

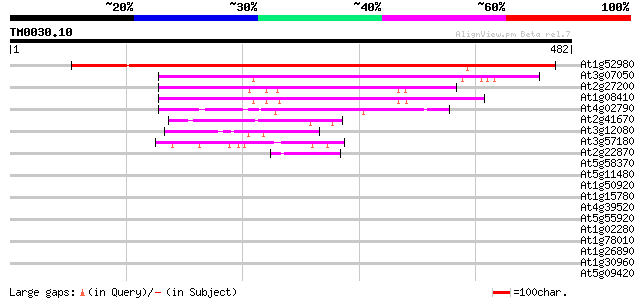
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0030.10
(482 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g52980 unknown protein 607 e-174
At3g07050 GTPase like protein 211 9e-55
At2g27200 putative nucleotide-binding protein 131 9e-31
At1g08410 unknown protein 130 2e-30
At4g02790 unknown protein 87 3e-17
At2g41670 unknown protein 49 8e-06
At3g12080 unknown protein 47 2e-05
At3g57180 unknown protein 45 9e-05
At2g22870 putative nucleotide-binding protein 42 8e-04
At5g58370 contains similarity to GTP-binding protein CGPA 40 0.003
At5g11480 putative GTP-binding protein 40 0.003
At1g50920 hypothetical protein 37 0.019
At1g15780 unknown protein 35 0.070
At4g39520 GTP-binding - like protein 35 0.12
At5g55920 nucleolar protein-like 34 0.16
At1g02280 unknown protein 34 0.16
At1g78010 unknown protein 34 0.20
At1g26890 hypothetical protein 34 0.20
At1g30960 F17F8.15 33 0.27
At5g09420 putative subunit of TOC complex 32 0.60
>At1g52980 unknown protein
Length = 576
Score = 607 bits (1565), Expect = e-174
Identities = 298/418 (71%), Positives = 354/418 (84%), Gaps = 3/418 (0%)
Query: 54 PFTPVFGPKTKRKRPCLLAFDYQSLAKKAHVAQEAFEEKYAAGASGTAEANEADGFIDLL 113
PF FG KTKRKRP L+A DY++L KKA +Q+AFEEK AG SG E DGF DL+
Sbjct: 134 PFQDAFGRKTKRKRPKLVASDYEALVKKAAESQDAFEEKNGAGPSGEG-GEEEDGFRDLV 192
Query: 114 GHTMFQKGQTKHISGELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLL 173
HTMF+KGQ+K I GELYKVIDSSDV+VQV+DARDPQGTRC+HLEK LKE+ K+KHM+LL
Sbjct: 193 RHTMFEKGQSKRIWGELYKVIDSSDVIVQVIDARDPQGTRCHHLEKTLKEHHKHKHMILL 252
Query: 174 LNKCDLIPAWATKGWLRVLSKEYPTLAFHASINKSFGKGSLLSVLRQFARLKSDKQAISV 233
LNKCDL+PAWATKGWLRVLSKEYPTLAFHAS+NKSFGKGSLLSVLRQFARLKSDKQAISV
Sbjct: 253 LNKCDLVPAWATKGWLRVLSKEYPTLAFHASVNKSFGKGSLLSVLRQFARLKSDKQAISV 312
Query: 234 GFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQNKDSE 293
GF+GYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQ++D+E
Sbjct: 313 GFVGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQSRDTE 372
Query: 294 ADVVLKGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYKIKEWENENDFLLQLCKLTGKLL 353
D+VLKGVVRVTNL+DA++ IGEVL+RV KEHL RAYKIK+WE+++DFLLQLCK +GKLL
Sbjct: 373 TDIVLKGVVRVTNLEDASEHIGEVLRRVKKEHLQRAYKIKDWEDDHDFLLQLCKSSGKLL 432
Query: 354 KGGEPDLMTAAKMVLHDWQRGKIPFFVPPPQQEDLSEEP--IVNGLDVDDGVDSNQASAA 411
KGGEPDLMT AKM+LHDWQRG+IPFFVPPP+ ++++ E IV G+D + D++QA+AA
Sbjct: 433 KGGEPDLMTGAKMILHDWQRGRIPFFVPPPKLDNVASESEVIVPGIDKEAIADNSQAAAA 492
Query: 412 IKAIANVLSSQQQSSVPVQKDLFTENELEEEADNLLLISGDSPDGDVLDSDSDTSEQD 469
+KAIA ++S+QQQ VPVQ+D + E +L+++ D+ +G + D D +D
Sbjct: 493 LKAIAGIMSTQQQKDVPVQRDFYDEKDLKDDKKAKESTETDAENGTDAEEDEDAVSED 550
Score = 29.6 bits (65), Expect = 3.9
Identities = 22/59 (37%), Positives = 31/59 (52%), Gaps = 4/59 (6%)
Query: 22 KKKESSGKSKQSLDANGNNDAKMEQSNSRVQRPFTPVFGPKTKRKRPC---LLAFDYQS 77
KK SGK K SLDAN D K + + +R + + KT+ KR +L+ +YQS
Sbjct: 6 KKANVSGKPKHSLDAN-RADGKKKTTETRSKSTVNRLKMYKTRPKRNAGGKILSNEYQS 63
>At3g07050 GTPase like protein
Length = 582
Score = 211 bits (536), Expect = 9e-55
Identities = 128/367 (34%), Positives = 202/367 (54%), Gaps = 40/367 (10%)
Query: 129 ELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGW 188
EL KVI+ SDV+++VLDARDP GTRC +E+ + + KH++LLLNK DL+P A + W
Sbjct: 129 ELVKVIELSDVILEVLDARDPLGTRCTDMERMVMQAGPNKHLVLLLNKIDLVPREAAEKW 188
Query: 189 LRVLSKEYPTLAFHASINKS-----------------------FGKGSLLSVLRQFARLK 225
L L +E+P +AF S + G +L+ +L+ ++R
Sbjct: 189 LMYLREEFPAVAFKCSTQEQRSNLGWKSSKASKPSNMLQTSDCLGADTLIKLLKNYSRSH 248
Query: 226 SDKQAISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGV 285
K++I+VG IG PNVGKSS+IN+L+ +V V PG T+ Q + L K + L+DCPGV
Sbjct: 249 ELKKSITVGIIGLPNVGKSSLINSLKRAHVVNVGATPGLTRSLQEVHLDKNVKLLDCPGV 308
Query: 286 V-YQNKDSEADVVLKGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYKIKEWENENDFLLQ 344
V ++ ++A + L+ R+ L D + E+LK K+ L YKI +E +DFL +
Sbjct: 309 VMLKSSGNDASIALRNCKRIEKLDDPVSPVKEILKLCPKDMLVTLYKIPSFEAVDDFLYK 368
Query: 345 LCKLTGKLLKGGEPDLMTAAKMVLHDWQRGKIPFFVPPPQQED--LSEEPIVNGLDVDDG 402
+ + GKL KGG D+ AA++VLHDW GKIP++ PP+++ +E IV L D
Sbjct: 369 VATVRGKLKKGGLVDIDAAARIVLHDWNEGKIPYYTMPPKRDQGGHAESKIVTELAKDFN 428
Query: 403 VD---SNQAS--AAIKAI---------ANVLSSQQQSSVPVQKDLFTENELEEEADNLLL 448
+D S ++S ++K + +N + ++ + + TE E E E+D+
Sbjct: 429 IDEVYSGESSFIGSLKTVNEFNPVIIPSNGPLNFDETMIEDESKTQTEEEAEHESDDDES 488
Query: 449 ISGDSPD 455
+ G+ +
Sbjct: 489 MGGEEEE 495
>At2g27200 putative nucleotide-binding protein
Length = 537
Score = 131 bits (329), Expect = 9e-31
Identities = 87/311 (27%), Positives = 149/311 (46%), Gaps = 55/311 (17%)
Query: 129 ELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGW 188
+L++V++ SD++V V+DARDP RC LE + +E ++K MLL+NK DL+P++ + W
Sbjct: 160 QLWRVLERSDLIVMVVDARDPLFYRCPDLEAYAQEIDEHKKTMLLVNKADLLPSYVREKW 219
Query: 189 LRVLSKEYPTLAFHAS----------------------------INKSFGKGSLLSVLR- 219
S+ F ++ K +G+ LL L+
Sbjct: 220 AEYFSRNNILFVFWSAKAATATLEGKPLKEQWRAPDTTQKTDNPAVKVYGRDDLLDRLKL 279
Query: 220 ---QFARLKSDK-----------QAISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGET 265
+ +++ + + + VGF+GYPNVGKSS IN L + V PG+T
Sbjct: 280 EALEIVKMRKSRGVSATSTESHCEQVVVGFVGYPNVGKSSTINALVGQKRTGVTSTPGKT 339
Query: 266 KVWQYITLTKRIFLIDCPGVVYQN-KDSEADVVLKGVVRVTNLKDAADRIGEVLKRVMKE 324
K +Q + +++ + L DCPG+V+ + S ++V GV+ + + + + I V + V +
Sbjct: 340 KHFQTLIISEDLMLCDCPGLVFPSFSSSRYEMVASGVLPIDRMTEHLEAIKVVAELVPRH 399
Query: 325 HLHRAYKI-----KEWENE------NDFLLQLCKLTGKLLKGGEPDLMTAAKMVLHDWQR 373
+ Y I K +E + ++ L C G + G PD AA+ +L D+
Sbjct: 400 AIEDVYNISLPKPKSYEPQSRPPLASELLRTYCLSRGYVASSGLPDETRAARQILKDYIE 459
Query: 374 GKIPFFVPPPQ 384
GK+P F PP+
Sbjct: 460 GKLPHFAMPPE 470
>At1g08410 unknown protein
Length = 589
Score = 130 bits (326), Expect = 2e-30
Identities = 89/339 (26%), Positives = 159/339 (46%), Gaps = 59/339 (17%)
Query: 129 ELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGW 188
+L++V++ SD++V V+DARDP RC LE + +E ++K +MLL+NK DL+P + W
Sbjct: 160 QLWRVLERSDLIVMVVDARDPLFYRCPDLEAYAQEIDEHKKIMLLVNKADLLPTDVREKW 219
Query: 189 LRVLSKEYPTLAFHASINKS----------------------------FGKGSLLSVLR- 219
F ++I + +G+ LLS L+
Sbjct: 220 AEYFRLNNILFVFWSAIAATATLEGKVLKEQWRQPDNLQKTDDPDIMIYGRDELLSRLQF 279
Query: 220 ---QFARLKSDKQA---------------ISVGFIGYPNVGKSSVINTLRTKNVCKVAPI 261
+ ++++ + A VGF+GYPNVGKSS IN L + V
Sbjct: 280 EAQEIVKVRNSRAASVSSQSWTGEYQRDQAVVGFVGYPNVGKSSTINALVGQKRTGVTST 339
Query: 262 PGETKVWQYITLTKRIFLIDCPGVVYQN-KDSEADVVLKGVVRVTNLKDAADRIGEVLKR 320
PG+TK +Q + ++ + L DCPG+V+ + S +++ GV+ + + + + I V +
Sbjct: 340 PGKTKHFQTLIISDELMLCDCPGLVFPSFSSSRYEMIASGVLPIDRMTEHREAIQVVADK 399
Query: 321 VMKEHLHRAYKI-----KEWENEN------DFLLQLCKLTGKLLKGGEPDLMTAAKMVLH 369
V + + Y I K +E ++ + L C G + G PD AA+++L
Sbjct: 400 VPRRVIESVYNISLPKPKTYERQSRPPHAAELLKSYCASRGYVASSGLPDETKAARLILK 459
Query: 370 DWQRGKIPFFVPPPQQEDLSEEPIVNGLDVDDGVDSNQA 408
D+ GK+P + PP E I + +++D ++ +++
Sbjct: 460 DYIGGKLPHYAMPPGMPQADEPDIEDTQELEDILEGSES 498
>At4g02790 unknown protein
Length = 372
Score = 86.7 bits (213), Expect = 3e-17
Identities = 73/266 (27%), Positives = 123/266 (45%), Gaps = 26/266 (9%)
Query: 129 ELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGW 188
EL + + DVV++V DAR P T ++ L + +L+LN+ D+I W
Sbjct: 109 ELREQLKLMDVVIEVRDARIPLSTTHPKMDAWLGN----RKRILVLNREDMISNDDRNDW 164
Query: 189 LRVLSKEYPTLAFHASINKSFGKGSLLSVLRQFARLKSD-----------KQAISVGFIG 237
R +K+ + F N G G++ + R L D +++ G IG
Sbjct: 165 ARYFAKQGIKVIF---TNGKLGMGAM-KLGRLAKSLAGDVNGKRREKGLLPRSVRAGIIG 220
Query: 238 YPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQNKDSEADVV 297
YPNVGKSS+IN L + +C AP PG T+ +++ L K + L+D PG++ D +A +
Sbjct: 221 YPNVGKSSLINRLLKRKICAAAPRPGVTREMKWVKLGKDLDLLDSPGMLPMRIDDQAAAI 280
Query: 298 LKGVV-----RVTNLKDAADRIGEVLKRVMKEHLHRAYKIKEWENENDFLLQLCKLTGKL 352
+ + + D A + ++L R+ + Y + + E + + K G
Sbjct: 281 KLAICDDIGEKAYDFTDVAGILVQMLARIPEVGAKALYNRYKIQLEGNCGKKFVKTLGLN 340
Query: 353 LKGGEPDLMTAAKMVLHDWQRGKIPF 378
L GG D AA +L D+++GK +
Sbjct: 341 LFGG--DSHQAAFRILTDFRKGKFGY 364
>At2g41670 unknown protein
Length = 386
Score = 48.5 bits (114), Expect = 8e-06
Identities = 45/165 (27%), Positives = 76/165 (45%), Gaps = 21/165 (12%)
Query: 137 SDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGWLRVL-SKE 195
SD+V++V DAR P + + L+ K ++ LNK DL W R S +
Sbjct: 47 SDLVIEVRDARIPLSSA----NEDLQSQMSAKRRIIALNKKDLANPNVLNKWTRHFESSK 102
Query: 196 YPTLAFHASINKSFGKGSLLSVLR-QFARLKSDKQAISVGFIGYPNVGKSSVINTLRTKN 254
+A +A S K LL ++ + + + + + V +G PNVGKS++IN++
Sbjct: 103 QDCIAINAHSRSSVMK--LLDLVELKLKEVIAREPTLLVMVVGVPNVGKSALINSIHQIA 160
Query: 255 VCK-----------VAPIPGETKVWQYITLTKR--IFLIDCPGVV 286
+ V P+PG T+ + R I+++D PGV+
Sbjct: 161 AARFPVQERLKRATVGPLPGVTQDIAGFKIAHRPSIYVLDSPGVL 205
>At3g12080 unknown protein
Length = 663
Score = 47.0 bits (110), Expect = 2e-05
Identities = 36/146 (24%), Positives = 68/146 (45%), Gaps = 19/146 (13%)
Query: 134 IDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGWLRVLS 193
+D S V++ V+D + + L++ +K+++L +NKC+ KG ++ +
Sbjct: 265 VDESAVIIFVVDGQAGPSGADVEIADWLRKYYSHKYIILAVNKCES----PRKGLMQ--A 318
Query: 194 KEYPTLAFHA---SINKSFGKGSLLS----------VLRQFARLKSDKQAISVGFIGYPN 240
E+ +L F S G G LL ++ + + ++ IG PN
Sbjct: 319 SEFWSLGFTPIPISALSGTGTGELLDLVCSGLIKLEIMENIEEEEEENYIPAIAIIGRPN 378
Query: 241 VGKSSVINTLRTKNVCKVAPIPGETK 266
VGKSS++N L ++ V+P+ G T+
Sbjct: 379 VGKSSILNALVREDRTIVSPVSGTTR 404
Score = 30.8 bits (68), Expect = 1.7
Identities = 15/34 (44%), Positives = 20/34 (58%)
Query: 233 VGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETK 266
V +G PNVGKS++ N L +N V PG T+
Sbjct: 161 VAIVGRPNVGKSALFNRLVGENRAIVVDEPGVTR 194
>At3g57180 unknown protein
Length = 637
Score = 45.1 bits (105), Expect = 9e-05
Identities = 54/190 (28%), Positives = 85/190 (44%), Gaps = 33/190 (17%)
Query: 126 ISGELYKVIDSSD--VVVQVLDARDPQGTRCYHLEKHL-----------KENCKYKHMML 172
IS L K + +S VVV V+D D G+ K L K + ++L
Sbjct: 249 ISTRLIKPMSNSSTTVVVMVVDCVDFDGSFPKRAAKSLFQVLQKAENDPKGSKNLPKLVL 308
Query: 173 LLNKCDLIPAWATKG----WLRVLSKE--YPTLA--FHASINKSFGKGSLLSVLRQFARL 224
+ K DL+P + W+R +K P L+ + S K G +LL+ +++ A
Sbjct: 309 VATKVDLLPTQISPARLDRWVRHRAKAGGAPKLSGVYMVSARKDIGVKNLLAYIKELAGP 368
Query: 225 KSDKQAISVGFIGYPNVGKSSVINTLRTKNVCKV-----APIPGETKVWQYI--TLTKRI 277
+ + V IG N GKS++IN L K+ KV AP+PG T I L+ +
Sbjct: 369 RGN-----VWVIGAQNAGKSTLINALSKKDGAKVTRLTEAPVPGTTLGILKIGGILSAKA 423
Query: 278 FLIDCPGVVY 287
+ D PG+++
Sbjct: 424 KMYDTPGLLH 433
>At2g22870 putative nucleotide-binding protein
Length = 300
Score = 42.0 bits (97), Expect = 8e-04
Identities = 23/61 (37%), Positives = 37/61 (59%), Gaps = 3/61 (4%)
Query: 225 KSDKQAISVGFIGYPNVGKSSVINTL-RTKNVCKVAPIPGETKVWQYITLTKRIFLIDCP 283
K D+ I++ +G NVGKSS+IN L R K V + PG+T++ + + K +++D P
Sbjct: 117 KDDRPEIAI--LGRSNVGKSSLINCLVRKKEVALTSKKPGKTQLINHFLVNKSWYIVDLP 174
Query: 284 G 284
G
Sbjct: 175 G 175
>At5g58370 contains similarity to GTP-binding protein CGPA
Length = 465
Score = 40.0 bits (92), Expect = 0.003
Identities = 27/82 (32%), Positives = 44/82 (52%), Gaps = 6/82 (7%)
Query: 233 VGFIGYPNVGKSSVINTL-RTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVY---- 287
+ F G NVGKSS++N L R V + + PG T+ + L ++ L+D PG +
Sbjct: 291 IAFAGRSNVGKSSLLNALTRQWGVVRTSDKPGLTQTINFFGLGPKVRLVDLPGYGFAFAK 350
Query: 288 -QNKDSEADVVLKGVVRVTNLK 308
+ K++ D+V + V T+LK
Sbjct: 351 DEVKEAWEDLVKEYVSTRTSLK 372
>At5g11480 putative GTP-binding protein
Length = 318
Score = 40.0 bits (92), Expect = 0.003
Identities = 20/59 (33%), Positives = 34/59 (56%), Gaps = 1/59 (1%)
Query: 236 IGYPNVGKSSVINTL-RTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQNKDSE 293
+G NVGKSS++N+L R K + + PG+T+ + + + +L+D PG Y + E
Sbjct: 141 VGRSNVGKSSLLNSLVRRKRLALTSKKPGKTQCINHFRINDKWYLVDLPGYGYASAPHE 199
>At1g50920 hypothetical protein
Length = 671
Score = 37.4 bits (85), Expect = 0.019
Identities = 62/272 (22%), Positives = 102/272 (36%), Gaps = 51/272 (18%)
Query: 53 RPFTPVFGPKTKRKRPCLLAFDY------QSLAKKAHVAQEAFEEKYAAGASGTAEANEA 106
+ F + +T+R+ P ++ Y Q +K Q F K +A +
Sbjct: 16 KEFVDIILSRTQRQTPTVVHKGYKINRLRQFYMRKVKYTQTNFHAKLSAIIDEFPRLEQI 75
Query: 107 DGFIDLLGHTMFQKGQTKHISGELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCK 166
F L H ++ K K G QV AR+ + K K+ K
Sbjct: 76 HPFYGDLLHVLYNKDHYKLALG-------------QVNTARNL-------ISKISKDYVK 115
Query: 167 YKHMMLLLNKCDLIPAWATKGWLRVLSKEYPTLAFHASINKSFGKGSLLSVLRQFARLKS 226
L +C + A VL + P+LA+ L + + ARL S
Sbjct: 116 LLKYGDSLYRCKCLKVAALGRMCTVLKRITPSLAY------------LEQIRQHMARLPS 163
Query: 227 -DKQAISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKV-------WQYITLTKRIF 278
D +V GYPNVGKSS +N + +V V P TK ++Y+ R
Sbjct: 164 IDPNTRTVLICGYPNVGKSSFMNKVTRADV-DVQPYAFTTKSLFVGHTDYKYL----RYQ 218
Query: 279 LIDCPGVVYQNKDSEADVVLKGVVRVTNLKDA 310
+ID PG++ + + + + + + +L+ A
Sbjct: 219 VIDTPGILDRPFEDRNIIEMCSITALAHLRAA 250
>At1g15780 unknown protein
Length = 1335
Score = 35.4 bits (80), Expect = 0.070
Identities = 29/107 (27%), Positives = 46/107 (42%), Gaps = 10/107 (9%)
Query: 372 QRGKIPFFVPPPQQEDLSEEPIVNGLDVDDGVDSNQASAAIKAIANVLSSQQQSSVPVQK 431
Q PF VP P L+ P+ VDS + ++ ++ N+ Q V +
Sbjct: 953 QPANSPFVVPSPSSTPLAPSPMQ--------VDSEKPGSSSLSMGNIARQQATGMQGVVQ 1004
Query: 432 DLFTENELEEEADNLLLISGDSPDGDVLDSDSDTSEQDPDTEVPYEQ 478
L + LL SPDG++L+S + TS + TE+P E+
Sbjct: 1005 SLAIGTPGISASP--LLQEFTSPDGNILNSSTITSGKPSATELPIER 1049
>At4g39520 GTP-binding - like protein
Length = 369
Score = 34.7 bits (78), Expect = 0.12
Identities = 21/71 (29%), Positives = 36/71 (50%), Gaps = 18/71 (25%)
Query: 233 VGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLT----------KRIFLIDC 282
VG +G+P+VGKS+++N K+ E +++ TLT +I L+D
Sbjct: 68 VGLVGFPSVGKSTLLN--------KLTGTFSEVASYEFTTLTCIPGVITYRGAKIQLLDL 119
Query: 283 PGVVYQNKDSE 293
PG++ KD +
Sbjct: 120 PGIIEGAKDGK 130
>At5g55920 nucleolar protein-like
Length = 682
Score = 34.3 bits (77), Expect = 0.16
Identities = 27/93 (29%), Positives = 42/93 (45%), Gaps = 5/93 (5%)
Query: 384 QQEDLSEEPIVNGLDVDD----GVDSNQASAAIKAIANVLSSQ-QQSSVPVQKDLFTENE 438
++E L + + + D DD G D + S A+++ S + S + D EN+
Sbjct: 59 EEEPLEDYEVTDDSDEDDEVSDGSDEDDISPAVESEEIDESDDGENGSNQLFSDDEEEND 118
Query: 439 LEEEADNLLLISGDSPDGDVLDSDSDTSEQDPD 471
E D+ L SGD + LD+DSD D D
Sbjct: 119 EETLGDDFLEGSGDEDEEGSLDADSDADSDDDD 151
>At1g02280 unknown protein
Length = 297
Score = 34.3 bits (77), Expect = 0.16
Identities = 24/70 (34%), Positives = 38/70 (54%), Gaps = 4/70 (5%)
Query: 221 FARLKS-DKQAISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTK---R 276
F +LK D +++V +G VGKSS +N+L + V +V+P E ++ T
Sbjct: 26 FGKLKQKDMNSMTVLVLGKGGVGKSSTVNSLIGEQVVRVSPFQAEGLRPVMVSRTMGGFT 85
Query: 277 IFLIDCPGVV 286
I +ID PG+V
Sbjct: 86 INIIDTPGLV 95
>At1g78010 unknown protein
Length = 613
Score = 33.9 bits (76), Expect = 0.20
Identities = 36/125 (28%), Positives = 50/125 (39%), Gaps = 10/125 (8%)
Query: 195 EYPTLAFHASINKSFGKGSLLSVLRQFARL-KSDKQAISVGFIGYPNVGKSSVINTLRTK 253
E P L + INK + A K + + + +G PNVGKSS++N
Sbjct: 317 EMPPLDIESVINKITSMSQDVESALDTANYDKLLQSGLQIAIVGRPNVGKSSLLNAWSKS 376
Query: 254 NVCKVAPIPGETKVWQYITLTKR---IFLIDCPGVVYQNKDSEADVVLK-GVVRVTNLKD 309
V + G T+ +T R I L+D G+ N D+V K GV R
Sbjct: 377 ERAIVTEVAGTTRDVVEANVTVRGVPITLLDTAGIRETN-----DIVEKIGVERSETAAK 431
Query: 310 AADRI 314
AD I
Sbjct: 432 VADVI 436
>At1g26890 hypothetical protein
Length = 280
Score = 33.9 bits (76), Expect = 0.20
Identities = 24/91 (26%), Positives = 45/91 (49%), Gaps = 6/91 (6%)
Query: 388 LSEEPIVNGLDVDDGVDSNQASAAIKAIA----NVLSSQQQSSVPVQKDLFTENELEEEA 443
LS P++N L VD D N +K + +++++ V ++D+ ENE+E++
Sbjct: 128 LSSSPVLNCLKVDRHKDDNLTKFTVKVSTLKQLHYNTTRRKDGVEDEEDVVEENEVEDDD 187
Query: 444 DNLLLISGDSPDGDVLDSDSDTSEQDPDTEV 474
D + D +G V + D E+D D ++
Sbjct: 188 D--VDEVEDDEEGVVEEVDVGEVEEDDDDDL 216
>At1g30960 F17F8.15
Length = 437
Score = 33.5 bits (75), Expect = 0.27
Identities = 25/80 (31%), Positives = 39/80 (48%), Gaps = 8/80 (10%)
Query: 225 KSDKQAISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTK---RIFLID 281
+ D+++++VG IG PN GKSS+ N + V + T LTK ++ D
Sbjct: 148 EEDQKSLNVGIIGPPNAGKSSLTNFMVGTKVAAASRKTNTTTHEVLGVLTKGDTQVCFFD 207
Query: 282 CPGVV-----YQNKDSEADV 296
PG++ Y KD +A V
Sbjct: 208 TPGLMLKKSGYGYKDIKARV 227
>At5g09420 putative subunit of TOC complex
Length = 616
Score = 32.3 bits (72), Expect = 0.60
Identities = 23/74 (31%), Positives = 34/74 (45%), Gaps = 8/74 (10%)
Query: 278 FLIDCPGVVYQNKDSEADVVLKGVVRVTNLKDAADRIGEV-LKRVMKEHLHRAYKIKEWE 336
FL+D VY + +A + +NL +D G + VMKE + AYK K+W
Sbjct: 453 FLLDTTLDVYASLQDQAKLA-------SNLAPVSDTNGNMEASEVMKEKGNAAYKGKQWN 505
Query: 337 NENDFLLQLCKLTG 350
+F + KL G
Sbjct: 506 KAVNFYTEAIKLNG 519
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.132 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,903,215
Number of Sequences: 26719
Number of extensions: 471514
Number of successful extensions: 1380
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 33
Number of HSP's that attempted gapping in prelim test: 1330
Number of HSP's gapped (non-prelim): 68
length of query: 482
length of database: 11,318,596
effective HSP length: 103
effective length of query: 379
effective length of database: 8,566,539
effective search space: 3246718281
effective search space used: 3246718281
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0030.10


