query: 526 RDLIKCARLKGIPID-EPYEEIFEEN-GQFRRAPPLVRVEKMFERIQKELPGAPSFLLCL 583
+L + + G+ + EP I+ Q +A V M + KEL LL +
Sbjct: 574 NELGQMCEVSGMEFNPEPVIPIYSARPDQVEKALKHVYHTSMNKTKGKEL----ELLLAI 629
Query: 584 LPERKNSDLYGPWKKKNLAEYGIVTQCISPT---RVNDQYLTNVLMKINAKLGGLNSVLG 640
LP+ N LYG K+ E G+++QC +++ QYL NV +KIN K+GG N+VL
Sbjct: 630 LPDN-NGSLYGDLKRICETELGLISQCCLTKHVFKISKQYLANVSLKINVKMGGRNTVLV 688
Query: 641 VEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPK 700
++ IP+VS +PTII G DV+H G+ PSIAAVV+S++WP ++KY V Q+ +
Sbjct: 689 DAISCRIPLVSDIPTIIFGADVTHPENGEESSPSIAAVVASQDWPEVTKYAGLVCAQAHR 748
Query: 701 VEMIDNLFKQ----VSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLN 756
E+I +L+K V G+IR+LLI F ++G+ KP II +RDGVSE QF QVL
Sbjct: 749 QELIQDLYKTWQDPVRGTVSGGMIRDLLISFRKATGQ-KPLRIIFYRDGVSEGQFYQVLL 807
Query: 757 IELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPD--------NVPPGTVIDNK 808
EL+ I +AC L+ + P IV QK HHT+ F D N+ PGTV+D K
Sbjct: 808 YELDAIRKACASLEPNYQPPVTFIVVQKRHHTRLFANNHRDKNSTDRSGNILPGTVVDTK 867
Query: 809 ICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISV 868
ICHP DFY+C+HAG+ GTSRP HYHVL D+ F+ D +Q L ++L Y Y R T ++S+
Sbjct: 868 ICHPTEFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADGIQSLTNNLCYTYARCTRSVSI 927
Query: 869 VAPICYAHLAATQIGQFMK---FEDKSDTSSSHGGLTAAGVAPVVPQLPKLQDSVSSSMF 925
V P YAHLAA + +++ +D + T G V P LP L+++V MF
Sbjct: 928 VPPAYYAHLAAFRARFYLEPEIMQDNGSPGKKNTKTTTVGDVGVKP-LPALKENVKRVMF 986
Query: 926 FC 927
+C
Sbjct: 987 YC 988
>At1g48410 Argonaute protein (AGO1)
Length = 870
Score = 479 bits (1233), Expect = e-135
Identities = 314/909 (34%), Positives = 478/909 (52%), Gaps = 81/909 (8%)
Query: 60 RKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQET 119
R G G G + + NHF + + D H Y V ++ E V +GV R V+ ++ +
Sbjct: 2 RPGKGQSGKRCIVKANHFFAELPDKDLH--HYDVTITPE----VTSRGVNRAVMKQLVDN 55
Query: 120 Y-GSELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDK 178
Y S L + AYDG K+L+T G L N EF + L D +
Sbjct: 56 YRDSHLGSRLPAYDGRKSLYTAGPLPFNSKEFRINLLD--------------EEVGAGGQ 101
Query: 179 KRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQ-HAAKQGC 237
+R R FKV I A+ L + L G++S+ QEA++VLDI+LR+ ++
Sbjct: 102 RRERE------FKVVIKLVARADLHHLGMFLEGKQSDAPQEALQVLDIVLRELPTSRIRY 155
Query: 238 LLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFL 297
+ V +SF+ D +G G+ RGF+ S R TQ GLSLNID+S+T I+ PV+ F+
Sbjct: 156 IPVGRSFYSPDIGKKQSLGDGLESWRGFYQSIRPTQMGLSLNIDMSSTAFIEANPVIQFV 215
Query: 298 --IANQNVRD-PFS-LDWAKAKRTLKNLRIKASPSN---QEYKITGLSELPCKEQTFTMK 350
+ N+++ P S D K K+ L+ ++++ + ++Y+I+GL+ + +E TF +
Sbjct: 216 CDLLNRDISSRPLSDADRVKIKKALRGVKVEVTHRGNMRRKYRISGLTAVATRELTFPV- 274
Query: 351 KKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSL 410
D + +V EYF ++++ LPC+ VG RP Y+P+E+C +V
Sbjct: 275 ---------DERNTQKSVVEYFHETYGFRIQHT-QLPCLQVGNSNRPNYLPMEVCKIVEG 324
Query: 411 QRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVE 470
QRY+K L Q ++L++ + Q+P++R + Q ++ ++Y + + GI I++ VE
Sbjct: 325 QRYSKRLNERQITALLKVTCQRPIDREKDILQTVQLNDYAKDNYAQEFGIKISTSLASVE 384
Query: 471 GRVLQAPRLKF---GNGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARCD---VRGL 524
R+L P LK+ G P+ G+WN+ NKK++ + +W +NFS + R
Sbjct: 385 ARILPPPWLKYHESGREGTCLPQVGQWNMMNKKMINGGTVNNWICINFSRQVQDNLARTF 444
Query: 525 VRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVRVEKMFERIQKELPGAPSFLLCLL 584
++L + + G+ + E + V + + K G LL ++
Sbjct: 445 CQELAQMCYVSGMAFNP--EPVLPPVSARPEQVEKVLKTRYHDATSKLSQGKEIDLLIVI 502
Query: 585 PERKNSDLYGPWKKKNLAEYGIVTQCISPTRV---NDQYLTNVLMKINAKLGGLNSVLGV 641
N LYG K+ E GIV+QC V + QY+ NV +KIN K+GG N+VL
Sbjct: 503 LPDNNGSLYGDLKRICETELGIVSQCCLTKHVFKMSKQYMANVALKINVKVGGRNTVLVD 562
Query: 642 EMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKV 701
++ IP+VS PTII G DV+H PG+ PSIAAVV+S++WP I+KY V Q+ +
Sbjct: 563 ALSRRIPLVSDRPTIIFGADVTHPHPGEDSSPSIAAVVASQDWPEITKYAGLVCAQAHRQ 622
Query: 702 EMIDNLFKQVSEKEDE----GIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNI 757
E+I +LFK+ + + G+I+ELLI F S+G KP II +RDGVSE QF QVL
Sbjct: 623 ELIQDLFKEWKDPQKGVVTGGMIKELLIAFRRSTG-HKPLRIIFYRDGVSEGQFYQVLLY 681
Query: 758 ELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPD--------NVPPGTVIDNKI 809
EL+ I +AC L+ + P +V QK HHT+ F D N+ PGTV+D+KI
Sbjct: 682 ELDAIRKACASLEAGYQPPVTFVVVQKRHHTRLFAQNHNDRHSVDRSGNILPGTVVDSKI 741
Query: 810 CHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISVV 869
CHP DFY+C+HAG+ GTSRP HYHVL D+ F+ D LQ L ++L Y Y R T ++S+V
Sbjct: 742 CHPTEFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADGLQSLTNNLCYTYARCTRSVSIV 801
Query: 870 APICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAG-----------VAPVVPQLPKLQD 918
P YAHLAA + +M+ E S + G + G V V LP L++
Sbjct: 802 PPAYYAHLAAFRARFYMEPETSDSGSMASGSMARGGGMAGRSTRGPNVNAAVRPLPALKE 861
Query: 919 SVSSSMFFC 927
+V MF+C
Sbjct: 862 NVKRVMFYC 870
>At1g69440 hypothetical protein
Length = 990
Score = 386 bits (992), Expect = e-107
Identities = 292/979 (29%), Positives = 476/979 (47%), Gaps = 137/979 (13%)
Query: 20 MPPPPPPPIVPADVEPVKVDLLDLPPEP----VKKKLPTRLPIARKGL------------ 63
+PPPPPP ++P + P LL LPP K LP + RK
Sbjct: 78 LPPPPPPHLLP--LSPPLPPLLPLPPPHSMTRFHKSLPVSQVVERKQQHQQKKKIQVSNN 135
Query: 64 ----------------------GSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGR 101
G G+ + LL NHF V +S + Y+V +S
Sbjct: 136 KVSGSIAIEEAALVVAKRPDFGGQDGSVIYLLANHFLVKFDSSQ-RIYHYNVEIS----- 189
Query: 102 PVEGKGVGRKVIDKVQETYGSELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNR 161
P K + R + K+ ET + +G A+DG + +++ ++LEF V L
Sbjct: 190 PQPSKEIARMIKQKLVETDRNSFSGVVPAFDGRQNIYSPVEFQGDRLEFFVNLP------ 243
Query: 162 NNGNCSPDGASTNDSDKKRMRRPYHS--KTFKVEISFAAKIPLQAIVNALRGQESENYQ- 218
P + + R ++P K F+V + +K + + +E E++
Sbjct: 244 -----IPSCKAVMNYGDLREKQPQKKIEKLFRVNMKLVSKFDGKE-----QRKEGEDWAP 293
Query: 219 ---EAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSG 275
E I LD+ILR++ ++ C + +SF+ + ++GGG +G RGF S R TQ G
Sbjct: 294 LPPEYIHALDVILRENPMEK-CTSIGRSFYSSSMGGSKEIGGGAVGLRGFFQSLRHTQQG 352
Query: 276 LSLNIDVSTTMIIQPGPVVDFL---------IANQNVRDPFSLDWAKAKRTLKNLRIKAS 326
L+LN+D+S T + V+ +L + R+ + + ++ LKN+R+
Sbjct: 353 LALNMDLSITAFHESIGVIAYLQKRLEFLTDLPRNKGRELSLEEKREVEKALKNIRVFVC 412
Query: 327 PSN--QEYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSA 384
Q Y++ GL+E + F D + + + YF ++ ++++
Sbjct: 413 HRETVQRYRVYGLTEEITENIWFP-----------DREGKYLRLMSYFKDHYGYEIQFK- 460
Query: 385 DLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQAL 444
+LPC+ + + RP Y+P+ELC + Q++ L+ Q + +++ QKP ER ++++ +
Sbjct: 461 NLPCLQISRA-RPCYLPMELCMICEGQKFLGKLSDDQAAKIMKMGCQKPNERKAIIDKVM 519
Query: 445 KTS---NYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKFGNGEDFNPRNGRWNLNNKKV 501
S + GN+ + + ++ T ++GR+LQ P+LK PRN L KV
Sbjct: 520 TGSVGPSSGNQT--REFNLEVSREMTLLKGRILQPPKLKLDR-----PRN----LKESKV 568
Query: 502 VRPAKIEHWAVVNFSARCDVRGLVRDLI-----KCARLKGIPIDEPYEEIFEENGQFRRA 556
+ +IE WA+++ D + + I KC L F E
Sbjct: 569 FKGTRIERWALMSIGGSSDQKSTIPKFINELTQKCEHLGVFLSKNTLSSTFFEPSHILNN 628
Query: 557 PPLVRVEKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCI---SP 613
L+ E + IQ+ ++C++ ++ YG K+ + G+VTQC +
Sbjct: 629 ISLL--ESKLKEIQRAASNNLQLIICVMEKKHKG--YGDLKRISETRIGVVTQCCLYPNI 684
Query: 614 TRVNDQYLTNVLMKINAKLGGLNSVLGVEMNPSIPIVSKV--PTIILGMDVSHGSPGQSD 671
T+++ Q+++N+ +KINAK+GG + L + IP + + P I +G DV+H P
Sbjct: 685 TKLSSQFVSNLALKINAKIGGSMTELYNSIPSHIPRLLRPDEPVIFMGADVTHPHPFDDC 744
Query: 672 IPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSS 731
PS+AAVV S WP ++Y + +R+Q+ + E+I +L + +++ELL DFY +
Sbjct: 745 SPSVAAVVGSINWPEANRYVSRMRSQTHRQEIIQDL---------DLMVKELLDDFYKAV 795
Query: 732 GKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEAC-KFLDETWNPKFLVIVAQKNHHTKF 790
K+ P+ II FRDGVSE+QF +VL EL I AC KF D +NP V QK HHT+
Sbjct: 796 -KKLPNRIIFFRDGVSETQFKKVLQEELQSIKTACSKFQD--YNPSITFAVVQKRHHTRL 852
Query: 791 FQPGSPD--NVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDEL 848
F+ PD N+PPGTV+D I HP+ DFY+C+H G+ GTSRPTHYH+L D+ F+ DEL
Sbjct: 853 FRC-DPDHENIPPGTVVDTVITHPKEFDFYLCSHLGVKGTSRPTHYHILWDENEFTSDEL 911
Query: 849 QELVHSLSYVYQRSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGVAP 908
Q LV++L Y + R T IS+V P YAHLAA + +++ +S+ S + + P
Sbjct: 912 QRLVYNLCYTFVRCTKPISIVPPAYYAHLAAYRGRLYIERSSESNGGSMNPSSVSRVGPP 971
Query: 909 VVPQLPKLQDSVSSSMFFC 927
LPKL D+V + MF+C
Sbjct: 972 KTIPLPKLSDNVKNLMFYC 990
>At1g31280 unknown protein
Length = 1013
Score = 326 bits (835), Expect = 4e-89
Identities = 268/904 (29%), Positives = 428/904 (46%), Gaps = 102/904 (11%)
Query: 15 GNEDLMPPPPPPPIVPADV--EPVKVDLLDLPPEPVKKKLPTRLPIARKGLGSKGT--KL 70
G + +P P V EPV+ ++++L P + P+ R G ++
Sbjct: 116 GRKPQVPSDSASPSTSTTVVSEPVRAEVMNLKPSVQVATSDRKEPMKRPDRGGVVAVRRV 175
Query: 71 PLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGR----KVIDKVQETYGSELNG 126
L NH+KV N + Y V + E + K V R V DKV E
Sbjct: 176 NLYVNHYKVNF-NPESVIRHYDVEIKGE----IPTKKVSRFELAMVRDKVFTDNPDEFPL 230
Query: 127 KDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDKKRMRRPYH 186
AYDG+K +F+ L P G+ + K R
Sbjct: 231 AMTAYDGQKNIFSAVEL------------------------PTGSYKVEYPKTEEMR--- 263
Query: 187 SKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCLL-VRQSFF 245
+++ I + L + + G+ S N ++ ++ +D+++++H +K C++ V +SFF
Sbjct: 264 GRSYTFTIKQVNVLKLGDLKEYMTGRSSFNPRDVLQGMDVVMKEHPSK--CMITVGKSFF 321
Query: 246 HNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFLIANQNVRD 305
+ + D GV+ +G+ + + T GLSL +D S + V+++L N D
Sbjct: 322 TRETEPDEDFRFGVIAAKGYRHTLKPTAQGLSLCLDYSVLAFRKAMSVIEYLKLYFNWSD 381
Query: 306 PFSLDWAKAKRTLKNLRIKASPSNQEYKIT--GLSELPCKEQTFTMKKKGGNNGEEDATE 363
+ L L++ + + K+T GLS K+ F + + GN
Sbjct: 382 MRQFRRRDVEEELIGLKVTVNHRKNKQKLTIVGLSMQNTKDIKFDLIDQEGNE-----PP 436
Query: 364 EEITVYEYF-VNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQR 422
+ ++ EYF + Y + + D+PC+++GK R +VP+E C LV Q Y K L +
Sbjct: 437 RKTSIVEYFRIKYGRHIVH--KDIPCLDLGKNGRQNFVPMEFCDLVEGQIYPK--DNLDK 492
Query: 423 SS---LVEKSRQKPLERMNVLNQALKTSN--YGNEPMLKNCGITIASGFTQVEGRVLQAP 477
S L + S P +R +++ +K N G E ++ N G+ + + T VEGRVL+AP
Sbjct: 493 DSALWLKKLSLVNPQQRQRNIDKMIKARNGPSGGE-IIGNFGLKVDTNMTPVEGRVLKAP 551
Query: 478 RLKFGNG-----EDFNPR-NGRWNLNNKKVVRPAKIEHWAVVNFSARCDVRGLVRDLI-- 529
LK E+ NPR N +WNL K V R + ++HWAV++F+A + D +
Sbjct: 552 SLKLAERGRVVREEPNPRQNNQWNLMKKGVTRGSIVKHWAVLDFTASERFNKMPNDFVDN 611
Query: 530 ---KCARLKGIPIDEP--YEEIFEENGQFRRAPPLVRVEKMFERIQKELPGAPSFLLCLL 584
+C RL G+ ++ P Y+ E A + + E +K P+ +LC +
Sbjct: 612 LIDRCWRL-GMQMEAPIVYKTSRMETLSNGNAIEELLRSVIDEASRKHGGARPTLVLCAM 670
Query: 585 PERKNSDLYGPWKKKNLAEYGIVTQCI---SPTRVNDQYLTNVLMKINAKLGGLNSVLGV 641
+ D Y K + G+VTQC T+ DQY N+ +K+NAK+GG N V
Sbjct: 671 SRK--DDGYKTLKWIAETKLGLVTQCFLTGPATKGGDQYRANLALKMNAKVGGSN----V 724
Query: 642 EMNPSIPIVSKVPTII-LGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPK 700
E+ + K ++ +G DV+H + PSI AVV + WP ++Y A V Q +
Sbjct: 725 ELMDTFSFFKKEDEVMFIGADVNHPAARDKMSPSIVAVVGTLNWPEANRYAARVIAQPHR 784
Query: 701 VEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELN 760
E I EL+ ++GKR P+ I+IFRDGVS++QF+ VLN+EL
Sbjct: 785 KEEIQGF---------GDACLELVKAHVQATGKR-PNKIVIFRDGVSDAQFDMVLNVELL 834
Query: 761 QIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPD-----NVPPGTVIDNKICHPRNN 815
+ F +NPK VIVAQK H T+FF + D NVP GTV+D K+ HP
Sbjct: 835 DV--KLTFEKNGYNPKITVIVAQKRHQTRFFPATNNDGSDKGNVPSGTVVDTKVIHPYEY 892
Query: 816 DFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISVVAPICYA 875
DFY+C+H G IGTS+PTHY+ L D++GF+ D++Q+L+ + + + R T +S+V P+ YA
Sbjct: 893 DFYLCSHHGGIGTSKPTHYYTLWDELGFTSDQVQKLIFEMCFTFTRCTKPVSLVPPVYYA 952
Query: 876 HLAA 879
+ A
Sbjct: 953 DMVA 956
>At1g31290 hypothetical protein
Length = 1194
Score = 320 bits (821), Expect = 2e-87
Identities = 274/942 (29%), Positives = 452/942 (47%), Gaps = 133/942 (14%)
Query: 45 PEPVKKKLPTRLPIARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVE 104
P KK P + P + KG + L NHF+V+ + ++ Y V + E+
Sbjct: 325 PSSSDKKEPVKRPDKGGNIKVKGV-INLSVNHFRVSFS-TESVIRHYDVDIKGENS---- 378
Query: 105 GKGVGRKVIDKVQETYGSELNGKDF-----AYDGEKTLFTIGSLARNKLEFTVVLEDVIS 159
K + R + V+E + N DF AYDG+K +F+ L
Sbjct: 379 SKKISRFELAMVKEKLFKDNN--DFPNAMTAYDGQKNIFSAVELP--------------- 421
Query: 160 NRNNGNCSPDGASTNDSDKKRMRRPYHSKTFKV-EISFAAKIPLQAIVNALRGQESENYQ 218
G+ D + T + + R S TF + ++ + LQA ++ G+ + +
Sbjct: 422 ---TGSFKVDFSETEEIMRGR------SYTFIIKQVKELKLLDLQAYID---GRSTFIPR 469
Query: 219 EAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSL 278
+ ++ +D+++++H +K+ + V + FF + D G GV +GFH + + T GLSL
Sbjct: 470 DVLQGMDVVMKEHPSKR-MITVGKRFFST--RLEIDFGYGVGAAKGFHHTLKPTVQGLSL 526
Query: 279 NIDVSTTMIIQPGPVVDFL---IANQNVRDPFSLDWAKAKRTLKNLRIKAS--PSNQEYK 333
++ S + V+++L +N+R + + L L++ + Q++
Sbjct: 527 CLNSSLLAFRKAISVIEYLKLYFGWRNIRQFKNCRPDDVVQELIGLKVTVDHRKTKQKFI 586
Query: 334 ITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGK 393
I GLS+ K+ F GN +I++ EYF D+ + D+PC+N+GK
Sbjct: 587 IMGLSKDDTKDIKFDFIDHAGNQ-----PPRKISIVEYFKEKYGRDIDHK-DIPCLNLGK 640
Query: 394 PKRPTYVPVELCSLVSLQRYTKALTTLQRSS---LVEKSRQKPLERMNVLNQALKTSN-- 448
R +VP+E C+LV Q + K L R S L E S P +R+ +N+ +K+S+
Sbjct: 641 KGRENFVPMEFCNLVEGQIFPKE--KLYRDSAAWLKELSLVTPQQRLENINKMIKSSDGP 698
Query: 449 YGNEPMLKNCGITIASGFTQVEGRVLQAPRLKF----GNG--EDFNPRNGRWNLNNKKVV 502
G + ++ N G+ + T VEGRVL+AP LK GN E + +WNL K V
Sbjct: 699 RGGD-IIGNFGLRVDPNMTTVEGRVLEAPTLKLTDRRGNPIHEKLMSESNQWNLTTKGVT 757
Query: 503 RPAKIEHWAVVNFSARCDVRG-----LVRDLIKCARLKGIPIDEPY-------EEIFEEN 550
+ + I+HWAV++F+A ++ V LI+ + G+ ++ P E +++ N
Sbjct: 758 KGSIIKHWAVLDFTASESLKKKMPGYFVNKLIERCKGLGMQMEAPIVCKTSSMETLYDGN 817
Query: 551 GQFRRAPPLVRVEKMFERIQKELP-----GAPSFLLCLLPERKNSDLYGPWKKKNLAEYG 605
+E++ + E P+ +LC + + D Y K + G
Sbjct: 818 A----------LEELLRSVIDEASHNHGGACPTLVLCAMTGKH--DGYKTLKWIAETKLG 865
Query: 606 IVTQC------ISPTRVNDQYLTNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILG 659
+VTQC I V+DQYL N+ +KINAK+GG N L + KV + +G
Sbjct: 866 LVTQCFLTISAIKGETVSDQYLANLALKINAKVGGTNVELVDNIFSFFKKEDKV--MFIG 923
Query: 660 MDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGI 719
DV+H + + PSI AVV + WP ++Y A V+ QS + E I + E
Sbjct: 924 ADVNHPAAHDNMSPSIVAVVGTLNWPEANRYAARVKAQSHRKEEIQGFGETCWE------ 977
Query: 720 IRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLV 779
LI+ +S + +++P+ I+IFRDGVS+ QF+ VLN+EL + + F +NP+ V
Sbjct: 978 ----LIEAHSQAPEKRPNKIVIFRDGVSDGQFDMVLNVELQNVKDV--FAKVGYNPQITV 1031
Query: 780 IVAQKNHHTKFFQPGSPD------NVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTH 833
IVAQK H T+FF + NVP GTV+D I HP DFY+C+ G IGTS+PTH
Sbjct: 1032 IVAQKRHQTRFFPATTSKDGRAKGNVPSGTVVDTTIIHPFEYDFYLCSQHGAIGTSKPTH 1091
Query: 834 YHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISVVAPICYAHLAATQ-----IGQFMKF 888
Y+VL D+IGF+ +++Q+L+ L + + R T +++V P+ YA AA++ MK
Sbjct: 1092 YYVLSDEIGFNSNQIQKLIFDLCFTFTRCTKPVALVPPVSYADKAASRGRVYYEASLMKK 1151
Query: 889 EDKSDTSSSHGGLTAAGVAPVV----PQLPKLQDSVSSSMFF 926
K +S + A + V ++ K+ + + MFF
Sbjct: 1152 NSKQSRGASSSSASVASSSSSVTMEDKEIFKVHAGIENFMFF 1193
>At4g03390 SRF3 (LRR receptor-like protein kinase like protein)
Length = 776
Score = 40.4 bits (93), Expect = 0.005
Identities = 20/38 (52%), Positives = 22/38 (57%)
Query: 21 PPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLPI 58
PPPPPPP P E V V + P PVKK P RLP+
Sbjct: 430 PPPPPPPPPPPLDEKVTVMPIISPERPVKKTSPKRLPL 467
>At3g19430 putative late embryogenesis abundant protein
Length = 550
Score = 38.5 bits (88), Expect = 0.018
Identities = 17/37 (45%), Positives = 19/37 (50%)
Query: 21 PPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLP 57
PPP P P VP+ PV D + PP PV PT P
Sbjct: 154 PPPTPTPSVPSPTPPVPTDPMPSPPPPVSPPPPTPTP 190
Score = 32.0 bits (71), Expect = 1.7
Identities = 18/57 (31%), Positives = 26/57 (45%), Gaps = 6/57 (10%)
Query: 3 PNEHENGNGNGNGNED--LMPPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLP 57
P+ ++G G+ +G +D PP P PP+ P P + P PV PT P
Sbjct: 54 PSPGDDGGGDDSGGDDGGYTPPAPVPPVSP----PPPTPSVPSPTPPVSPPPPTPTP 106
>At1g67770 terminal ear1-like protein
Length = 527
Score = 35.4 bits (80), Expect = 0.15
Identities = 18/50 (36%), Positives = 23/50 (46%), Gaps = 2/50 (4%)
Query: 4 NEHENGNGNGNGNEDLMPPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLP 53
N+H+N N + +P PPPPP P P+ LPP P LP
Sbjct: 25 NQHQNQNPSLIPTRFFLPHPPPPP--PPPPPPLYFSYFSLPPPPPPPHLP 72
Score = 30.0 bits (66), Expect = 6.3
Identities = 14/27 (51%), Positives = 17/27 (62%), Gaps = 1/27 (3%)
Query: 19 LMPPPPPPPIVPADVEPVK-VDLLDLP 44
L PPPPPP + P V P + V LL +P
Sbjct: 62 LPPPPPPPHLPPTSVTPTRAVMLLQVP 88
>At5g57070 putative protein
Length = 607
Score = 33.9 bits (76), Expect = 0.44
Identities = 16/34 (47%), Positives = 20/34 (58%), Gaps = 2/34 (5%)
Query: 4 NEHENGNGNGNGNEDLMPPPPPPPIVPADVEPVK 37
N +E NG G+ + PPPPPPP P V P+K
Sbjct: 456 NFNEENNGQGSPLIQITPPPPPPP--PFRVPPLK 487
>At5g61270 bHLH transcription factor like protein
Length = 366
Score = 33.5 bits (75), Expect = 0.57
Identities = 12/19 (63%), Positives = 15/19 (78%)
Query: 11 GNGNGNEDLMPPPPPPPIV 29
G GNG L+PPPPPPP++
Sbjct: 281 GGGNGYGGLVPPPPPPPMM 299
>At4g14750 unknown protein
Length = 387
Score = 33.5 bits (75), Expect = 0.57
Identities = 14/33 (42%), Positives = 20/33 (60%)
Query: 17 EDLMPPPPPPPIVPADVEPVKVDLLDLPPEPVK 49
+D PPPPPPP P +P V+++D E +K
Sbjct: 64 KDSPPPPPPPPPPPPLQQPFVVEIVDNEDEQIK 96
>At5g58160 strong similarity to unknown protein (gb|AAD23008.1)
Length = 1307
Score = 32.7 bits (73), Expect = 0.97
Identities = 15/37 (40%), Positives = 17/37 (45%)
Query: 21 PPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLP 57
PPPPPPP PA P + + P P RLP
Sbjct: 728 PPPPPPPPPPAPPTPQSNGISAMKSSPPAPPAPPRLP 764
>At2g02070 putative C2H2-type zinc finger protein
Length = 602
Score = 32.7 bits (73), Expect = 0.97
Identities = 22/68 (32%), Positives = 30/68 (43%), Gaps = 6/68 (8%)
Query: 16 NEDLMPPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLPIARKGLGSKGTKLPLLTN 75
N PPPPPP A + P L+ PP+ K+ P R P + + + K + TN
Sbjct: 26 NSSAAAPPPPPPHHQAPLPP-----LEAPPQKKKRNQP-RTPNSDAEVIALSPKTLMATN 79
Query: 76 HFKVTVAN 83
F V N
Sbjct: 80 RFICEVCN 87
>At4g30150 hypothetical protein
Length = 1966
Score = 32.3 bits (72), Expect = 1.3
Identities = 21/65 (32%), Positives = 31/65 (47%), Gaps = 5/65 (7%)
Query: 89 FQYSVALSYEDGRPV-----EGKGVGRKVIDKVQETYGSELNGKDFAYDGEKTLFTIGSL 143
+Q+S AL + D + + EG G +I+ + E + LN + EKT F SL
Sbjct: 1323 YQFSKALPFSDEKLISSEISEGTGQANLIIENLTEQAETLLNALRATFRDEKTAFKCESL 1382
Query: 144 ARNKL 148
NKL
Sbjct: 1383 ILNKL 1387
>At1g59910 hypothetical protein
Length = 929
Score = 32.3 bits (72), Expect = 1.3
Identities = 20/52 (38%), Positives = 22/52 (41%), Gaps = 5/52 (9%)
Query: 21 PPPPPPP-----IVPADVEPVKVDLLDLPPEPVKKKLPTRLPIARKGLGSKG 67
PPPPPPP P P K PP P+ KK P + P KG G
Sbjct: 395 PPPPPPPKKGPAAPPPPPPPGKKGAGPPPPPPMSKKGPPKPPGNPKGPTKSG 446
Score = 30.4 bits (67), Expect = 4.8
Identities = 19/48 (39%), Positives = 22/48 (45%), Gaps = 6/48 (12%)
Query: 21 PPPPPPPIV---PADVEPVKVDLLDLPPEPVKKK---LPTRLPIARKG 62
PPPPPPP P P K PP P KK P P+++KG
Sbjct: 384 PPPPPPPSAAAPPPPPPPKKGPAAPPPPPPPGKKGAGPPPPPPMSKKG 431
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,515,606
Number of Sequences: 26719
Number of extensions: 1077134
Number of successful extensions: 5757
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 4932
Number of HSP's gapped (non-prelim): 325
length of query: 927
length of database: 11,318,596
effective HSP length: 108
effective length of query: 819
effective length of database: 8,432,944
effective search space: 6906581136
effective search space used: 6906581136
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0031.13


