

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0030.2
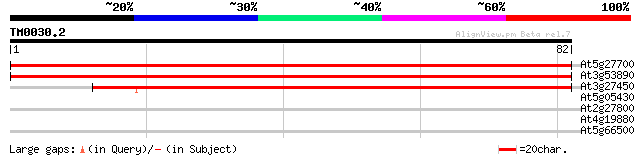
(82 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g27700 ribosomal protein S21 - like 148 4e-37
At3g53890 40S ribosomal protein S21 homolog 144 6e-36
At3g27450 40S ribosomal subunit protein S21, putative 107 1e-24
At5g05430 putative protein 26 4.0
At2g27800 hypothetical protein 26 4.0
At4g19880 unknown protein 25 5.3
At5g66500 unknown protein 25 9.0
>At5g27700 ribosomal protein S21 - like
Length = 85
Score = 148 bits (374), Expect = 4e-37
Identities = 66/82 (80%), Positives = 77/82 (93%)
Query: 1 MQNEEGQITELYIPRKCSATNRLITAKDHASVQINIGHLDESGVYNGQFSTYALCGYIRA 60
MQNEEGQ+TELYIPRKCSATNRLIT+KDHASVQ+NIGHLD +G+Y GQF+T+ALCG++RA
Sbjct: 1 MQNEEGQVTELYIPRKCSATNRLITSKDHASVQLNIGHLDANGLYTGQFTTFALCGFVRA 60
Query: 61 QGDADSAIDRLWQKKKAEIKQQ 82
QGDADS +DRLWQKKK E KQ+
Sbjct: 61 QGDADSGVDRLWQKKKVEAKQK 82
>At3g53890 40S ribosomal protein S21 homolog
Length = 82
Score = 144 bits (364), Expect = 6e-36
Identities = 63/82 (76%), Positives = 76/82 (91%)
Query: 1 MQNEEGQITELYIPRKCSATNRLITAKDHASVQINIGHLDESGVYNGQFSTYALCGYIRA 60
M+N+ GQ+TELYIPRKCSATNR+IT+KDHASVQ+NIGHLD +G+Y GQF+T+ALCG++RA
Sbjct: 1 MENDAGQVTELYIPRKCSATNRMITSKDHASVQLNIGHLDANGLYTGQFTTFALCGFVRA 60
Query: 61 QGDADSAIDRLWQKKKAEIKQQ 82
QGDADS +DRLWQKKK E KQQ
Sbjct: 61 QGDADSGVDRLWQKKKVEAKQQ 82
>At3g27450 40S ribosomal subunit protein S21, putative
Length = 79
Score = 107 bits (266), Expect = 1e-24
Identities = 51/74 (68%), Positives = 61/74 (81%), Gaps = 4/74 (5%)
Query: 13 IPRKC----SATNRLITAKDHASVQINIGHLDESGVYNGQFSTYALCGYIRAQGDADSAI 68
IPR+ S TNR IT+KDHASVQ+NIGHLD +G+Y GQF+T+ALCG++RAQGDADS +
Sbjct: 6 IPRQSMHIRSTTNRFITSKDHASVQLNIGHLDANGLYTGQFTTFALCGFVRAQGDADSGV 65
Query: 69 DRLWQKKKAEIKQQ 82
DRL QKKK E KQQ
Sbjct: 66 DRLRQKKKVEAKQQ 79
>At5g05430 putative protein
Length = 158
Score = 25.8 bits (55), Expect = 4.0
Identities = 13/41 (31%), Positives = 21/41 (50%), Gaps = 9/41 (21%)
Query: 6 GQITELYIPRKCSATNRLITA---------KDHASVQINIG 37
G+I E+YIP K TN ++ ++S Q+N+G
Sbjct: 51 GEIWEIYIPLKFDETNTILNRFWWLLPPPFLSNSSAQVNVG 91
>At2g27800 hypothetical protein
Length = 427
Score = 25.8 bits (55), Expect = 4.0
Identities = 18/70 (25%), Positives = 27/70 (37%), Gaps = 6/70 (8%)
Query: 4 EEGQITELYIPRKCSATNRLITAKDHASVQINIGHLDESGVYNGQFSTYALCGYIRAQGD 63
E T Y+ A R I A++ S G + YN + +AL G+
Sbjct: 306 EPNSFTYDYLIHGLCAQGRTINARELLSEMKGKGFVPNGKSYNSLVNAFAL------SGE 359
Query: 64 ADSAIDRLWQ 73
D A+ LW+
Sbjct: 360 IDDAVKCLWE 369
>At4g19880 unknown protein
Length = 239
Score = 25.4 bits (54), Expect = 5.3
Identities = 10/37 (27%), Positives = 23/37 (62%), Gaps = 2/37 (5%)
Query: 39 LDESG--VYNGQFSTYALCGYIRAQGDADSAIDRLWQ 73
+DE+ +YNG + CG+ + QG + A++++++
Sbjct: 90 IDETNEWIYNGINNGVYRCGFAKKQGPYEEAVEQVYE 126
>At5g66500 unknown protein
Length = 532
Score = 24.6 bits (52), Expect = 9.0
Identities = 13/42 (30%), Positives = 18/42 (41%)
Query: 17 CSATNRLITAKDHASVQINIGHLDESGVYNGQFSTYALCGYI 58
CS + L K V + G + +S + NG Y CG I
Sbjct: 291 CSDNSDLWIGKQIHCVALRNGFVSDSKLCNGLMDMYGKCGQI 332
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.131 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,793,229
Number of Sequences: 26719
Number of extensions: 59862
Number of successful extensions: 115
Number of sequences better than 10.0: 7
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 110
Number of HSP's gapped (non-prelim): 7
length of query: 82
length of database: 11,318,596
effective HSP length: 58
effective length of query: 24
effective length of database: 9,768,894
effective search space: 234453456
effective search space used: 234453456
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0030.2


