

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0030.6
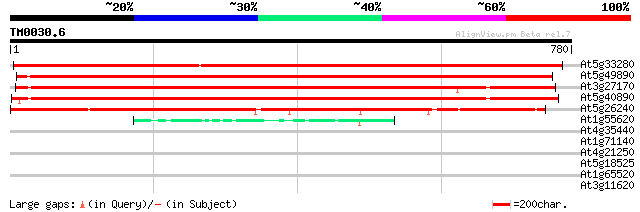
(780 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g33280 chloride channel-like protein 1148 0.0
At5g49890 chloride channel (emb|CAA70310.1) 946 0.0
At3g27170 CLC-b chloride channel protein 822 0.0
At5g40890 anion channel protein (gb|AAC05742.1) 815 0.0
At5g26240 CLC-d chloride channel protein 640 0.0
At1g55620 CLC-f chloride channel protein 49 1e-05
At4g35440 CLC-e chloride channel protein 37 0.042
At1g71140 hypothetical protein 32 1.8
At4g21250 hypothetical protein 31 2.3
At5g18525 unknown protein 30 4.0
At1g65520 hypothetical protein 30 5.2
At3g11620 unknown protein 29 8.8
>At5g33280 chloride channel-like protein
Length = 763
Score = 1148 bits (2970), Expect = 0.0
Identities = 562/763 (73%), Positives = 654/763 (85%), Gaps = 2/763 (0%)
Query: 6 LSNGDSEHRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGS 65
+ N +E + PLL S R NS+SQVAIVGANVCPIESLDYEI EN+FFKQDWR R
Sbjct: 1 MPNSTTEDSVAVPLLPSLRRATNSTSQVAIVGANVCPIESLDYEIAENDFFKQDWRGRSK 60
Query: 66 LQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSLAFVTFF 125
++IFQYV MKWLLCF IG I+ LIGF NNLAVENLAGVKFVVTSNMM+ RF++ FV F
Sbjct: 61 VEIFQYVFMKWLLCFCIGIIVSLIGFANNLAVENLAGVKFVVTSNMMIAGRFAMGFVVFS 120
Query: 126 ASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVSS 185
+NL LTLFA++ITA +APAAAGSGI EVKAYLNGVDAP IF+L TLI+KIIG+I+AVS+
Sbjct: 121 VTNLILTLFASVITAFVAPAAAGSGIPEVKAYLNGVDAPEIFSLRTLIIKIIGNISAVSA 180
Query: 186 SLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAAA 245
SL IGKAGPM+HTGACVA++LGQGGSKRY LTW+WLR+FKNDRDRRDL+ CG+AAGIAA+
Sbjct: 181 SLLIGKAGPMVHTGACVASILGQGGSKRYRLTWRWLRFFKNDRDRRDLVTCGAAAGIAAS 240
Query: 246 FRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLIM 305
FRAPVGGVLFALEEM+SW +ALLWR FF+TA+VAI LRA+IDVCLSGKCGLFGKGGLIM
Sbjct: 241 FRAPVGGVLFALEEMSSW--SALLWRIFFSTAVVAIVLRALIDVCLSGKCGLFGKGGLIM 298
Query: 306 FDAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLACV 365
FD YS + YH DV V +LGV+GGILGSLYN+L +KVLR YN I EKG KILLAC
Sbjct: 299 FDVYSENASYHLGDVLPVLLLGVVGGILGSLYNFLLDKVLRAYNYIYEKGVTWKILLACA 358
Query: 366 ISMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDLASLIFNTND 425
IS+FTSCLLFGLP+LASCQPCP D +E CPTIGRSG +KK+QCPPGHYNDLASLIFNTND
Sbjct: 359 ISIFTSCLLFGLPFLASCQPCPVDALEECPTIGRSGNFKKYQCPPGHYNDLASLIFNTND 418
Query: 426 DAIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGM 485
DAI+NLFSKNTD EF Y S+ +FF+T FFLSI S G+V PAG+FVP+IVTGASYGRFVGM
Sbjct: 419 DAIKNLFSKNTDFEFHYFSVLVFFVTCFFLSIFSYGIVAPAGLFVPVIVTGASYGRFVGM 478
Query: 486 LFGKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVA 545
L G SNLNHGL+AVLGAASFLGG+MR TVS CVI+LELTNNLLLLP++M+VLL+SK+VA
Sbjct: 479 LLGSNSNLNHGLFAVLGAASFLGGTMRMTVSTCVILLELTNNLLLLPMMMVVLLISKTVA 538
Query: 546 DVFNANIYDLIMKSKGLPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTT 605
D FNANIY+LIMK KG PYL +HAEPYMRQL VGDVVTGPLQ+F+G EKV +V VLKTT
Sbjct: 539 DGFNANIYNLIMKLKGFPYLYSHAEPYMRQLLVGDVVTGPLQVFNGIEKVETIVHVLKTT 598
Query: 606 SHNGFPVIDEPPFSEAPVLFGIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKR 665
+HNGFPV+D PP + APVL G+ILR H+LTLL+K+VFM +P+A + L QF A++FAK+
Sbjct: 599 NHNGFPVVDGPPLAAAPVLHGLILRAHILTLLKKRVFMPSPVACDSNTLSQFKAEEFAKK 658
Query: 666 GSDRGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIP 725
GS R ++IED+ L++EE++M++DLHPF+NASPYTVVE+MSL KALILFRE+G+RHLLVIP
Sbjct: 659 GSGRSDKIEDVELSEEELNMYLDLHPFSNASPYTVVETMSLAKALILFREVGIRHLLVIP 718
Query: 726 KIPSRSPVVGILTRHDFTAEHILGTHPYLVRSRWKRLRFWQPF 768
K +R PVVGILTRHDF EHILG HP + RS+WKRLR PF
Sbjct: 719 KTSNRPPVVGILTRHDFMPEHILGLHPSVSRSKWKRLRIRLPF 761
>At5g49890 chloride channel (emb|CAA70310.1)
Length = 779
Score = 946 bits (2446), Expect = 0.0
Identities = 460/746 (61%), Positives = 563/746 (74%), Gaps = 3/746 (0%)
Query: 10 DSEHRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGSLQIF 69
D RQPLL+ R N++SQ+AIVGAN CPIESLDYEIFEN+FFKQDWRSR ++I
Sbjct: 32 DGSVGFRQPLLARNRK--NTTSQIAIVGANTCPIESLDYEIFENDFFKQDWRSRKKIEIL 89
Query: 70 QYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSLAFVTFFASNL 129
QY +KW L FLIG GL+GF NNL VEN+AG K ++ N+ML +++ AF F NL
Sbjct: 90 QYTFLKWALAFLIGLATGLVGFLNNLGVENIAGFKLLLIGNLMLKEKYFQAFFAFAGCNL 149
Query: 130 ALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVSSSLHI 189
L A + A IAPAAAGSGI EVKAYLNG+DA I TL VKI GSI V++ +
Sbjct: 150 ILATAAASLCAFIAPAAAGSGIPEVKAYLNGIDAYSILAPSTLFVKIFGSIFGVAAGFVV 209
Query: 190 GKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAAAFRAP 249
GK GPM+HTGAC+A LLGQGGSK+Y LTWKWLR+FKNDRDRRDLI CG+AAG+AAAFRAP
Sbjct: 210 GKEGPMVHTGACIANLLGQGGSKKYRLTWKWLRFFKNDRDRRDLITCGAAAGVAAAFRAP 269
Query: 250 VGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLIMFDAY 309
VGGVLFALEE ASWWR ALLWR FFTTA+VA+ LR++I+ C SG+CGLFGKGGLIMFD
Sbjct: 270 VGGVLFALEEAASWWRNALLWRTFFTTAVVAVVLRSLIEFCRSGRCGLFGKGGLIMFDVN 329
Query: 310 SGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLACVISMF 369
SG ++Y D+ + LGVIGG+LGSLYNYL +KVLR Y+ INEKG KI+L +S+
Sbjct: 330 SGPVLYSTPDLLAIVFLGVIGGVLGSLYNYLVDKVLRTYSIINEKGPRFKIMLVMAVSIL 389
Query: 370 TSCLLFGLPWLASCQPCPADPVE-PCPTIGRSGIYKKFQCPPGHYNDLASLIFNTNDDAI 428
+SC FGLPWL+ C PCP E CP++GRS IYK FQCPP HYNDL+SL+ NTNDDAI
Sbjct: 390 SSCCAFGLPWLSQCTPCPIGIEEGKCPSVGRSSIYKSFQCPPNHYNDLSSLLLNTNDDAI 449
Query: 429 RNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGMLFG 488
RNLF+ +++EF S++ IFF+ + L I++ G+ +P+G+F+P+I+ GASYGR VG L G
Sbjct: 450 RNLFTSRSENEFHISTLAIFFVAVYCLGIITYGIAIPSGLFIPVILAGASYGRLVGRLLG 509
Query: 489 KKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVF 548
S L+ GL+++LGAASFLGG+MR TVSLCVI+LELTNNLL+LPL+M+VLL+SK+VAD F
Sbjct: 510 PVSQLDVGLFSLLGAASFLGGTMRMTVSLCVILLELTNNLLMLPLVMLVLLISKTVADCF 569
Query: 549 NANIYDLIMKSKGLPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTTSHN 608
N +YD I+ KGLPY+E HAEPYMR L DVV+G L F EKV + LK T HN
Sbjct: 570 NRGVYDQIVTMKGLPYMEDHAEPYMRNLVAKDVVSGALISFSRVEKVGVIWQALKMTRHN 629
Query: 609 GFPVIDEPPFSEAPVLFGIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSD 668
GFPVIDEPPF+EA L GI LR HLL LL+ K F G +LR A DF K G
Sbjct: 630 GFPVIDEPPFTEASELCGIALRSHLLVLLQGKKFSKQRTTFGSQILRSCKARDFGKAGLG 689
Query: 669 RGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIP 728
+G +IED+ L++EEM+M++DLHP TN SPYTV+E++SL KA ILFR+LGLRHL V+PK P
Sbjct: 690 KGLKIEDLDLSEEEMEMYVDLHPITNTSPYTVLETLSLAKAAILFRQLGLRHLCVVPKTP 749
Query: 729 SRSPVVGILTRHDFTAEHILGTHPYL 754
R P+VGILTRHDF EH+LG +P++
Sbjct: 750 GRPPIVGILTRHDFMPEHVLGLYPHI 775
>At3g27170 CLC-b chloride channel protein
Length = 780
Score = 822 bits (2122), Expect = 0.0
Identities = 409/762 (53%), Positives = 546/762 (70%), Gaps = 18/762 (2%)
Query: 9 GDSE-HRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGSLQ 67
GD E + L QPL+ + R++ SS+ +A+VGA V IESLDYEI EN+ FK DWR R Q
Sbjct: 20 GDPESNTLNQPLVKANRTL--SSTPLALVGAKVSHIESLDYEINENDLFKHDWRKRSKAQ 77
Query: 68 IFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSLAFVTFFAS 127
+ QYV +KW L L+G GLI NLAVEN+AG K + + + +R+ + +
Sbjct: 78 VLQYVFLKWTLACLVGLFTGLIATLINLAVENIAGYKLLAVGHFLTQERYVTGLMVLVGA 137
Query: 128 NLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVSSSL 187
NL LTL A+++ AP AAG GI E+KAYLNGVD P +F T+IVKI+GSI AV++ L
Sbjct: 138 NLGLTLVASVLCVCFAPTAAGPGIPEIKAYLNGVDTPNMFGATTMIVKIVGSIGAVAAGL 197
Query: 188 HIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAAAFR 247
+GK GP++H G+C+A+LLGQGG+ + + W+WLRYF NDRDRRDLI CGSAAG+ AAFR
Sbjct: 198 DLGKEGPLVHIGSCIASLLGQGGTDNHRIKWRWLRYFNNDRDRRDLITCGSAAGVCAAFR 257
Query: 248 APVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLIMFD 307
+PVGGVLFALEE+A+WWR+ALLWR FF+TA+V + LR I++C SGKCGLFGKGGLIMFD
Sbjct: 258 SPVGGVLFALEEVATWWRSALLWRTFFSTAVVVVVLREFIEICNSGKCGLFGKGGLIMFD 317
Query: 308 AYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLACVIS 367
+ YH D+ V ++GVIGGILGSLYN+L +KVLR+YN INEKG I K+LL+ +S
Sbjct: 318 VSHVTYTYHVTDIIPVMLIGVIGGILGSLYNHLLHKVLRLYNLINEKGKIHKVLLSLTVS 377
Query: 368 MFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDLASLIFNTNDDA 427
+FTS L+GLP+LA C+PC E CPT GRSG +K+F CP G+YNDLA+L+ TNDDA
Sbjct: 378 LFTSVCLYGLPFLAKCKPCDPSIDEICPTNGRSGNFKQFHCPKGYYNDLATLLLTTNDDA 437
Query: 428 IRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGMLF 487
+RNLFS NT +EF S++IFF+ L + + G+ P+G+F+PII+ GA+YGR +G
Sbjct: 438 VRNLFSSNTPNEFGMGSLWIFFVLYCILGLFTFGIATPSGLFLPIILMGAAYGRMLGAAM 497
Query: 488 GKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADV 547
G ++++ GLYAVLGAA+ + GSMR TVSLCVI LELTNNLLLLP+ M+VLL++K+V D
Sbjct: 498 GSYTSIDQGLYAVLGAAALMAGSMRMTVSLCVIFLELTNNLLLLPITMIVLLIAKTVGDS 557
Query: 548 FNANIYDLIMKSKGLPYLETHAEPYMRQLTVGDV--VTGPLQIFHGYEKVRNVVFVLKTT 605
FN +IYD+I+ KGLP+LE + EP+MR LTVG++ P+ G EKV N+V VLK T
Sbjct: 558 FNPSIYDIILHLKGLPFLEANPEPWMRNLTVGELGDAKPPVVTLQGVEKVSNIVDVLKNT 617
Query: 606 SHNGFPVIDEPPFSE------APVLFGIILRHHLLTLLRKKVFMSAP-MAMGGDVLRQFS 658
+HN FPV+DE + A L G+ILR HL+ +L+K+ F++ +V +F
Sbjct: 618 THNAFPVLDEAEVPQVGLATGATELHGLILRAHLVKVLKKRWFLTEKRRTEEWEVREKFP 677
Query: 659 ADDFAKRGSDRGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGL 718
D+ A +R + +D+ +T EM+M++DLHP TN +PYTV+E+MS+ KAL+LFR++GL
Sbjct: 678 WDELA----EREDNFDDVAITSAEMEMYVDLHPLTNTTPYTVMENMSVAKALVLFRQVGL 733
Query: 719 RHLLVIPKIPSRS--PVVGILTRHDFTAEHILGTHPYLVRSR 758
RHLL++PKI + PVVGILTR D A +IL P L +S+
Sbjct: 734 RHLLIVPKIQASGMCPVVGILTRQDLRAYNILQAFPLLEKSK 775
>At5g40890 anion channel protein (gb|AAC05742.1)
Length = 775
Score = 815 bits (2106), Expect = 0.0
Identities = 407/772 (52%), Positives = 547/772 (70%), Gaps = 17/772 (2%)
Query: 3 TNHLSNGDSE------HRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFF 56
+N NG+ E + L QPLL R++ SS+ +A+VGA V IESLDYEI EN+ F
Sbjct: 10 SNSNYNGEEEGEDPENNTLNQPLLKRHRTL--SSTPLALVGAKVSHIESLDYEINENDLF 67
Query: 57 KQDWRSRGSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKR 116
K DWRSR Q+FQY+ +KW L L+G GLI NLAVEN+AG K + + R
Sbjct: 68 KHDWRSRSKAQVFQYIFLKWTLACLVGLFTGLIATLINLAVENIAGYKLLAVGYYIAQDR 127
Query: 117 FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKI 176
F + F +NL LTL AT++ AP AAG GI E+KAYLNG+D P +F T++VKI
Sbjct: 128 FWTGLMVFTGANLGLTLVATVLVVYFAPTAAGPGIPEIKAYLNGIDTPNMFGFTTMMVKI 187
Query: 177 IGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVC 236
+GSI AV++ L +GK GP++H G+C+A+LLGQGG + + W+WLRYF NDRDRRDLI C
Sbjct: 188 VGSIGAVAAGLDLGKEGPLVHIGSCIASLLGQGGPDNHRIKWRWLRYFNNDRDRRDLITC 247
Query: 237 GSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCG 296
GSA+G+ AAFR+PVGGVLFALEE+A+WWR+ALLWR FF+TA+V + LRA I++C SGKCG
Sbjct: 248 GSASGVCAAFRSPVGGVLFALEEVATWWRSALLWRTFFSTAVVVVVLRAFIEICNSGKCG 307
Query: 297 LFGKGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGT 356
LFG GGLIMFD + YHA D+ V ++GV GGILGSLYN+L +KVLR+YN IN+KG
Sbjct: 308 LFGSGGLIMFDVSHVEVRYHAADIIPVTLIGVFGGILGSLYNHLLHKVLRLYNLINQKGK 367
Query: 357 ICKILLACVISMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDL 416
I K+LL+ +S+FTS LFGLP+LA C+PC E CPT GRSG +K+F CP G+YNDL
Sbjct: 368 IHKVLLSLGVSLFTSVCLFGLPFLAECKPCDPSIDEICPTNGRSGNFKQFNCPNGYYNDL 427
Query: 417 ASLIFNTNDDAIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTG 476
++L+ TNDDA+RN+FS NT +EF S++IFF L +++ G+ P+G+F+PII+ G
Sbjct: 428 STLLLTTNDDAVRNIFSSNTPNEFGMVSLWIFFGLYCILGLITFGIATPSGLFLPIILMG 487
Query: 477 ASYGRFVGMLFGKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMM 536
++YGR +G G +N++ GLYAVLGAAS + GSMR TVSLCVI LELTNNLLLLP+ M
Sbjct: 488 SAYGRMLGTAMGSYTNIDQGLYAVLGAASLMAGSMRMTVSLCVIFLELTNNLLLLPITMF 547
Query: 537 VLLVSKSVADVFNANIYDLIMKSKGLPYLETHAEPYMRQLTVGDV--VTGPLQIFHGYEK 594
VLL++K+V D FN +IY++I+ KGLP+LE + EP+MR LTVG++ P+ +G EK
Sbjct: 548 VLLIAKTVGDSFNLSIYEIILHLKGLPFLEANPEPWMRNLTVGELNDAKPPVVTLNGVEK 607
Query: 595 VRNVVFVLKTTSHNGFPVIDEPPFSEAPVLFGIILRHHLLTLLRKKVFMSAP-MAMGGDV 653
V N+V VL+ T+HN FPV+D + L G+ILR HL+ +L+K+ F++ +V
Sbjct: 608 VANIVDVLRNTTHNAFPVLDGADQNTGTELHGLILRAHLVKVLKKRWFLNEKRRTEEWEV 667
Query: 654 LRQFSADDFAKRGSDRGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILF 713
+F+ + A +R + +D+ +T EM +++DLHP TN +PYTVV+SMS+ KAL+LF
Sbjct: 668 REKFTPVELA----EREDNFDDVAITSSEMQLYVDLHPLTNTTPYTVVQSMSVAKALVLF 723
Query: 714 RELGLRHLLVIPKIPS--RSPVVGILTRHDFTAEHILGTHPYLVRSRWKRLR 763
R +GLRHLLV+PKI + SPV+GILTR D A +IL P+L + + + R
Sbjct: 724 RSVGLRHLLVVPKIQASGMSPVIGILTRQDLRAYNILQAFPHLDKHKSGKAR 775
>At5g26240 CLC-d chloride channel protein
Length = 792
Score = 640 bits (1650), Expect = 0.0
Identities = 362/768 (47%), Positives = 482/768 (62%), Gaps = 38/768 (4%)
Query: 1 MSTNHLSNG-DSEHRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQD 59
M +NHL NG +S++ L + S + + + + + SLDYE+ EN ++++
Sbjct: 1 MLSNHLQNGIESDNLLWSRVPESDDTSTDDITLLNSHRDGDGGVNSLDYEVIENYAYREE 60
Query: 60 WRSRGSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSL 119
RG L + YV +KW LIG GL NL+VEN AG KF +T ++ K +
Sbjct: 61 QAHRGKLYVGYYVAVKWFFSLLIGIGTGLAAVFINLSVENFAGWKFALTF-AIIQKSYFA 119
Query: 120 AFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGS 179
F+ + NL L + I APAAAGSGI E+K YLNG+D PG TLI KI GS
Sbjct: 120 GFIVYLLINLVLVFSSAYIITQFAPAAAGSGIPEIKGYLNGIDIPGTLLFRTLIGKIFGS 179
Query: 180 ITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSA 239
I +V L +GK GP++HTGAC+A+LLGQGGS +Y L +W + FK+DRDRRDL+ CG A
Sbjct: 180 IGSVGGGLALGKEGPLVHTGACIASLLGQGGSTKYHLNSRWPQLFKSDRDRRDLVTCGCA 239
Query: 240 AGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFG 299
AG+AAAFRAPVGGVLFALEE+ SWWR+ L+WR FFT+AIVA+ +R + C SG CG FG
Sbjct: 240 AGVAAAFRAPVGGVLFALEEVTSWWRSQLMWRVFFTSAIVAVVVRTAMGWCKSGICGHFG 299
Query: 300 KGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYL------WNKVLRIYNAINE 353
GG I++D G Y+ K++ + ++GVIGG+LG+L+N L W + N++++
Sbjct: 300 GGGFIIWDVSDGQDDYYFKELLPMAVIGVIGGLLGALFNQLTLYMTSWRR-----NSLHK 354
Query: 354 KGTICKILLACVISMFTSCLLFGLPWLASCQPCP---ADPVEPCP-TIGRSGIYKKFQC- 408
KG KI+ AC+IS TS + FGLP L C PCP D CP G G Y F C
Sbjct: 355 KGNRVKIIEACIISCITSAISFGLPLLRKCSPCPESVPDSGIECPRPPGMYGNYVNFFCK 414
Query: 409 PPGHYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGV 468
YNDLA++ FNT DDAIRNLFS T EF S+ F + L++++ G VPAG
Sbjct: 415 TDNEYNDLATIFFNTQDDAIRNLFSAKTMREFSAQSLLTFLAMFYTLAVVTFGTAVPAGQ 474
Query: 469 FVPIIVTGASYGRFVGML---FGKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELT 525
FVP I+ G++YGR VGM F KK N+ G YA+LGAASFLGGSMR TVSLCVI++E+T
Sbjct: 475 FVPGIMIGSTYGRLVGMFVVRFYKKLNIEEGTYALLGAASFLGGSMRMTVSLCVIMVEIT 534
Query: 526 NNLLLLPLIMMVLLVSKSVADVFNANIYDLIMKSKGLPYLETHAEPYMRQLTVGD----- 580
NNL LLPLIM+VLL+SK+V D FN +Y++ + KG+P LE+ + +MRQ+ +
Sbjct: 535 NNLKLLPLIMLVLLISKAVGDAFNEGLYEVQARLKGIPLLESRPKYHMRQMIAKEACQSQ 594
Query: 581 -VVTGPLQIFHGYEKVRNVVFVLKTTSHNGFPVIDEPPFSEAPVLFGIILRHHLLTLLRK 639
V++ P I +V +V +L + HNGFPVID E V+ G++LR HLL LL+
Sbjct: 595 KVISLPRVI-----RVADVASILGSNKHNGFPVIDHTRSGETLVI-GLVLRSHLLVLLQS 648
Query: 640 KV-FMSAPMAMGGDVLR-QFSADDFAKRGSDRGERIEDIHLTQEEMDMFIDLHPFTNASP 697
KV F +P+ + S +FAK S +G IEDIHLT ++++M+IDL PF N SP
Sbjct: 649 KVDFQHSPLPCDPSARNIRHSFSEFAKPVSSKGLCIEDIHLTSDDLEMYIDLAPFLNPSP 708
Query: 698 YTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILTRHDFTAE 745
Y V E MSL K LFR+LGLRHL V+P+ PSR V+G++TR D E
Sbjct: 709 YVVPEDMSLTKVYNLFRQLGLRHLFVVPR-PSR--VIGLITRKDLLIE 753
>At1g55620 CLC-f chloride channel protein
Length = 781
Score = 48.5 bits (114), Expect = 1e-05
Identities = 79/373 (21%), Positives = 147/373 (39%), Gaps = 73/373 (19%)
Query: 173 IVKIIGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRR- 231
++K I + + + +G GP + +G+ + + L +N+R+RR
Sbjct: 228 VIKAIQAAVTLGTGCSLGPEGPSVD--------IGKSCANGFALM------MENNRERRI 273
Query: 232 DLIVCGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCL 291
L G+A+GIA+ F A V G FA+E + R FTTA+ I L ++I +
Sbjct: 274 ALTAAGAASGIASGFNAAVAGCFFAIETVLRPLRAEN--SPPFTTAM--IILASVISSTV 329
Query: 292 SGKCGLFGKGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAI 351
S L G +Y + A ++PL ILG++ G + +++ L + ++ I
Sbjct: 330 SN--ALLGTQSAFTVPSYD---LKSAAELPLYLILGMLCGAVSVVFSRLVTWFTKSFDFI 384
Query: 352 NEKGTICKILLACVISMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPG 411
+K FGLP + CP +G G PG
Sbjct: 385 KDK--------------------FGLPAIV------------CPALGGLGAGIIALKYPG 412
Query: 412 HYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVP 471
+ F ++ + S + + + + + + L G + G++ P
Sbjct: 413 ----ILYWGFTNVEEILHTGKSASAPGIWLLAQLAAAKVVA---TALCKGSGLVGGLYAP 465
Query: 472 IIVTGASYGRFVG---------MLFGKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIIL 522
++ GA+ G G + G + YA++G A+ L ++ +++
Sbjct: 466 SLMIGAAVGAVFGGSAAEIINRAIPGNAAVAQPQAYALVGMAATLASMCSVPLTSVLLLF 525
Query: 523 ELTNNL-LLLPLI 534
ELT + +LLPL+
Sbjct: 526 ELTKDYRILLPLM 538
>At4g35440 CLC-e chloride channel protein
Length = 710
Score = 37.0 bits (84), Expect = 0.042
Identities = 36/186 (19%), Positives = 79/186 (42%), Gaps = 19/186 (10%)
Query: 174 VKIIGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDL 233
+K + + + + +G GP + GA +A +G + + K+ + L
Sbjct: 172 LKTVAACVTLGTGNSLGPEGPSVEIGASIA----KGVNSLFN---------KSPQTGFSL 218
Query: 234 IVCGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSG 293
+ GSAAGI++ F A V G FA+E + W ++ + ++ + + + +
Sbjct: 219 LAAGSAAGISSGFNAAVAGCFFAVESVL--WPSSSTDSSTSLPNTTSMVILSAVTASVVS 276
Query: 294 KCGLFGKGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINE 353
+ GL + + D S ++PL +LG + G++ + + + +++N+
Sbjct: 277 EIGLGSEPAFKVPDYDFRS----PGELPLYLLLGALCGLVSLALSRCTSSMTSAVDSLNK 332
Query: 354 KGTICK 359
I K
Sbjct: 333 DAGIPK 338
Score = 33.5 bits (75), Expect = 0.47
Identities = 17/68 (25%), Positives = 37/68 (54%), Gaps = 9/68 (13%)
Query: 476 GASYGRFVGMLFGKKSNLNHGL--------YAVLGAASFLGGSMRTTVSLCVIILELTNN 527
G +YG+F+G+ + + N + Y ++G A+ L G + ++ +++ ELT +
Sbjct: 420 GMAYGKFIGLALAQNPDFNLSILEVASPQAYGLVGMAATLAGVCQVPLTAVLLLFELTQD 479
Query: 528 L-LLLPLI 534
++LPL+
Sbjct: 480 YRIVLPLL 487
>At1g71140 hypothetical protein
Length = 485
Score = 31.6 bits (70), Expect = 1.8
Identities = 26/86 (30%), Positives = 43/86 (49%), Gaps = 7/86 (8%)
Query: 121 FVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSI 180
+V A L+L++ + A ++ A GSG ++ AY+N + A +F +PT I+ G
Sbjct: 368 YVKSMAPLLSLSVIFDALHAALSGVARGSGRQDIGAYVN-LAAYYLFGIPTAILLAFG-F 425
Query: 181 TAVSSSLHIGKAGPMLHTGACVAALL 206
L IG + G+CV A+L
Sbjct: 426 KMRGRGLWIG-----ITVGSCVQAVL 446
>At4g21250 hypothetical protein
Length = 449
Score = 31.2 bits (69), Expect = 2.3
Identities = 28/103 (27%), Positives = 44/103 (42%), Gaps = 20/103 (19%)
Query: 429 RNLFSKNTDSEFQYSSMFIFFITGFFLSIL--SCGVVVPAGVFVPII------------- 473
R +++ +E + SS I FL+ L S G + G+F+PI+
Sbjct: 41 RTSLKESSAAELKLSSAIIMAGVLCFLAALISSAGGIGGGGLFIPIMTIVAGVDLKTASS 100
Query: 474 -----VTGASYGRFVGMLFGKKSNLNHGLYAVLGAASFLGGSM 511
VTG S + LFG K+ L++ L +L LG S+
Sbjct: 101 FSAFMVTGGSIANVISNLFGGKALLDYDLALLLEPCMLLGVSI 143
>At5g18525 unknown protein
Length = 909
Score = 30.4 bits (67), Expect = 4.0
Identities = 27/84 (32%), Positives = 40/84 (47%), Gaps = 17/84 (20%)
Query: 695 ASPYTVVESMSLGKAL--ILFRELGLRHLLVIPKI-----PSRSPVVGILTRHD------ 741
AS + +++ S G+A L LGL L K+ P+ SPV+G+LT D
Sbjct: 178 ASLHRLIDGKSSGQATHSFLCLLLGLPLLEEKSKLRCLRHPNLSPVLGLLTSSDCLVSVL 237
Query: 742 ----FTAEHILGTHPYLVRSRWKR 761
+T E+IL P ++S W R
Sbjct: 238 PKAPYTLENILYYSPSAIKSEWHR 261
>At1g65520 hypothetical protein
Length = 240
Score = 30.0 bits (66), Expect = 5.2
Identities = 30/108 (27%), Positives = 45/108 (40%), Gaps = 3/108 (2%)
Query: 9 GDSEHRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGSLQI 68
GD EHRL LL S RS IN +V I + D + F N + S SL +
Sbjct: 17 GDGEHRLNPTLLDSLRSTINQIRSDPSFSQSVL-ITTSDGKFFSNGYDLALAESNPSLSV 75
Query: 69 FQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKR 116
++ L+ LI + I A + AG ++ + +L +R
Sbjct: 76 VMDAKLRSLVADLISLPMPTIAAVTGHA--SAAGCILAMSHDYVLMRR 121
>At3g11620 unknown protein
Length = 312
Score = 29.3 bits (64), Expect = 8.8
Identities = 28/86 (32%), Positives = 37/86 (42%), Gaps = 9/86 (10%)
Query: 468 VFVPIIVTGASYGRFVGM----LFGKKSNLNHGLYAVL-----GAASFLGGSMRTTVSLC 518
V VPII+ G S G ++ + F K GLY L L G + + L
Sbjct: 110 VKVPIILVGHSIGSYISLELLKKFSDKVVYCIGLYPFLTLNQQSTKQSLIGKLAASSVLS 169
Query: 519 VIILELTNNLLLLPLIMMVLLVSKSV 544
L +L LLP+ LLVSKS+
Sbjct: 170 ATASFLIASLRLLPMSAARLLVSKSI 195
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.142 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,396,610
Number of Sequences: 26719
Number of extensions: 759378
Number of successful extensions: 2042
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 2012
Number of HSP's gapped (non-prelim): 15
length of query: 780
length of database: 11,318,596
effective HSP length: 107
effective length of query: 673
effective length of database: 8,459,663
effective search space: 5693353199
effective search space used: 5693353199
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0030.6


