

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0029b.5
(313 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
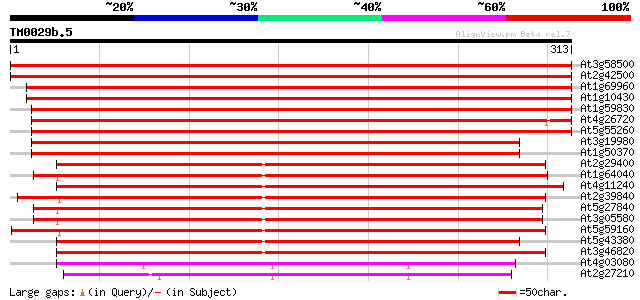
At3g58500 phosphoprotein phosphatase 2A isoform 4 634 0.0
At2g42500 putative serine/threonine protein phosphatase catalyti... 630 0.0
At1g69960 serine/threonine protein phosphatase (type 2A) 530 e-151
At1g10430 serine/threonine protein phosphatase, PP2A, catalytic ... 529 e-151
At1g59830 unknown protein 528 e-150
At4g26720 phosphoprotein phosphatase (PPX-1) 427 e-120
At5g55260 protein phosphatase X isoform 2 (gb|AAB86419.1) 420 e-118
At3g19980 serine/threonine protein phosphatase, putative 370 e-103
At1g50370 phosphoprotein phosphatase 367 e-102
At2g29400 phosphoprotein phosphatase, type 1 catalytic subunit 289 2e-78
At1g64040 phosphoprotein phosphatase 1 288 2e-78
At4g11240 unknown protein 286 1e-77
At2g39840 putative serine/threonine protein phosphatase PP1 isoz... 283 1e-76
At5g27840 TOPP8 serine/threonine protein phosphatase type one 281 2e-76
At3g05580 putative serine/threonine protein phosphatase type one 281 3e-76
At5g59160 phosphoprotein phosphatase 1 catalytic chain 278 4e-75
At5g43380 protein phosphatase 1 catalytic subunit 273 9e-74
At3g46820 phosphoprotein phosphatase 272 2e-73
At4g03080 phospho-ser/thr phosphatase like protein 208 3e-54
At2g27210 putative phosphoprotein phosphatase 202 2e-52
>At3g58500 phosphoprotein phosphatase 2A isoform 4
Length = 313
Score = 634 bits (1635), Expect = 0.0
Identities = 299/313 (95%), Positives = 307/313 (97%)
Query: 1 MGANSLPSESTHDLDEQISQLMQCKPLSEQQVKELCEKAKEILMEESNVQPVKSPVTICG 60
MGANSLP+++T DLDEQISQLMQCKPLSEQQV+ LCEKAKEILM+ESNVQPVKSPVTICG
Sbjct: 1 MGANSLPTDATLDLDEQISQLMQCKPLSEQQVRALCEKAKEILMDESNVQPVKSPVTICG 60
Query: 61 DIHGQFHDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRG 120
DIHGQFHDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLV LKVRY QRITILRG
Sbjct: 61 DIHGQFHDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVGLKVRYPQRITILRG 120
Query: 121 NHESRQITQVYGFYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIET 180
NHESRQITQVYGFYDECLRKYGNANVWK FTDLFD+FPLTALVESEIFCLHGGLSPSIET
Sbjct: 121 NHESRQITQVYGFYDECLRKYGNANVWKIFTDLFDYFPLTALVESEIFCLHGGLSPSIET 180
Query: 181 LDNIRNFDRVQEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDISEQFNHTNSLKL 240
LDNIRNFDRVQEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDISEQFNHTN+LKL
Sbjct: 181 LDNIRNFDRVQEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDISEQFNHTNNLKL 240
Query: 241 IARAHQLVMDGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAPRRG 300
IARAHQLVMDGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDC+ HTFIQFEPAPRRG
Sbjct: 241 IARAHQLVMDGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCRNHTFIQFEPAPRRG 300
Query: 301 EPDVTRRTPDYFL 313
EPDVTRRTPDYFL
Sbjct: 301 EPDVTRRTPDYFL 313
>At2g42500 putative serine/threonine protein phosphatase catalytic
subunit, PP2A-3
Length = 313
Score = 630 bits (1625), Expect = 0.0
Identities = 296/313 (94%), Positives = 307/313 (97%)
Query: 1 MGANSLPSESTHDLDEQISQLMQCKPLSEQQVKELCEKAKEILMEESNVQPVKSPVTICG 60
MGANS+P+++T DLDEQISQLMQCKPLSEQQV+ LCEKAKEILM+ESNVQPVKSPVTICG
Sbjct: 1 MGANSIPTDATIDLDEQISQLMQCKPLSEQQVRALCEKAKEILMDESNVQPVKSPVTICG 60
Query: 61 DIHGQFHDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRG 120
DIHGQFHDLAELFRIGG CPDTNYLFMGDYVDRGYYSVETVTLLVALK+RY QRITILRG
Sbjct: 61 DIHGQFHDLAELFRIGGMCPDTNYLFMGDYVDRGYYSVETVTLLVALKMRYPQRITILRG 120
Query: 121 NHESRQITQVYGFYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIET 180
NHESRQITQVYGFYDECLRKYGNANVWK FTDLFD+FPLTALVESEIFCLHGGLSPSIET
Sbjct: 121 NHESRQITQVYGFYDECLRKYGNANVWKIFTDLFDYFPLTALVESEIFCLHGGLSPSIET 180
Query: 181 LDNIRNFDRVQEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDISEQFNHTNSLKL 240
LDNIRNFDRVQEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDISEQFNHTN+LKL
Sbjct: 181 LDNIRNFDRVQEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDISEQFNHTNNLKL 240
Query: 241 IARAHQLVMDGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAPRRG 300
IARAHQLVMDG+NWAHEQKVVTIFSAPNYCYRCGNMASILEVDDC+ HTFIQFEPAPRRG
Sbjct: 241 IARAHQLVMDGYNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCRNHTFIQFEPAPRRG 300
Query: 301 EPDVTRRTPDYFL 313
EPDVTRRTPDYFL
Sbjct: 301 EPDVTRRTPDYFL 313
>At1g69960 serine/threonine protein phosphatase (type 2A)
Length = 307
Score = 530 bits (1366), Expect = e-151
Identities = 242/304 (79%), Positives = 272/304 (88%)
Query: 10 STHDLDEQISQLMQCKPLSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDL 69
+T D+D QI QLM+CK LSE +VK LCE AK IL+EE NVQPVK PVT+CGDIHGQF+DL
Sbjct: 4 ATGDIDRQIEQLMECKALSETEVKMLCEHAKTILVEEYNVQPVKCPVTVCGDIHGQFYDL 63
Query: 70 AELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQ 129
ELFRIGG PDTNYLFMGDYVDRGYYSVETV+LLVALKVRY R+TILRGNHESRQITQ
Sbjct: 64 IELFRIGGSSPDTNYLFMGDYVDRGYYSVETVSLLVALKVRYRDRLTILRGNHESRQITQ 123
Query: 130 VYGFYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDR 189
VYGFYDECLRKYGNANVWK FTDLFD+ PLTAL+ES++FCLHGGLSPS++TLDNIR+ DR
Sbjct: 124 VYGFYDECLRKYGNANVWKHFTDLFDYLPLTALIESQVFCLHGGLSPSLDTLDNIRSLDR 183
Query: 190 VQEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQLVM 249
+QEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDI+ QFNHTN L LI+RAHQLVM
Sbjct: 184 IQEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDIATQFNHTNGLSLISRAHQLVM 243
Query: 250 DGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAPRRGEPDVTRRTP 309
+GFNW E+ VVT+FSAPNYCYRCGNMA+ILE+ + F+QF+PAPR+ EP+ TR+TP
Sbjct: 244 EGFNWCQEKNVVTVFSAPNYCYRCGNMAAILEIGENMDQNFLQFDPAPRQVEPETTRKTP 303
Query: 310 DYFL 313
DYFL
Sbjct: 304 DYFL 307
>At1g10430 serine/threonine protein phosphatase, PP2A, catalytic
subunit
Length = 306
Score = 529 bits (1362), Expect = e-151
Identities = 240/304 (78%), Positives = 271/304 (88%)
Query: 10 STHDLDEQISQLMQCKPLSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDL 69
S DLD QI QLM+CKPLSE V+ LC++A+ IL+EE NVQPVK PVT+CGDIHGQF+DL
Sbjct: 3 SNGDLDRQIEQLMECKPLSEADVRTLCDQARAILVEEYNVQPVKCPVTVCGDIHGQFYDL 62
Query: 70 AELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQ 129
ELFRIGG PDTNYLFMGDYVDRGYYSVETV+LLVALKVRY R+TILRGNHESRQITQ
Sbjct: 63 IELFRIGGNAPDTNYLFMGDYVDRGYYSVETVSLLVALKVRYRDRLTILRGNHESRQITQ 122
Query: 130 VYGFYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDR 189
VYGFYDECLRKYGNANVWK FTDLFD+ PLTAL+ES++FCLHGGLSPS++TLDNIR+ DR
Sbjct: 123 VYGFYDECLRKYGNANVWKYFTDLFDYLPLTALIESQVFCLHGGLSPSLDTLDNIRSLDR 182
Query: 190 VQEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQLVM 249
+QEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDI+ QFNH N L LI+RAHQLVM
Sbjct: 183 IQEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDIAAQFNHNNGLSLISRAHQLVM 242
Query: 250 DGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAPRRGEPDVTRRTP 309
+GFNW ++ VVT+FSAPNYCYRCGNMA+ILE+ + F+QF+PAPR+ EPD TR+TP
Sbjct: 243 EGFNWCQDKNVVTVFSAPNYCYRCGNMAAILEIGENMEQNFLQFDPAPRQVEPDTTRKTP 302
Query: 310 DYFL 313
DYFL
Sbjct: 303 DYFL 306
>At1g59830 unknown protein
Length = 306
Score = 528 bits (1360), Expect = e-150
Identities = 240/301 (79%), Positives = 269/301 (88%)
Query: 13 DLDEQISQLMQCKPLSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDLAEL 72
DLD QI QLM+CKPL E VK LC++AK IL+EE NVQPVK PVT+CGDIHGQF+DL EL
Sbjct: 6 DLDRQIEQLMECKPLGEADVKILCDQAKAILVEEYNVQPVKCPVTVCGDIHGQFYDLIEL 65
Query: 73 FRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYG 132
FRIGG PDTNYLFMGDYVDRGYYSVETV+LLVALKVRY R+TILRGNHESRQITQVYG
Sbjct: 66 FRIGGNAPDTNYLFMGDYVDRGYYSVETVSLLVALKVRYRDRLTILRGNHESRQITQVYG 125
Query: 133 FYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDRVQE 192
FYDECLRKYGNANVWK FTDLFD+ PLTAL+ES++FCLHGGLSPS++TLDNIR+ DR+QE
Sbjct: 126 FYDECLRKYGNANVWKYFTDLFDYLPLTALIESQVFCLHGGLSPSLDTLDNIRSLDRIQE 185
Query: 193 VPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQLVMDGF 252
VPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDI+ QFNH N L LI+RAHQLVM+G+
Sbjct: 186 VPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDIATQFNHNNGLSLISRAHQLVMEGY 245
Query: 253 NWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAPRRGEPDVTRRTPDYF 312
NW E+ VVT+FSAPNYCYRCGNMA+ILE+ + F+QF+PAPR+ EPD TR+TPDYF
Sbjct: 246 NWCQEKNVVTVFSAPNYCYRCGNMAAILEIGEKMEQNFLQFDPAPRQVEPDTTRKTPDYF 305
Query: 313 L 313
L
Sbjct: 306 L 306
>At4g26720 phosphoprotein phosphatase (PPX-1)
Length = 305
Score = 427 bits (1098), Expect = e-120
Identities = 200/304 (65%), Positives = 246/304 (80%), Gaps = 4/304 (1%)
Query: 13 DLDEQISQLMQCKPLSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDLAEL 72
DLD QI QL +C+PLSE +VK LC KA EIL+EESNVQ V +PVT+CGDIHGQF+D+ EL
Sbjct: 3 DLDRQIGQLKRCEPLSESEVKALCLKAMEILVEESNVQRVDAPVTLCGDIHGQFYDMMEL 62
Query: 73 FRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYG 132
F++GG CP TNYLFMGD+VDRGYYSVET LL+ALKVRY RIT++RGNHESRQITQVYG
Sbjct: 63 FKVGGDCPKTNYLFMGDFVDRGYYSVETFLLLLALKVRYPDRITLIRGNHESRQITQVYG 122
Query: 133 FYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDRVQE 192
FYDECLRKYG++NVW+ TD+FD+ L+A+VE++IFC+HGGLSP+I TLD IR DR QE
Sbjct: 123 FYDECLRKYGSSNVWRYCTDIFDYMSLSAVVENKIFCVHGGLSPAIMTLDQIRTIDRKQE 182
Query: 193 VPHEGPMCDLLWSDPDDRC-GWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQLVMDG 251
VPH+G MCDLLWSDP+D GWG+SPRGAG+ FG + FNH+N++ IARAHQLVM+G
Sbjct: 183 VPHDGAMCDLLWSDPEDIVDGWGLSPRGAGFLFGGSVVTSFNHSNNIDYIARAHQLVMEG 242
Query: 252 FNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAPR--RGEPDVTRRTP 309
+ W + ++VT++SAPNYCYRCGN+ASILE+D+ F F+ A + RG P + P
Sbjct: 243 YKWMFDSQIVTVWSAPNYCYRCGNVASILELDENLNKEFRVFDAAQQDSRG-PPAKKPAP 301
Query: 310 DYFL 313
DYFL
Sbjct: 302 DYFL 305
>At5g55260 protein phosphatase X isoform 2 (gb|AAB86419.1)
Length = 305
Score = 420 bits (1079), Expect = e-118
Identities = 194/303 (64%), Positives = 242/303 (79%), Gaps = 2/303 (0%)
Query: 13 DLDEQISQLMQCKPLSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDLAEL 72
DLD+QI QL +C+ L E +VK LC KA EIL+EESNVQ V +PVTICGDIHGQF+D+ EL
Sbjct: 3 DLDKQIEQLKRCEALKESEVKALCLKAMEILVEESNVQRVDAPVTICGDIHGQFYDMKEL 62
Query: 73 FRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYG 132
F++GG CP TNYLF+GD+VDRG+YSVET LL+ALKVRY RIT++RGNHESRQITQVYG
Sbjct: 63 FKVGGDCPKTNYLFLGDFVDRGFYSVETFLLLLALKVRYPDRITLIRGNHESRQITQVYG 122
Query: 133 FYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDRVQE 192
FYDECLRKYG+ NVW+ TD+FD+ L+ALVE++IFC+HGGLSP+I TLD IR DR QE
Sbjct: 123 FYDECLRKYGSVNVWRYCTDIFDYLSLSALVENKIFCVHGGLSPAIMTLDQIRAIDRKQE 182
Query: 193 VPHEGPMCDLLWSDPDDRC-GWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQLVMDG 251
VPH+G MCDLLWSDP+D GWG+SPRGAG+ FG + FNH+N++ I RAHQLVM+G
Sbjct: 183 VPHDGAMCDLLWSDPEDIVDGWGLSPRGAGFLFGGSVVTSFNHSNNIDYICRAHQLVMEG 242
Query: 252 FNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAPRRGEPDVTRR-TPD 310
+ W ++VT++SAPNYCYRCGN+A+ILE+D+ F F+ AP+ + ++ PD
Sbjct: 243 YKWMFNSQIVTVWSAPNYCYRCGNVAAILELDENLNKEFRVFDAAPQESRGALAKKPAPD 302
Query: 311 YFL 313
YFL
Sbjct: 303 YFL 305
>At3g19980 serine/threonine protein phosphatase, putative
Length = 303
Score = 370 bits (949), Expect = e-103
Identities = 166/273 (60%), Positives = 213/273 (77%), Gaps = 1/273 (0%)
Query: 13 DLDEQISQLMQCKPLSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDLAEL 72
DLD+ IS++ + LSE +++ LCE KEIL+EESNVQPV SPVT+CGDIHGQFHDL +L
Sbjct: 2 DLDQWISKVKDGQHLSEDELQLLCEYVKEILIEESNVQPVNSPVTVCGDIHGQFHDLMKL 61
Query: 73 FRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYG 132
F+ GG PDTNY+FMGD+VDRGY S+E T+L+ LK RY IT+LRGNHESRQ+TQVYG
Sbjct: 62 FQTGGHVPDTNYIFMGDFVDRGYNSLEVFTILLLLKARYPANITLLRGNHESRQLTQVYG 121
Query: 133 FYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDRVQE 192
FYDEC RKYGNAN W+ TD+FD+ L+A+++ + C+HGGLSP + T+D IR +R E
Sbjct: 122 FYDECQRKYGNANAWRYCTDVFDYLTLSAIIDGTVLCVHGGLSPDVRTIDQIRLIERNCE 181
Query: 193 VPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQLVMDGF 252
+PHEGP CDL+WSDP+D W +SPRGAG+ FG ++ +FNH N L L+ RAHQLV +G
Sbjct: 182 IPHEGPFCDLMWSDPEDIETWAVSPRGAGWLFGSRVTTEFNHINKLDLVCRAHQLVQEGL 241
Query: 253 NWAHEQK-VVTIFSAPNYCYRCGNMASILEVDD 284
+ + K +VT++SAPNYCYRCGN+ASIL +D
Sbjct: 242 KYMFQDKGLVTVWSAPNYCYRCGNVASILSFND 274
>At1g50370 phosphoprotein phosphatase
Length = 303
Score = 367 bits (941), Expect = e-102
Identities = 164/273 (60%), Positives = 214/273 (78%), Gaps = 1/273 (0%)
Query: 13 DLDEQISQLMQCKPLSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDLAEL 72
DLD+ IS++ + LSE +++ LCE KEIL+EESNVQPV SPVT+CGDIHGQFHDL +L
Sbjct: 2 DLDQWISKVKDGQHLSEDELQLLCEYVKEILIEESNVQPVNSPVTVCGDIHGQFHDLMKL 61
Query: 73 FRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYG 132
F+ GG P+TNY+FMGD+VDRGY S+E T+L+ LK R+ IT+LRGNHESRQ+TQVYG
Sbjct: 62 FQTGGHVPETNYIFMGDFVDRGYNSLEVFTILLLLKARHPANITLLRGNHESRQLTQVYG 121
Query: 133 FYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDRVQE 192
FYDEC RKYGNAN W+ TD+FD+ L+A+++ + C+HGGLSP + T+D IR +R E
Sbjct: 122 FYDECQRKYGNANAWRYCTDVFDYLTLSAIIDGTVLCVHGGLSPDVRTIDQIRLIERNCE 181
Query: 193 VPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQLVMDGF 252
+PHEGP CDL+WSDP+D W +SPRGAG+ FG ++ +FNH N+L L+ RAHQLV +G
Sbjct: 182 IPHEGPFCDLMWSDPEDIETWAVSPRGAGWLFGSRVTTEFNHINNLDLVCRAHQLVQEGL 241
Query: 253 NWAHEQK-VVTIFSAPNYCYRCGNMASILEVDD 284
+ + K +VT++SAPNYCYRCGN+ASIL +D
Sbjct: 242 KYMFQDKGLVTVWSAPNYCYRCGNVASILSFND 274
>At2g29400 phosphoprotein phosphatase, type 1 catalytic subunit
Length = 318
Score = 289 bits (739), Expect = 2e-78
Identities = 127/273 (46%), Positives = 189/273 (68%), Gaps = 1/273 (0%)
Query: 27 LSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDLAELFRIGGKCPDTNYLF 86
LSE ++++LC +KEI +++ N+ +++P+ ICGDIHGQ+ DL LF GG P+ NYLF
Sbjct: 43 LSEGEIRQLCAVSKEIFLQQPNLLELEAPIKICGDIHGQYSDLLRLFEYGGFPPEANYLF 102
Query: 87 MGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYGFYDECLRKYGNANV 146
+GDYVDRG S+ET+ LL+A K++Y + +LRGNHES I ++YGFYDEC R++ N +
Sbjct: 103 LGDYVDRGKQSLETICLLLAYKIKYPENFFLLRGNHESASINRIYGFYDECKRRF-NVRL 161
Query: 147 WKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDRVQEVPHEGPMCDLLWSD 206
WK FTD F+ P+ AL++ I C+HGG+SP +++LD IRN R ++P G +CDLLWSD
Sbjct: 162 WKIFTDCFNCLPVAALIDDRILCMHGGISPELKSLDQIRNIARPMDIPESGLVCDLLWSD 221
Query: 207 PDDRCGWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQLVMDGFNWAHEQKVVTIFSA 266
P GWG++ RG YTFG D +F + + LI RAHQ+V DG+ + E+++VT+FSA
Sbjct: 222 PSGDVGWGMNDRGVSYTFGADKVAEFLEKHDMDLICRAHQVVEDGYEFFAERQLVTVFSA 281
Query: 267 PNYCYRCGNMASILEVDDCKGHTFIQFEPAPRR 299
PNYC N +++ +D+ +F +P+ ++
Sbjct: 282 PNYCGEFDNAGAMMSIDESLMCSFQILKPSEKK 314
>At1g64040 phosphoprotein phosphatase 1
Length = 322
Score = 288 bits (738), Expect = 2e-78
Identities = 139/296 (46%), Positives = 194/296 (64%), Gaps = 10/296 (3%)
Query: 14 LDEQISQLMQCK--------PLSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQ 65
+D+ I +L+ K L+E ++K LC AK+I + + N+ +++P+ ICGD HGQ
Sbjct: 6 VDDVIKRLLGAKNGKTTKQVQLTEAEIKHLCSTAKQIFLTQPNLLELEAPIKICGDTHGQ 65
Query: 66 FHDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESR 125
F DL LF GG P NYLF+GDYVDRG SVET+ LL+A K++Y + +LRGNHE
Sbjct: 66 FSDLLRLFEYGGYPPAANYLFLGDYVDRGKQSVETICLLLAYKIKYKENFFLLRGNHECA 125
Query: 126 QITQVYGFYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIR 185
I ++YGFYDEC ++Y + VWK FTD F+ P+ AL++ +I C+HGGLSP ++ LD IR
Sbjct: 126 SINRIYGFYDECKKRY-SVRVWKIFTDCFNCLPVAALIDEKILCMHGGLSPELKHLDEIR 184
Query: 186 NFDRVQEVPHEGPMCDLLWSDPD-DRCGWGISPRGAGYTFGQDISEQFNHTNSLKLIARA 244
N R ++P G +CDLLWSDPD D GWG + RG YTFG D E+F T+ L LI RA
Sbjct: 185 NIPRPADIPDHGLLCDLLWSDPDKDIEGWGENDRGVSYTFGADKVEEFLQTHDLDLICRA 244
Query: 245 HQLVMDGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAPRRG 300
HQ+V DG+ + +++VTIFSAPNYC N +++ VDD +F + + ++G
Sbjct: 245 HQVVEDGYEFFANRQLVTIFSAPNYCGEFDNAGAMMSVDDSLTCSFQILKASEKKG 300
>At4g11240 unknown protein
Length = 322
Score = 286 bits (731), Expect = 1e-77
Identities = 133/284 (46%), Positives = 191/284 (66%), Gaps = 2/284 (0%)
Query: 27 LSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDLAELFRIGGKCPDTNYLF 86
++E ++++LC +KE+ + + N+ +++P+ ICGD+HGQF DL LF GG P NYLF
Sbjct: 27 ITETEIRQLCLASKEVFLSQPNLLELEAPIKICGDVHGQFPDLLRLFEYGGYPPAANYLF 86
Query: 87 MGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYGFYDECLRKYGNANV 146
+GDYVDRG S+ET+ LL+A KV+Y +LRGNHE I +VYGFYDEC R+Y N +
Sbjct: 87 LGDYVDRGKQSIETICLLLAYKVKYKFNFFLLRGNHECASINRVYGFYDECKRRY-NVRL 145
Query: 147 WKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDRVQEVPHEGPMCDLLWSD 206
WKTFT+ F+ P++AL++ +I C+HGGLSP I++LD+IR R +VP +G +CDLLW+D
Sbjct: 146 WKTFTECFNCLPVSALIDDKILCMHGGLSPDIKSLDDIRRIPRPIDVPDQGILCDLLWAD 205
Query: 207 PDDRC-GWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQLVMDGFNWAHEQKVVTIFS 265
PD GWG + RG YTFG D +F T+ L LI RAHQ+V DG+ + ++++VTIFS
Sbjct: 206 PDREIQGWGENDRGVSYTFGADKVAEFLQTHDLDLICRAHQVVEDGYEFFAKRQLVTIFS 265
Query: 266 APNYCYRCGNMASILEVDDCKGHTFIQFEPAPRRGEPDVTRRTP 309
APNYC N +++ VDD +F + + ++G P
Sbjct: 266 APNYCGEFDNAGALMSVDDSLTCSFQILKASEKKGRFGFNNNVP 309
>At2g39840 putative serine/threonine protein phosphatase PP1 isozyme
4 (TOPP4)
Length = 321
Score = 283 bits (723), Expect = 1e-76
Identities = 133/301 (44%), Positives = 198/301 (65%), Gaps = 7/301 (2%)
Query: 5 SLPSESTHDLDEQISQLMQCKP-----LSEQQVKELCEKAKEILMEESNVQPVKSPVTIC 59
++ S D+ +++++ +P LSE ++K+LC A++I +++ N+ +++P+ IC
Sbjct: 13 AIDSAVLDDIIRRLTEVRLARPGKQVQLSEAEIKQLCTTARDIFLQQPNLLELEAPIKIC 72
Query: 60 GDIHGQFHDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILR 119
GDIHGQ+ DL LF GG P NYLF+GDYVDRG S+ET+ LL+A K++Y +LR
Sbjct: 73 GDIHGQYSDLLRLFEYGGFPPSANYLFLGDYVDRGKQSLETICLLLAYKIKYPGNFFLLR 132
Query: 120 GNHESRQITQVYGFYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIE 179
GNHE I ++YGFYDEC R++ N VWK FTD F+ P+ AL++ +I C+HGGLSP ++
Sbjct: 133 GNHECASINRIYGFYDECKRRF-NVRVWKVFTDCFNCLPVAALIDDKILCMHGGLSPDLD 191
Query: 180 TLDNIRNFDRVQEVPHEGPMCDLLWSDP-DDRCGWGISPRGAGYTFGQDISEQFNHTNSL 238
LD IRN R +P G +CDLLWSDP D GWG++ RG YTFG D +F + L
Sbjct: 192 HLDEIRNLPRPTMIPDTGLLCDLLWSDPGKDVKGWGMNDRGVSYTFGPDKVSEFLTKHDL 251
Query: 239 KLIARAHQLVMDGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAPR 298
L+ RAHQ+V DG+ + ++++VT+FSAPNYC N +++ VD+ +F +PA +
Sbjct: 252 DLVCRAHQVVEDGYEFFADRQLVTVFSAPNYCGEFDNAGAMMSVDENLMCSFQILKPAEK 311
Query: 299 R 299
+
Sbjct: 312 K 312
>At5g27840 TOPP8 serine/threonine protein phosphatase type one
Length = 318
Score = 281 bits (720), Expect = 2e-76
Identities = 136/290 (46%), Positives = 191/290 (64%), Gaps = 7/290 (2%)
Query: 14 LDEQISQLMQCK-----PLSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHD 68
LD+ I +L++ K LSE ++++LC A++I + + N+ + +P+ ICGDIHGQ+ D
Sbjct: 14 LDDIIRRLLEGKGGKQVQLSESEIRQLCFNARQIFLSQPNLLDLHAPIRICGDIHGQYQD 73
Query: 69 LAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQIT 128
L LF GG P NYLF+GDYVDRG S+ET+ LL+A K+RY +I +LRGNHE +I
Sbjct: 74 LLRLFEYGGYPPSANYLFLGDYVDRGKQSLETICLLLAYKIRYPSKIYLLRGNHEDAKIN 133
Query: 129 QVYGFYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFD 188
++YGFYDEC R++ N +WK FTD F+ P+ AL++ +I C+HGGLSP ++ L+ IR
Sbjct: 134 RIYGFYDECKRRF-NVRLWKVFTDCFNCLPVAALIDEKILCMHGGLSPDLDNLNQIREIQ 192
Query: 189 RVQEVPHEGPMCDLLWSDPDDRC-GWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQL 247
R E+P G +CDLLWSDPD + GW S RG TFG D +F N L LI R HQ+
Sbjct: 193 RPIEIPDSGLLCDLLWSDPDQKIEGWADSDRGISCTFGADKVAEFLDKNDLDLICRGHQV 252
Query: 248 VMDGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAP 297
V DG+ + ++++VTIFSAPNY N ++L VD+ +F +PAP
Sbjct: 253 VEDGYEFFAKRRLVTIFSAPNYGGEFDNAGALLSVDESLVCSFEIMKPAP 302
>At3g05580 putative serine/threonine protein phosphatase type one
Length = 318
Score = 281 bits (719), Expect = 3e-76
Identities = 137/290 (47%), Positives = 189/290 (64%), Gaps = 7/290 (2%)
Query: 14 LDEQISQLMQCK-----PLSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHD 68
LD+ I +L++ K LSE ++++LC A++I + + N+ + +P+ ICGDIHGQ+ D
Sbjct: 14 LDDIIRRLLEGKGGKQVQLSEVEIRQLCVNARQIFLSQPNLLELHAPIRICGDIHGQYQD 73
Query: 69 LAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQIT 128
L LF GG P NYLF+GDYVDRG S+ET+ LL+A K+RY +I +LRGNHE +I
Sbjct: 74 LLRLFEYGGYPPSANYLFLGDYVDRGKQSLETICLLLAYKIRYPSKIFLLRGNHEDAKIN 133
Query: 129 QVYGFYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFD 188
++YGFYDEC R++ N +WK FTD F+ P+ AL++ +I C+HGGLSP +E L IR
Sbjct: 134 RIYGFYDECKRRF-NVRLWKIFTDCFNCLPVAALIDEKILCMHGGLSPELENLGQIREIQ 192
Query: 189 RVQEVPHEGPMCDLLWSDPDDRC-GWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQL 247
R E+P G +CDLLWSDPD + GW S RG TFG D+ F N L LI R HQ+
Sbjct: 193 RPTEIPDNGLLCDLLWSDPDQKNEGWTDSDRGISCTFGADVVADFLDKNDLDLICRGHQV 252
Query: 248 VMDGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAP 297
V DG+ + ++++VTIFSAPNY N ++L VD +F +PAP
Sbjct: 253 VEDGYEFFAKRRLVTIFSAPNYGGEFDNAGALLSVDQSLVCSFEILKPAP 302
>At5g59160 phosphoprotein phosphatase 1 catalytic chain
Length = 312
Score = 278 bits (710), Expect = 4e-75
Identities = 131/305 (42%), Positives = 197/305 (63%), Gaps = 8/305 (2%)
Query: 2 GANSLPSESTHDLDEQISQLMQCKP------LSEQQVKELCEKAKEILMEESNVQPVKSP 55
G S+ + D+ ++ KP L+E ++++LC ++EI +++ N+ +++P
Sbjct: 5 GQGSMDPAALDDIIRRLLDYRNPKPGTKQAMLNESEIRQLCIVSREIFLQQPNLLELEAP 64
Query: 56 VTICGDIHGQFHDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRI 115
+ ICGDIHGQ+ DL LF GG P NYLF+GDYVDRG S+ET+ LL+A K++Y +
Sbjct: 65 IKICGDIHGQYSDLLRLFEYGGFPPTANYLFLGDYVDRGKQSLETICLLLAYKIKYPENF 124
Query: 116 TILRGNHESRQITQVYGFYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLS 175
+LRGNHE I ++YGFYDEC R++ + +WK FTD F+ P+ A+++ +I C+HGGLS
Sbjct: 125 FLLRGNHECASINRIYGFYDECKRRF-SVRLWKVFTDSFNCLPVAAVIDDKILCMHGGLS 183
Query: 176 PSIETLDNIRNFDRVQEVPHEGPMCDLLWSDPD-DRCGWGISPRGAGYTFGQDISEQFNH 234
P + ++ I+N R +VP G +CDLLWSDP D GWG++ RG YTFG D +F
Sbjct: 184 PDLTNVEQIKNIKRPTDVPDSGLLCDLLWSDPSKDVKGWGMNDRGVSYTFGPDKVAEFLI 243
Query: 235 TNSLKLIARAHQLVMDGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFE 294
N + LI RAHQ+V DG+ + ++++VTIFSAPNYC N +++ VD+ +F +
Sbjct: 244 KNDMDLICRAHQVVEDGYEFFADRQLVTIFSAPNYCGEFDNAGAMMSVDESLMCSFQILK 303
Query: 295 PAPRR 299
PA R+
Sbjct: 304 PADRK 308
>At5g43380 protein phosphatase 1 catalytic subunit
Length = 331
Score = 273 bits (698), Expect = 9e-74
Identities = 127/259 (49%), Positives = 179/259 (69%), Gaps = 2/259 (0%)
Query: 27 LSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDLAELFRIGGKCPDTNYLF 86
LSE ++K+LC +++I + + N+ +++PV ICGDIHGQ+ DL LF GG P++NYLF
Sbjct: 26 LSETEIKQLCFVSRDIFLRQPNLLELEAPVKICGDIHGQYPDLLRLFEHGGYPPNSNYLF 85
Query: 87 MGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYGFYDECLRKYGNANV 146
+GDYVDRG S+ET+ LL+A K+++ + +LRGNHES I ++YGFYDEC R++ + +
Sbjct: 86 LGDYVDRGKQSLETICLLLAYKIKFPENFFLLRGNHESASINRIYGFYDECKRRF-SVKI 144
Query: 147 WKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDRVQEVPHEGPMCDLLWSD 206
W+ FTD F+ P+ AL++ IFC+HGGLSP + +L IR+ R ++P G +CDLLWSD
Sbjct: 145 WRIFTDCFNCLPVAALIDERIFCMHGGLSPELLSLRQIRDIRRPTDIPDRGLLCDLLWSD 204
Query: 207 PD-DRCGWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQLVMDGFNWAHEQKVVTIFS 265
PD D GWG + RG YTFG DI F L LI RAHQ+V DGF + +++VTIFS
Sbjct: 205 PDKDVRGWGPNDRGVSYTFGSDIVSGFLKRLDLDLICRAHQVVEDGFEFFANKQLVTIFS 264
Query: 266 APNYCYRCGNMASILEVDD 284
APNYC N +++ V +
Sbjct: 265 APNYCGEFDNAGAMMSVSE 283
>At3g46820 phosphoprotein phosphatase
Length = 312
Score = 272 bits (695), Expect = 2e-73
Identities = 124/274 (45%), Positives = 185/274 (67%), Gaps = 2/274 (0%)
Query: 27 LSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDLAELFRIGGKCPDTNYLF 86
L++ ++++LC ++EI +++ + + +PV ICGDIHGQ+ DL LF GG P NYLF
Sbjct: 36 LNDSEIRQLCFVSREIFLQQPCLLELAAPVKICGDIHGQYSDLLRLFEYGGFPPAANYLF 95
Query: 87 MGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYGFYDECLRKYGNANV 146
+GDYVDRG S+ET+ LL+A K++Y + +LRGNHE I ++YGFYDEC R++ N +
Sbjct: 96 LGDYVDRGKQSLETICLLLAYKIKYPENFFLLRGNHECASINRIYGFYDECKRRF-NVKL 154
Query: 147 WKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDRVQEVPHEGPMCDLLWSD 206
WK FTD F+ P+ A+++ +I C+HGGLSP + ++ I+N +R +VP G +CDLLWSD
Sbjct: 155 WKVFTDTFNCLPVAAVIDEKILCMHGGLSPELINVEQIKNIERPTDVPDAGLLCDLLWSD 214
Query: 207 PD-DRCGWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQLVMDGFNWAHEQKVVTIFS 265
P D GWG++ RG YTFG D +F N + L+ RAHQ+V DG+ + ++++VT+FS
Sbjct: 215 PSKDVKGWGMNDRGVSYTFGADKVAEFLIKNDMDLVCRAHQVVEDGYEFFADRQLVTMFS 274
Query: 266 APNYCYRCGNMASILEVDDCKGHTFIQFEPAPRR 299
APNYC N +++ VD+ +F +P RR
Sbjct: 275 APNYCGEFDNAGALMSVDESLMCSFQILKPVDRR 308
>At4g03080 phospho-ser/thr phosphatase like protein
Length = 881
Score = 208 bits (529), Expect = 3e-54
Identities = 111/270 (41%), Positives = 159/270 (58%), Gaps = 14/270 (5%)
Query: 27 LSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDLAELF------RIGGKCP 80
L +V ELC A++I M E V +K+P+ + GD+HGQF DL LF G
Sbjct: 550 LDSYEVGELCYAAEQIFMHEQTVLQLKAPIKVFGDLHGQFGDLMRLFDEYGFPSTAGDIT 609
Query: 81 DTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYGFYDECLRK 140
+YLF+GDYVDRG +S+ET+TLL+ALK+ Y + + ++RGNHE+ I ++GF EC+ +
Sbjct: 610 YIDYLFLGDYVDRGQHSLETITLLLALKIEYPENVHLIRGNHEAADINALFGFRLECIER 669
Query: 141 YGNAN---VWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDR-VQEVPHE 196
G + W F LF++ PL AL+E++I C+HGG+ SI T++ I +R +
Sbjct: 670 MGENDGIWAWTRFNQLFNYLPLAALIENKIICMHGGIGRSISTVEQIEKIERPITMDAGS 729
Query: 197 GPMCDLLWSDPDDRCG-WGISPRGAG---YTFGQDISEQFNHTNSLKLIARAHQLVMDGF 252
+ DLLWSDP + G+ P G TFG D +F N L+LI RAH+ VMDGF
Sbjct: 730 LVLMDLLWSDPTENDSIEGLRPNARGPGLVTFGPDRVTEFCKRNKLQLIIRAHECVMDGF 789
Query: 253 NWAHEQKVVTIFSAPNYCYRCGNMASILEV 282
+ +++T+FSA NYC N +IL V
Sbjct: 790 ERFAQGQLITLFSATNYCGTANNAGAILVV 819
>At2g27210 putative phosphoprotein phosphatase
Length = 715
Score = 202 bits (513), Expect = 2e-52
Identities = 110/265 (41%), Positives = 155/265 (57%), Gaps = 16/265 (6%)
Query: 31 QVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDLAELFRIGGKCPDT-------N 83
++ +LC+ A+ I E V +K+P+ I GD+HGQF DL LF G P T +
Sbjct: 408 EIADLCDSAERIFSSEPTVLQLKAPIKIFGDLHGQFGDLMRLFDEYGS-PSTAGDISYID 466
Query: 84 YLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYGFYDECLRKYGN 143
YLF+GDYVDRG +S+ET+TLL+ALKV Y + ++RGNHE+ I ++GF EC+ + G
Sbjct: 467 YLFLGDYVDRGQHSLETITLLLALKVEYQHNVHLIRGNHEAADINALFGFRIECIERMGE 526
Query: 144 AN---VWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDR-VQEVPHEGPM 199
+ VW LF++ PL AL+E +I C+HGG+ SI ++ I N R + +
Sbjct: 527 RDGIWVWHRINRLFNWLPLAALIEKKIICMHGGIGRSINHVEQIENIQRPITMEAGSIVL 586
Query: 200 CDLLWSDPDDRCG-WGISPRGAG---YTFGQDISEQFNHTNSLKLIARAHQLVMDGFNWA 255
DLLWSDP + G+ P G TFG D +F + N L+LI RAH+ VMDGF
Sbjct: 587 MDLLWSDPTENDSVEGLRPNARGPGLVTFGPDRVMEFCNNNDLQLIVRAHECVMDGFERF 646
Query: 256 HEQKVVTIFSAPNYCYRCGNMASIL 280
+ ++T+FSA NYC N +IL
Sbjct: 647 AQGHLITLFSATNYCGTANNAGAIL 671
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.139 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,604,557
Number of Sequences: 26719
Number of extensions: 328253
Number of successful extensions: 874
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 796
Number of HSP's gapped (non-prelim): 33
length of query: 313
length of database: 11,318,596
effective HSP length: 99
effective length of query: 214
effective length of database: 8,673,415
effective search space: 1856110810
effective search space used: 1856110810
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0029b.5


