

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0029b.2
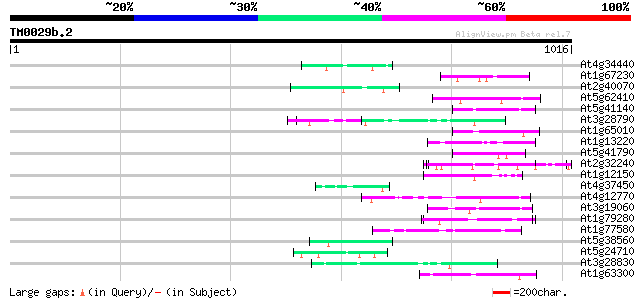
(1016 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g34440 serine/threonine protein kinase - like 51 4e-06
At1g67230 unknown protein 51 4e-06
At2g40070 En/Spm-like transposon protein 50 5e-06
At5g62410 chromosomal protein - like 50 6e-06
At5g41140 putative protein 50 6e-06
At3g28790 unknown protein 50 8e-06
At1g65010 hypothetical protein 50 8e-06
At1g13220 putative nuclear matrix constituent protein 49 2e-05
At5g41790 myosin heavy chain-like protein 48 3e-05
At2g32240 putative myosin heavy chain 47 4e-05
At1g12150 hypothetical protein 47 7e-05
At4g37450 arabinogalactan protein AGP18 46 9e-05
At4g12770 auxilin-like protein 46 9e-05
At3g19060 hypothetical protein 46 9e-05
At1g79280 hypothetical protein 46 9e-05
At1g77580 unknown protein 46 9e-05
At5g38560 putative protein 45 2e-04
At5g24710 unknown protein 45 2e-04
At3g28830 hypothetical protein 45 2e-04
At1g63300 hypothetical protein 45 2e-04
>At4g34440 serine/threonine protein kinase - like
Length = 670
Score = 50.8 bits (120), Expect = 4e-06
Identities = 43/181 (23%), Positives = 65/181 (35%), Gaps = 24/181 (13%)
Query: 529 AENQAESTKAPKRRRLTKTSSGAGTSNPGAQPTTAAAPKGKNVA------EASIAAATEP 582
A++ +S+ AP+ T S+G SN + PT ++P +++ AS + P
Sbjct: 2 ADSPVDSSPAPETSNGTPPSNGTSPSNESSPPTPPSSPPPSSISAPPPDISASFSPPPAP 61
Query: 583 TAVPASTPASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEKEKENETPKSPPH 642
S P S ++ V A S N + A TP+ P
Sbjct: 62 PTQETSPPTSPSSSPPVVANPS-----PQTPENPSPPAPEGSTPVTPPAPPQTPSNQSPE 116
Query: 643 QEAPPSPPPTRDA-----------GSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGS 691
+ PPSP D GS PSPP G ++ + S I +P G
Sbjct: 117 RPTPPSPGANDDRNRTNGGNNNRDGSTPSPPSSGNRTSGDGGSPSPPRSI--SPPQNSGD 174
Query: 692 S 692
S
Sbjct: 175 S 175
>At1g67230 unknown protein
Length = 1132
Score = 50.8 bits (120), Expect = 4e-06
Identities = 51/188 (27%), Positives = 82/188 (43%), Gaps = 32/188 (17%)
Query: 780 DKLATQAGKDKVYADKMIGTAGIKIGELED-------QLALMKEEADELDASLQACKKEK 832
DK+ Q GK+ A K I A + + +LED LAL ++E D L S++ +E
Sbjct: 264 DKIIKQKGKELEEAQKKIDAANLAVKKLEDDVSSRIKDLALREQETDVLKKSIETKAREL 323
Query: 833 EQAEKDLIARGEALIAK-----ESELAVLCAELEL------------VKKALAEQEKKSA 875
+ ++ L AR + + + +++L E EL +K +AE EK+ A
Sbjct: 324 QALQEKLEAREKMAVQQLVDEHQAKLDSTQREFELEMEQKRKSIDDSLKSKVAEVEKREA 383
Query: 876 ESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAE---IASQLAKIKSLEDELATEK 932
E ME + + + + E H E D I+ + +KS E L TEK
Sbjct: 384 E-----WKHMEEKVAKREQALDRKLEKHKEKENDFDLRLKGISGREKALKSEEKALETEK 438
Query: 933 AKAIEARE 940
K +E +E
Sbjct: 439 KKLLEDKE 446
Score = 35.8 bits (81), Expect = 0.13
Identities = 32/146 (21%), Positives = 69/146 (46%), Gaps = 10/146 (6%)
Query: 803 KIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEAL--IAKESE-----LAV 855
+I + Q L+++EA++L A ++ +KE E+ ++ G L I + E + +
Sbjct: 496 QIEKCRSQQELLQKEAEDLKAQRESFEKEWEELDERKAKIGNELKNITDQKEKLERHIHL 555
Query: 856 LCAELELVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIA 915
L+ K+A E ++ E+L +AK+ M+ + K E+ L +I
Sbjct: 556 EEERLKKEKQAANENMERELETLEVAKASFAETMEYERSMLSKKAESERSQLL---HDIE 612
Query: 916 SQLAKIKSLEDELATEKAKAIEAREQ 941
+ K++S + EK + ++A+++
Sbjct: 613 MRKRKLESDMQTILEEKERELQAKKK 638
Score = 30.8 bits (68), Expect = 4.1
Identities = 52/276 (18%), Positives = 113/276 (40%), Gaps = 50/276 (18%)
Query: 716 VIEKEVLSRGLNDTKEETLACLLRAGCIFAHTFEKFNAANVEAERLKAESAKHQEAAAAW 775
V + +++ + D E+ + + G +K +AAN+ ++L+ + + + A
Sbjct: 246 VAKSQMIVKQREDRANESDKIIKQKGKELEEAQKKIDAANLAVKKLEDDVSSRIKDLALR 305
Query: 776 EKRFDKLA----TQA-----------GKDKVYADKMIGTAGIKIGELEDQLAL-MKEEAD 819
E+ D L T+A ++K+ +++ K+ + + L M+++
Sbjct: 306 EQETDVLKKSIETKARELQALQEKLEAREKMAVQQLVDEHQAKLDSTQREFELEMEQKRK 365
Query: 820 ELDASLQA--CKKEKEQAE----KDLIARGEALI--------AKESELAVLCAELELVKK 865
+D SL++ + EK +AE ++ +A+ E + KE++ + + +K
Sbjct: 366 SIDDSLKSKVAEVEKREAEWKHMEEKVAKREQALDRKLEKHKEKENDFDLRLKGISGREK 425
Query: 866 ALAEQEKKSAESLALAKSDMEAVM-------------QATSEEIKK------ATETHAEA 906
AL +EK D E ++ QA EI K TE
Sbjct: 426 ALKSEEKALETEKKKLLEDKEIILNLKALVEKVSGENQAQLSEINKEKDELRVTEEERSE 485
Query: 907 LATKDAEIASQLAKIKSLEDELATEKAKAIEAREQA 942
E+ Q+ K +S + EL ++A+ ++A+ ++
Sbjct: 486 YLRLQTELKEQIEKCRS-QQELLQKEAEDLKAQRES 520
>At2g40070 En/Spm-like transposon protein
Length = 510
Score = 50.4 bits (119), Expect = 5e-06
Identities = 50/214 (23%), Positives = 85/214 (39%), Gaps = 23/214 (10%)
Query: 509 RMARFSPKFNTEGDAKKR---GGAENQAESTKAPKRRRLTKTSSGAGTSNPGAQPTTAAA 565
R SP ++ A +R G +T + LT S + S P ++ T ++A
Sbjct: 131 RQQTSSPGLSSSSGASRRPSSSGGPGSRPATPTGRSSTLTANSKSSRPSTPTSRATVSSA 190
Query: 566 PKGKNV-AEASIAAATEPTAVPASTPASAGATAAVAAES------SVGATAASAGVNATK 618
+ + ++++A T+PT + ST S+ A++ S G+ S TK
Sbjct: 191 TRPSLTNSRSTVSATTKPTPMSRSTSLSSSRLTPTASKPTTSTARSAGSVTRSTPSTTTK 250
Query: 619 AA--ASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPGAATT- 675
+A + S TP+ + TP S P+ PP++ +P + S A TT
Sbjct: 251 SAGPSRSTTPLSRSTARSSTPTS------RPTLPPSKTISRSSTPTRRPIASASAATTTA 304
Query: 676 ----SEAAQIEQAPAPEVGSSSYYNMLPNAIEPS 705
S+ APA + + S L A P+
Sbjct: 305 NPTISQIKPSSPAPAKPMPTPSKNPALSRAASPT 338
>At5g62410 chromosomal protein - like
Length = 1175
Score = 50.1 bits (118), Expect = 6e-06
Identities = 52/214 (24%), Positives = 106/214 (49%), Gaps = 20/214 (9%)
Query: 767 KHQEAAAAWEKRFDKLATQAGK-DKVYADKMIG-TAGIKIGELEDQLALMK-------EE 817
K +E AA ++RF +L+T + +K + + G ++G + LEDQL K E
Sbjct: 354 KSEEGAADLKQRFQELSTTLEECEKEHQGVLAGKSSGDEEKCLEDQLRDAKIAVGTAGTE 413
Query: 818 ADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKSAES 877
+L ++ C+KE ++ + L+++ E I E+EL ++E VKKAL +
Sbjct: 414 LKQLKTKIEHCEKELKERKSQLMSKLEEAIEVENELGARKNDVEHVKKALESIPYNEGQM 473
Query: 878 LALAK---SDMEAV------MQATSEEIKKATETHAEALATKD-AEIASQLAKIKSLEDE 927
AL K +++E V ++ S ++ T+++ + D +++ +AK+ ++D
Sbjct: 474 EALEKDRGAELEVVQRLEDKVRGLSAQLANFQFTYSDPVRNFDRSKVKGVVAKLIKVKDR 533
Query: 928 LATEKAKAIEAREQAADIALDNRERGFYLAKDQA 961
++ A + A + D+ +D+ + G L ++ A
Sbjct: 534 -SSMTALEVTAGGKLYDVVVDSEDTGKQLLQNGA 566
Score = 33.5 bits (75), Expect = 0.63
Identities = 50/243 (20%), Positives = 96/243 (38%), Gaps = 61/243 (25%)
Query: 759 ERLKAESAKHQEAAAAWEKRFDKLA--------TQAGKDKVYADKMIGTAGIKIGELEDQ 810
E+L+ E +++ + A D+L QA K + A +G K+G+++ +
Sbjct: 208 EKLRKEKSQYMQWANG-NAELDRLRRFCIAFEYVQAEKIRDNAVLGVGEMKAKLGKIDAE 266
Query: 811 LALMKEEADELDASLQACKKEKE----------------------QAEKDLIARGEALIA 848
+EE E + ++A + KE + L + + L+
Sbjct: 267 TEKTQEEIQEFEKQIKALTQAKEASMGGEVKTLSEKVDSLAQEMTRESSKLNNKEDTLLG 326
Query: 849 KESELAVLCAELELVKKALAEQE---KKSAESLALAKSDMEAVMQATSEEIKKATETHAE 905
++ + + +E +KK++ E+ KKS E A D++ Q S +++ + H
Sbjct: 327 EKENVEKIVHSIEDLKKSVKERAAAVKKSEEGAA----DLKQRFQELSTTLEECEKEHQG 382
Query: 906 ALATK--------------DAEIASQLA---------KIKSLEDELATEKAKAIEAREQA 942
LA K DA+IA A KI+ E EL K++ + E+A
Sbjct: 383 VLAGKSSGDEEKCLEDQLRDAKIAVGTAGTELKQLKTKIEHCEKELKERKSQLMSKLEEA 442
Query: 943 ADI 945
++
Sbjct: 443 IEV 445
Score = 31.2 bits (69), Expect = 3.1
Identities = 36/175 (20%), Positives = 74/175 (41%), Gaps = 10/175 (5%)
Query: 759 ERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEA 818
E L+ ++ +E A++ FD ++ K + G ++ +LE + +K +
Sbjct: 748 EELEEAKSQIKEKELAYKNCFDAVSKLENSIKDHDKNREG----RLKDLEKNIKTIKAQM 803
Query: 819 DELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELEL-VKKALAEQEKKSAES 877
L++ + EKE+ L+ EA+ ++S L LE + +E +++ A+
Sbjct: 804 QAASKDLKSHENEKEK----LVMEEEAMKQEQSSLESHLTSLETQISTLTSEVDEQRAKV 859
Query: 878 LALAKSDMEAVMQATSEEIK-KATETHAEALATKDAEIASQLAKIKSLEDELATE 931
AL K E++ + K K +T T + +L+ +K +L E
Sbjct: 860 DALQKIHDESLAELKLIHAKMKECDTQISGFVTDQEKCLQKLSDMKLERKKLENE 914
>At5g41140 putative protein
Length = 983
Score = 50.1 bits (118), Expect = 6e-06
Identities = 40/156 (25%), Positives = 75/156 (47%), Gaps = 10/156 (6%)
Query: 803 KIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELEL 862
K+ EL + L +E + A L+ K++KE DL + ++ E+ +L +LE
Sbjct: 693 KLNELSGKTDLKTKEMKRMSADLEYQKRQKEDVNADLT---HEITRRKDEIEILRLDLEE 749
Query: 863 VKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKA---TETHAEALATKDAEIAS--- 916
+K+ E E +E L + EAV+ A +++ A + +L+ ++EI +
Sbjct: 750 TRKSSMETEASLSEELQRIIDEKEAVITALKSQLETAIAPCDNLKHSLSNNESEIENLRK 809
Query: 917 QLAKIKSLEDELATEKAKAIEAREQAADIALDNRER 952
Q+ +++S E E E+ +E RE +AD +R
Sbjct: 810 QVVQVRS-ELEKKEEEMANLENREASADNITKTEQR 844
Score = 36.6 bits (83), Expect = 0.074
Identities = 54/232 (23%), Positives = 96/232 (41%), Gaps = 17/232 (7%)
Query: 700 NAIEPSEFLLAGLNRDVIEKEVLSRGLNDTKEETLACLLRAGCIFAHTFEKFNAANVEAE 759
+++E L L R + EKE + L E +A ++ + +
Sbjct: 753 SSMETEASLSEELQRIIDEKEAVITALKSQLETAIAPCDNLKHSLSNNESEIENLRKQVV 812
Query: 760 RLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEAD 819
++++E K +E A E R +A D + + +I +LE Q+ L KE A
Sbjct: 813 QVRSELEKKEEEMANLENR------EASADNITKTEQRSNED-RIKQLEGQIKL-KENA- 863
Query: 820 ELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKSAESLA 879
L+A K + EKDL R E L K +E++ E + + + E L
Sbjct: 864 -----LEASSKIFIEKEKDLKNRIEELQTKLNEVSQNSQETDETLQGPEAIAMQYTEVLP 918
Query: 880 LAKSD--MEAVMQATS-EEIKKATETHAEALATKDAEIASQLAKIKSLEDEL 928
L+KSD + V + S E ET + + + +EI+ + A+++ +L
Sbjct: 919 LSKSDNLQDLVNEVASLREQNGLMETELKEMQERYSEISLRFAEVEGERQQL 970
Score = 33.5 bits (75), Expect = 0.63
Identities = 35/155 (22%), Positives = 73/155 (46%), Gaps = 9/155 (5%)
Query: 803 KIGELEDQLALMKEEADE----LDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCA 858
+I ELE Q+ M+EE ++ + ++A + K + E+ I EAL + A +
Sbjct: 572 RIKELETQIKGMEEELEKQAQIFEGDIEAVTRAKVEQEQRAIEAEEALRKTRWKNASVAG 631
Query: 859 ELELVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQL 918
+++ K ++EQ S+ A K M+A+ + + E++ E L + E+
Sbjct: 632 KIQDEFKRISEQ--MSSTLAANEKVTMKAMTE--TRELRMQKRQLEELLMNANDELRVNR 687
Query: 919 AKIKSLEDELATE-KAKAIEAREQAADIALDNRER 952
+ ++ +EL+ + K E + +AD+ R++
Sbjct: 688 VEYEAKLNELSGKTDLKTKEMKRMSADLEYQKRQK 722
>At3g28790 unknown protein
Length = 608
Score = 49.7 bits (117), Expect = 8e-06
Identities = 84/395 (21%), Positives = 147/395 (36%), Gaps = 41/395 (10%)
Query: 519 TEGDAKKRGGAENQAESTKAPKRRRLTKTSSGAGTSNPGAQPTTAAAPKGKNVAEASIAA 578
TE +K A+ + S + T S +G S G+ T + P ++
Sbjct: 239 TEAGSKSSSSAKTKEVSGGSSGNTYKDTTGSSSGASPSGSPTPTPSTPTPSTPTPSTPTP 298
Query: 579 ATEPTAVPA-STPASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEKEKENETP 637
+T + P STPA + T A S G+ +AS + + S G K ET
Sbjct: 299 STPTPSTPTPSTPAPS--TPAAGKTSEKGSESASMKKESNSKSESESAASGSVSKTKETN 356
Query: 638 KSPPHQ-------EAPPSPPPTRDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVG 690
K + SP + PS G+ S G+A+ S A + + G
Sbjct: 357 KGSSGDTYKDTTGTSSGSPSGSPSGSPTPSTSTDGKASSKGSASASAGA----SASASAG 412
Query: 691 SSSYYNMLPNAIEPSEFLLAGLNRDVIEKEVLSRGLNDTKEETLACLLRAGCIFAHTFEK 750
+S+ +E A ++ K S + +E F EK
Sbjct: 413 ASA----------SAEESAASQKKESNSKSSSSSSSTTSVKEVETQTSSEVNSFISNLEK 462
Query: 751 FNAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQ 810
N E ++ E K +A+A KL+T K+ V + +A KI E
Sbjct: 463 KYTGNSEL-KVFFEKLKTSMSASA------KLSTSNAKELVTG---MRSAASKIAEAMMF 512
Query: 811 LALMKEEADELDASLQACKKEKEQAEKDL------IARGEALIA-KESELAVLCAELELV 863
++ +++E S+ +C++E Q+ K+L I G+ + + +++EL + E V
Sbjct: 513 VSSRFSKSEETKTSMASCQQEVMQSLKELQDINSQIVSGKTVTSTQQTELKQTITKWEQV 572
Query: 864 KKALAEQEKKSAESLALAKSDMEAVMQATSEEIKK 898
E S+ S + + S + Q +++ K
Sbjct: 573 TTQFVETAASSSSSSSSSSSSSSSSSQGSAKMAMK 607
Score = 43.5 bits (101), Expect = 6e-04
Identities = 36/140 (25%), Positives = 61/140 (42%), Gaps = 17/140 (12%)
Query: 504 KARERRMARFSPKFNTEGDAKKRGGAENQAESTKAPKR-------RRLTKTSSGAGTSNP 556
K E+ S K + ++ A TK + + T TSSG+ + +P
Sbjct: 320 KTSEKGSESASMKKESNSKSESESAASGSVSKTKETNKGSSGDTYKDTTGTSSGSPSGSP 379
Query: 557 GAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAGVNA 616
PT + + GK ++ S +A+ A ASA A A+ +AE S A+ +
Sbjct: 380 SGSPTPSTSTDGKASSKGSASAS-------AGASASASAGASASAEES---AASQKKESN 429
Query: 617 TKAAASSDTPIGEKEKENET 636
+K+++SS + KE E +T
Sbjct: 430 SKSSSSSSSTTSVKEVETQT 449
Score = 42.0 bits (97), Expect = 0.002
Identities = 81/400 (20%), Positives = 136/400 (33%), Gaps = 61/400 (15%)
Query: 545 TKTSSGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESS 604
+ +S +SN +Q T++ + G SI T S +S+ T V+ SS
Sbjct: 202 SSSSDNESSSNTKSQGTSSKS--GSESTAGSIETNTGSKTEAGSKSSSSAKTKEVSGGSS 259
Query: 605 VGATAASAGVNATKAAASSDTPIGEKEKENETPKSP-PHQEAPPSPPPTRDAGSMPSPPH 663
+ G ++ + + S TP TP +P P P +P P+ S P+P
Sbjct: 260 GNTYKDTTGSSSGASPSGSPTP----TPSTPTPSTPTPSTPTPSTPTPSTPTPSTPAP-- 313
Query: 664 QGEKSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFLLAGLNRDVIEKEVLS 723
S P A TSE + E S S SE +G E S
Sbjct: 314 ----STPAAGKTSEKGSESASMKKESNSKS----------ESESAASGSVSKTKETNKGS 359
Query: 724 RGLNDTKEETLACLLRAGCIFAHTFEKFNAANVEAERLKAESAKHQEAAAAWEKRFDKLA 783
G DT ++T T + + + S D A
Sbjct: 360 SG--DTYKDTTG-----------TSSGSPSGSPSGSPTPSTST-------------DGKA 393
Query: 784 TQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEADELDASLQACKKEKEQAE------- 836
+ G A + E+ A K+E++ +S + ++ E
Sbjct: 394 SSKGSASASAGASASASAGASASAEESAASQKKESNSKSSSSSSSTTSVKEVETQTSSEV 453
Query: 837 KDLIARGEALIAKESELAVLCAELELVKKALAEQEKKSAESLAL----AKSDMEAVMQAT 892
I+ E SEL V +L+ A A+ +A+ L A S + M
Sbjct: 454 NSFISNLEKKYTGNSELKVFFEKLKTSMSASAKLSTSNAKELVTGMRSAASKIAEAMMFV 513
Query: 893 SEEIKKATETHAEALATKDAEIASQLAKIKSLEDELATEK 932
S K+ ET ++A+ E+ L +++ + ++ + K
Sbjct: 514 SSRFSKSEETKT-SMASCQQEVMQSLKELQDINSQIVSGK 552
>At1g65010 hypothetical protein
Length = 1318
Score = 49.7 bits (117), Expect = 8e-06
Identities = 50/168 (29%), Positives = 80/168 (46%), Gaps = 21/168 (12%)
Query: 802 IKIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELE 861
+K EL+ QL ++E+ + D ++ KK+K +A DL KESE V A E
Sbjct: 51 VKGTELQTQLNQIQEDLKKADEQIELLKKDKAKAIDDL---------KESEKLVEEAN-E 100
Query: 862 LVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKI 921
+K+ALA Q K++ ES + K + QA E ++K T L + ++ A ++ +
Sbjct: 101 KLKEALAAQ-KRAEESFEVEKFRAVELEQAGLEAVQKKDVTSKNELESIRSQHALDISAL 159
Query: 922 KSLEDEL----------ATEKAKAIEAREQAADIALDNRERGFYLAKD 959
S +EL A K KA+ E+A IA + E+ LA +
Sbjct: 160 LSTTEELQRVKHELSMTADAKNKALSHAEEATKIAEIHAEKAEILASE 207
Score = 34.3 bits (77), Expect = 0.37
Identities = 30/118 (25%), Positives = 54/118 (45%), Gaps = 15/118 (12%)
Query: 816 EEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKSA 875
++ EL L+ CK E+E+++KD+ + AL +E + A L + ++ L E +
Sbjct: 406 DQRTELSIELERCKVEEEKSKKDMESLTLALQEASTESSEAKATLLVCQEELKNCESQ-V 464
Query: 876 ESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELATEKA 933
+SL LA K+ E + + L EI S + + S+++E KA
Sbjct: 465 DSLKLAS--------------KETNEKYEKMLEDARNEIDSLKSTVDSIQNEFENSKA 508
Score = 33.9 bits (76), Expect = 0.48
Identities = 50/193 (25%), Positives = 82/193 (41%), Gaps = 30/193 (15%)
Query: 749 EKFNAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELE 808
E A+ + E LK + AK + EK ++ A + K+ + A K E
Sbjct: 65 EDLKKADEQIELLKKDKAKAIDDLKESEKLVEE-ANEKLKEALAAQK----------RAE 113
Query: 809 DQLALMKEEADELD-ASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKAL 867
+ + K A EL+ A L+A +K+ ++ +L + S L EL+ VK L
Sbjct: 114 ESFEVEKFRAVELEQAGLEAVQKKDVTSKNELESIRSQHALDISALLSTTEELQRVKHEL 173
Query: 868 AEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDE 927
+ ++L+ A EE K E HAE K +AS+L ++K+L
Sbjct: 174 SMTADAKNKALSHA------------EEATKIAEIHAE----KAEILASELGRLKALLG- 216
Query: 928 LATEKAKAIEARE 940
+ E+ +AIE E
Sbjct: 217 -SKEEKEAIEGNE 228
>At1g13220 putative nuclear matrix constituent protein
Length = 1128
Score = 48.5 bits (114), Expect = 2e-05
Identities = 44/198 (22%), Positives = 86/198 (43%), Gaps = 22/198 (11%)
Query: 757 EAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKM--IGTAGIKIGELEDQLALM 814
E E + K +E WEK+ GK++ ++ + K+ E+E +L L
Sbjct: 250 ERESYEGTFQKQREYLNEWEKKLQ------GKEESITEQKRNLNQREEKVNEIEKKLKLK 303
Query: 815 KEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKS 874
++E +E + + + ++ E+D+ R E L KE E L ++ L+ K E E ++
Sbjct: 304 EKELEEWNRKVDLSMSKSKETEEDITKRLEELTTKEKEAHTL--QITLLAK---ENELRA 358
Query: 875 AESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELATEKAK 934
E +A+ EI+K + E L +K E + +I+ D+ K +
Sbjct: 359 FEEKLIARE---------GTEIQKLIDDQKEVLGSKMLEFELECEEIRKSLDKELQRKIE 409
Query: 935 AIEAREQAADIALDNRER 952
+E ++ D + + E+
Sbjct: 410 ELERQKVEIDHSEEKLEK 427
Score = 30.0 bits (66), Expect = 6.9
Identities = 40/185 (21%), Positives = 84/185 (44%), Gaps = 30/185 (16%)
Query: 766 AKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEAD-ELDAS 824
AK E A EK + T+ K D G K+ E E + +++ D EL
Sbjct: 351 AKENELRAFEEKLIAREGTEIQK---LIDDQKEVLGSKMLEFELECEEIRKSLDKELQRK 407
Query: 825 LQACKKEK---EQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKSAESLALA 881
++ +++K + +E+ L R +A+ K + +LE K + E+EK
Sbjct: 408 IEELERQKVEIDHSEEKLEKRNQAMNKKFDRVNEKEMDLEAKLKTIKEREK--------- 458
Query: 882 KSDMEAVMQATSEEIKKATETHAEALATKDA--EIASQLAKIK---SLEDELATEKAKAI 936
++QA E K+ + + L+ K++ ++ ++ KI+ + ++E+ E+ K++
Sbjct: 459 ------IIQA---EEKRLSLEKQQLLSDKESLEDLQQEIEKIRAEMTKKEEMIEEECKSL 509
Query: 937 EAREQ 941
E +++
Sbjct: 510 EIKKE 514
Score = 29.6 bits (65), Expect = 9.1
Identities = 45/205 (21%), Positives = 83/205 (39%), Gaps = 21/205 (10%)
Query: 749 EKFNAANVEAERLKAESAK---HQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIG 805
E+ E +RL E + +E+ ++ +K+ + K + ++ + IK
Sbjct: 455 EREKIIQAEEKRLSLEKQQLLSDKESLEDLQQEIEKIRAEMTKKEEMIEEECKSLEIKKE 514
Query: 806 ELEDQLALMKEEADELDAS----------LQACKKEKEQAEKDLIARGEALIAKESELAV 855
E E+ L L E +++ S ++ K+EKE+ EK+ E E
Sbjct: 515 EREEYLRLQSELKSQIEKSRVHEEFLSKEVENLKQEKERFEKEWEILDEKQAVYNKERIR 574
Query: 856 LCAELELVKK-ALAEQEKKSAESLALAKSDMEAV----MQATSEEIKKATETHA--EALA 908
+ E E ++ L E E+ E AL M+ + +Q S E E A E +
Sbjct: 575 ISEEKEKFERFQLLEGERLKKEESALRVQIMQELDDIRLQRESFEANMEHERSALQEKVK 634
Query: 909 TKDAEIASQLAKI-KSLEDELATEK 932
+ +++ L + ++LE EL K
Sbjct: 635 LEQSKVIDDLEMMRRNLEIELQERK 659
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 47.8 bits (112), Expect = 3e-05
Identities = 32/148 (21%), Positives = 78/148 (52%), Gaps = 16/148 (10%)
Query: 803 KIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELEL 862
++ ELE QL L+++ +L ASL A ++EK+ ++ + L +S++ L EL
Sbjct: 486 RLSELETQLKLLEQRVVDLSASLNAAEEEKKSLSSMILEITDELKQAQSKVQELVTELAE 545
Query: 863 VKKALAEQEKKSAESLALAKS----------DMEAVMQATSEEIKK------ATETHAEA 906
K L ++E + + + + ++ ++EA +++ E++K+ ++E +
Sbjct: 546 SKDTLTQKENELSSFVEVHEAHKRDSSSQVKELEARVESAEEQVKELNQNLNSSEEEKKI 605
Query: 907 LATKDAEIASQLAKIKSLEDELATEKAK 934
L+ + +E++ ++ + +S EL++E +
Sbjct: 606 LSQQISEMSIKIKRAESTIQELSSESER 633
Score = 42.4 bits (98), Expect = 0.001
Identities = 56/284 (19%), Positives = 120/284 (41%), Gaps = 45/284 (15%)
Query: 675 TSEAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFLLA----GLNRDVIEKEVLSRGLNDTK 730
+ ++ +E E SSS L IE SE L+A LN EK++LS+ + +
Sbjct: 48 SEHSSLVELHKTHERESSSQVKELEAHIESSEKLVADFTQSLNNAEEEKKLLSQKIAELS 107
Query: 731 EETLACLLRAGCIFAHTFEKFNAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDK 790
E A + L +ES + +E+ + E+ L +
Sbjct: 108 NE------------------IQEAQNTMQELMSESGQLKESHSVKERELFSL-------R 142
Query: 791 VYADKMIGTAGIKIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKE 850
+ + + ELE QL K++ +L ASL+A ++E + + L +
Sbjct: 143 DIHEIHQRDSSTRASELEAQLESSKQQVSDLSASLKAAEEENKAISSKNVETMNKLEQTQ 202
Query: 851 SELAVLCAELELVKKALAEQEKKSAESLALAKS----------DMEAVMQATSEEIKKAT 900
+ + L AEL +K + E+E + + + + ++ ++E ++++ + + +
Sbjct: 203 NTIQELMAELGKLKDSHREKESELSSLVEVHETHQRDSSIHVKELEEQVESSKKLVAELN 262
Query: 901 ET------HAEALATKDAEIASQLAKIKSLEDELATEKAKAIEA 938
+T + L+ K AE+++++ + ++ EL +E + E+
Sbjct: 263 QTLNNAEEEKKVLSQKIAELSNEIKEAQNTIQELVSESGQLKES 306
Score = 41.6 bits (96), Expect = 0.002
Identities = 44/207 (21%), Positives = 92/207 (44%), Gaps = 26/207 (12%)
Query: 759 ERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVY-----ADKMIGTAGIKIGELEDQLAL 813
+ LK +K QE + D L + + + A K ++ +K ELE ++
Sbjct: 527 DELKQAQSKVQELVTELAESKDTLTQKENELSSFVEVHEAHKRDSSSQVK--ELEARVES 584
Query: 814 MKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKK 873
+E+ EL+ +L + ++EK+ + + + ES + L +E E +K + AE++ +
Sbjct: 585 AEEQVKELNQNLNSSEEEKKILSQQISEMSIKIKRAESTIQELSSESERLKGSHAEKDNE 644
Query: 874 ----------SAESLALAKSDMEAVMQATS------EEIKKATETHAEALATKDAEIASQ 917
L+ +EA ++++ E KA E + ++TK +E + +
Sbjct: 645 LFSLRDIHETHQRELSTQLRGLEAQLESSEHRVLELSESLKAAEEESRTMSTKISETSDE 704
Query: 918 LAKIKSLEDELATEKAKAIEAREQAAD 944
L + + + EL + +K +EQ A+
Sbjct: 705 LERTQIMVQELTADSSK---LKEQLAE 728
Score = 39.7 bits (91), Expect = 0.009
Identities = 49/219 (22%), Positives = 89/219 (40%), Gaps = 17/219 (7%)
Query: 756 VEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKV-YADKMIGTAGIKIGELEDQLALM 814
++ L+ E+ Q + E R K DK+ A I I L+++L +
Sbjct: 937 IKGRELELETLGKQRSELDEELRTKKEENVQMHDKINVASSEIMALTELINNLKNELDSL 996
Query: 815 KEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEK-- 872
+ + E +A L+ K+EK + + +AL+ +E+ L E + + + E E
Sbjct: 997 QVQKSETEAELEREKQEKSELSNQITDVQKALVEQEAAYNTLEEEHKQINELFKETEATL 1056
Query: 873 -------KSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLE 925
K A+ L + +T ++ E+ L K EI + + KI ++E
Sbjct: 1057 NKVTVDYKEAQRLLEERGKEVTSRDSTIGVHEETMESLRNELEMKGDEIETLMEKISNIE 1116
Query: 926 DELATEKAKAIEAREQAADIALDNRERGFYLAKDQAQHL 964
+L K + EQ L +E F K++A+HL
Sbjct: 1117 VKLRLSNQK-LRVTEQ----VLTEKEEAF--RKEEAKHL 1148
Score = 37.7 bits (86), Expect = 0.033
Identities = 42/185 (22%), Positives = 83/185 (44%), Gaps = 33/185 (17%)
Query: 800 AGIKIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAE 859
+ I + ELE+Q+ K+ EL+ +L ++EK+ + + + ++ + L +E
Sbjct: 240 SSIHVKELEEQVESSKKLVAELNQTLNNAEEEKKVLSQKIAELSNEIKEAQNTIQELVSE 299
Query: 860 LELVKK----------ALAEQEKKSAESLALAKSDMEAVMQATSEEIK------KATETH 903
+K+ +L + + + S++EA ++++ + I K E
Sbjct: 300 SGQLKESHSVKDRDLFSLRDIHETHQRESSTRVSELEAQLESSEQRISDLTVDLKDAEEE 359
Query: 904 AEALATKDAEIASQLAK----IKSLEDELATEKAKAIE-----------AREQAADI--A 946
+A+++K+ EI +L + IK L DEL K + E A +Q AD+ +
Sbjct: 360 NKAISSKNLEIMDKLEQAQNTIKELMDELGELKDRHKEKESELSSLVKSADQQVADMKQS 419
Query: 947 LDNRE 951
LDN E
Sbjct: 420 LDNAE 424
Score = 35.4 bits (80), Expect = 0.17
Identities = 69/299 (23%), Positives = 124/299 (41%), Gaps = 55/299 (18%)
Query: 691 SSSYYNMLPNAIEPSEFLLAGLNRDVI----EKEVLSRGL----NDTKE--ETLACLLRA 740
SS + L +E S+ L+A LN+ + EK+VLS+ + N+ KE T+ L+
Sbjct: 240 SSIHVKELEEQVESSKKLVAELNQTLNNAEEEKKVLSQKIAELSNEIKEAQNTIQELVSE 299
Query: 741 GCIFAHTF-----EKFNAANVEAERLKAESAKHQEAAAAWEKRFDKLA--TQAGKDKVYA 793
+ + F+ ++ + S + E A E +++ T KD
Sbjct: 300 SGQLKESHSVKDRDLFSLRDIHETHQRESSTRVSELEAQLESSEQRISDLTVDLKDAEEE 359
Query: 794 DKMIGTAGIKI-GELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESE 852
+K I + ++I +LE +KE DEL L+ KEKE L+ + +A
Sbjct: 360 NKAISSKNLEIMDKLEQAQNTIKELMDEL-GELKDRHKEKESELSSLVKSADQQVAD--- 415
Query: 853 LAVLCAELELVKKAL--AEQEKKSAESLALAKSDMEAVMQAT-------SEEIKKA---- 899
+K++L AE+EKK L S+ Q T SE++K++
Sbjct: 416 ----------MKQSLDNAEEEKKMLSQRILDISNEIQEAQKTIQEHMSESEQLKESHGVK 465
Query: 900 ----------TETHAEALATKDAEIASQLAKIKSLEDELATEKAKAIEAREQAADIALD 948
ETH +T+ +E+ +QL ++ +L+ A E ++ + + L+
Sbjct: 466 ERELTGLRDIHETHQRESSTRLSELETQLKLLEQRVVDLSASLNAAEEEKKSLSSMILE 524
Score = 30.8 bits (68), Expect = 4.1
Identities = 51/260 (19%), Positives = 102/260 (38%), Gaps = 21/260 (8%)
Query: 708 LLAGLNRDVIEKEVLSRGLNDTKEETLACLLRAGCIFAHTFEKFNAANVEAERLKAESAK 767
L A L+ ++KE + + + EE + R ++ + + + L+ + K
Sbjct: 838 LRAELDSMSVQKEEVEKQMVCKSEEASVKIKRLDDEVNGLRQQVASLDSQRAELEIQLEK 897
Query: 768 HQEAAAAWEKRFDKLATQA-GKDKVYADKMIGTAGI--KIGELEDQLALMKEEADELDAS 824
E + + + L + K KV+ + G+ KI E +L + ++ ELD
Sbjct: 898 KSEEISEYLSQITNLKEEIINKVKVHESILEEINGLSEKIKGRELELETLGKQRSELDEE 957
Query: 825 LQACKKEKEQ-------AEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKSAE- 876
L+ K+E Q A +++A E + ++EL L + + L ++++ +E
Sbjct: 958 LRTKKEENVQMHDKINVASSEIMALTELINNLKNELDSLQVQKSETEAELEREKQEKSEL 1017
Query: 877 -----SLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELATE 931
+ A + EA EE K+ E E AT + + LE E
Sbjct: 1018 SNQITDVQKALVEQEAAYNTLEEEHKQINELFKETEATLNKVTVDYKEAQRLLE-----E 1072
Query: 932 KAKAIEAREQAADIALDNRE 951
+ K + +R+ + + E
Sbjct: 1073 RGKEVTSRDSTIGVHEETME 1092
>At2g32240 putative myosin heavy chain
Length = 1333
Score = 47.4 bits (111), Expect = 4e-05
Identities = 67/279 (24%), Positives = 117/279 (41%), Gaps = 22/279 (7%)
Query: 749 EKFNAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELE 808
E+F + + EA L + + ++E ++LA +GK +K+ T G ++ E
Sbjct: 813 EEFTSRDSEASSLTEKLRDLEGKIKSYE---EQLAEASGKSSSLKEKLEQTLG-RLAAAE 868
Query: 809 DQLALMKEEADEL-DASLQACKKEKEQAE-----KDLIARGEALIAKES-ELAVLCAELE 861
+K+E D+ + SLQ+ + + AE K I E LI S E LE
Sbjct: 869 SVNEKLKQEFDQAQEKSLQSSSESELLAETNNQLKIKIQELEGLIGSGSVEKETALKRLE 928
Query: 862 LVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKI 921
+ ++E +S++ + K+ + EE KK + T+ E+ L+K+
Sbjct: 929 EAIERFNQKETESSDLVEKLKTHENQI-----EEYKKLAHEASGVADTRKVELEDALSKL 983
Query: 922 KSLEDELATEKAKAIEAREQAADIALDNRERGFYLAKDQAQHLYPSFDFSAMGVMKEITA 981
K+LE + AK +++ D+A N + LA ++ SA+ KE TA
Sbjct: 984 KNLESTIEELGAKCQGLEKESGDLAEVNLKLNLELANHGSEANELQTKLSALEAEKEQTA 1043
Query: 982 AGLVGPDDPPLIDQNLWTATEEEEEEEEQ----EKENNE 1016
L I+ T E E+ + Q +ENN+
Sbjct: 1044 NELEA--SKTTIEDLTKQLTSEGEKLQSQISSHTEENNQ 1080
Score = 44.3 bits (103), Expect = 4e-04
Identities = 71/274 (25%), Positives = 122/274 (43%), Gaps = 32/274 (11%)
Query: 749 EKFNAANVEAERLKAESAKHQEA---AAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIG 805
E F+A +E E + + + +E +A ++F++L Q+ +AD A ++
Sbjct: 184 EAFDALGIELESSRKKLIELEEGLKRSAEEAQKFEELHKQSAS---HADSESQKA-LEFS 239
Query: 806 ELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKK 865
EL +E +E ASLQ KE + + AL + ELA + EL L K
Sbjct: 240 ELLKSTKESAKEMEEKMASLQQEIKELNEKMSENEKVEAALKSSAGELAAVQEELALSKS 299
Query: 866 ALAEQEKKSAESLALAKSDMEAVMQATSE-EIKKATETHAEALATKDAEIASQLAKIKSL 924
L E E+K + + AL + + T E E KKA+E + +L+ ++ L
Sbjct: 300 RLLETEQKVSSTEAL-------IDELTQELEQKKASE----------SRFKEELSVLQDL 342
Query: 925 EDELATEKAKAIEAREQAADIALDNRERGFY--LAKDQAQHLYPSFDFSAMGVMKEITAA 982
+ + +AK E + +A + +E+ L+KDQ + L + + A V+KE A
Sbjct: 343 DAQTKGLQAKLSEQEGINSKLAEELKEKELLESLSKDQEEKLRTANEKLA-EVLKEKEAL 401
Query: 983 GLVGPDDPPLIDQNLWTATEEEEEEEEQEKENNE 1016
+ + N+ T TE E EE+ K ++E
Sbjct: 402 ----EANVAEVTSNVATVTEVCNELEEKLKTSDE 431
Score = 44.3 bits (103), Expect = 4e-04
Identities = 60/280 (21%), Positives = 119/280 (42%), Gaps = 44/280 (15%)
Query: 759 ERLKAESAKHQEA----AAAWEKRFD-----KLATQAGKDKVYADKMIGTAGIKIGELED 809
++LK+ H EA AAA +K + + ++QA ++ A +I ELE
Sbjct: 452 QKLKSLEELHSEAGSAAAAATQKNLELEDVVRSSSQAAEE----------AKSQIKELET 501
Query: 810 QLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAE 869
+ +++ EL+ L + + AE++L E ++ + V E + + E
Sbjct: 502 KFTAAEQKNAELEQQLNLLQLKSSDAERELKELSEKSSELQTAIEVAEEEKKQATTQMQE 561
Query: 870 QEKKSAE-SLALAKS---------DMEAVMQATSEEIKKATETHAEALATKDAEIASQL- 918
++K++E L+L +S D+ +Q +E +A TH ++ + +SQ
Sbjct: 562 YKQKASELELSLTQSSARNSELEEDLRIALQKGAEHEDRANTTHQRSIELEGLCQSSQSK 621
Query: 919 -----AKIKSLEDELATEKAKAIEAREQAADIALDNRE-----RGFYLAKDQAQHLYPSF 968
++K LE L TEK + E EQ + + + E +G+ + Q +F
Sbjct: 622 HEDAEGRLKDLELLLQTEKYRIQELEEQVSSLEKKHGETEADSKGYLGQVAELQSTLEAF 681
Query: 969 DFSAMGVMKEITAAGLVGPDDPPLIDQNLWTATEEEEEEE 1008
+ + AA + ++ + +NL T E+++ E
Sbjct: 682 QVKS----SSLEAALNIATENEKELTENLNAVTSEKKKLE 717
Score = 43.9 bits (102), Expect = 5e-04
Identities = 41/200 (20%), Positives = 93/200 (46%), Gaps = 11/200 (5%)
Query: 755 NVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALM 814
N+E +E+ + Q +A E ++ A + K + + + +L+ Q++
Sbjct: 1015 NLELANHGSEANELQTKLSALEAEKEQTANELEASKTTIEDLTKQLTSEGEKLQSQISSH 1074
Query: 815 KEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKS 874
EE ++++A Q+ K+E + L E L + S+ L +E+E ++ AE+
Sbjct: 1075 TEENNQVNAMFQSTKEELQSVIAKL---EEQLTVESSKADTLVSEIEKLRAVAAEKSVLE 1131
Query: 875 AESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSL--EDELATEK 932
+ ++E + ++K+ E A A + K AE+ S+L + + + E ++ E+
Sbjct: 1132 SHF-----EELEKTLSEVKAQLKENVENAATA-SVKVAELTSKLQEHEHIAGERDVLNEQ 1185
Query: 933 AKAIEAREQAADIALDNRER 952
++ QAA ++D +++
Sbjct: 1186 VLQLQKELQAAQSSIDEQKQ 1205
Score = 42.0 bits (97), Expect = 0.002
Identities = 41/193 (21%), Positives = 89/193 (45%), Gaps = 16/193 (8%)
Query: 754 ANVEAERLKAESAKHQ-EAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLA 812
+ +EA ++K+ S + A EK + +K + + +KI E E+ L
Sbjct: 676 STLEAFQVKSSSLEAALNIATENEKELTENLNAVTSEKKKLEATVDEYSVKISESENLLE 735
Query: 813 LMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEK 872
++ E + L E E DL A G +ESE+ +L+ +++L EQ+
Sbjct: 736 SIRNELNVTQGKL-------ESIENDLKAAG----LQESEVM---EKLKSAEESL-EQKG 780
Query: 873 KSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELATEK 932
+ + + ++EA+ Q+ S + + + E ++D+E +S K++ LE ++ + +
Sbjct: 781 REIDEATTKRMELEALHQSLSIDSEHRLQKAMEEFTSRDSEASSLTEKLRDLEGKIKSYE 840
Query: 933 AKAIEAREQAADI 945
+ EA +++ +
Sbjct: 841 EQLAEASGKSSSL 853
Score = 39.7 bits (91), Expect = 0.009
Identities = 41/167 (24%), Positives = 70/167 (41%), Gaps = 19/167 (11%)
Query: 807 LEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKA 866
+E+Q +++ + L +++ ++ E +L L ESE L EL K+
Sbjct: 70 VEEQKEVIERSSSGSQRELHESQEKAKELELELERVAGELKRYESENTHLKDELLSAKEK 129
Query: 867 LAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLED 926
L E EKK D+E V + E+I + E H+ L + + + S AK K L +
Sbjct: 130 LEETEKK--------HGDLEVVQKKQQEKIVEGEERHSSQLKSLEDALQSHDAKDKELTE 181
Query: 927 ----------ELATEKAKAIEAREQAADIALDNRERGFYLAKDQAQH 963
EL + + K IE E+ + + ++ L K A H
Sbjct: 182 VKEAFDALGIELESSRKKLIEL-EEGLKRSAEEAQKFEELHKQSASH 227
Score = 37.7 bits (86), Expect = 0.033
Identities = 54/250 (21%), Positives = 99/250 (39%), Gaps = 29/250 (11%)
Query: 677 EAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFLLAGLNRDVIEKEVLSRGLNDTKEETLAC 736
EA + + A E ++ ++ E L + ++ E ++ TKEE +
Sbjct: 1036 EAEKEQTANELEASKTTIEDLTKQLTSEGEKLQSQISSHTEENNQVNAMFQSTKEELQS- 1094
Query: 737 LLRAGCIFAHTFEKFNAANVEAERLKAESAKHQEAAA---AWEKRFDKLATQAGKDKVYA 793
+ A E+ + +A+ L +E K + AA E F++L + K
Sbjct: 1095 ------VIAKLEEQLTVESSKADTLVSEIEKLRAVAAEKSVLESHFEELEKTLSEVKAQL 1148
Query: 794 DKMI---GTAGIKIGELEDQLALMKE---EADELDASLQACKKEKEQAEKDLIARGEALI 847
+ + TA +K+ EL +L + E D L+ + +KE + A+ + + +A
Sbjct: 1149 KENVENAATASVKVAELTSKLQEHEHIAGERDVLNEQVLQLQKELQAAQSSIDEQKQAHS 1208
Query: 848 AKESELAVLCAELELVKKALAEQ---EKKSAESLALAKSDMEAVMQATSEEIKKATETHA 904
K+SEL E K E+ +KK+ D+E +Q + K ET A
Sbjct: 1209 QKQSEL-------ESALKKSQEEIEAKKKAVTEFESMVKDLEQKVQLADAKTK---ETEA 1258
Query: 905 EALATKDAEI 914
+ K +I
Sbjct: 1259 MDVGVKSRDI 1268
Score = 36.2 bits (82), Expect = 0.097
Identities = 46/207 (22%), Positives = 84/207 (40%), Gaps = 15/207 (7%)
Query: 757 EAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKE 816
E++ E E A KR++ T + + A + + K G+LE +E
Sbjct: 90 ESQEKAKELELELERVAGELKRYESENTHLKDELLSAKEKLEETEKKHGDLEVVQKKQQE 149
Query: 817 EADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQE---KK 873
+ E + + K E A + A+ + L + L ELE +K L E E K+
Sbjct: 150 KIVEGEERHSSQLKSLEDALQSHDAKDKELTEVKEAFDALGIELESSRKKLIELEEGLKR 209
Query: 874 SAESL---------ALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSL 924
SAE + + +D E+ E+ K+T+ A+ + K +AS +IK L
Sbjct: 210 SAEEAQKFEELHKQSASHADSESQKALEFSELLKSTKESAKEMEEK---MASLQQEIKEL 266
Query: 925 EDELATEKAKAIEAREQAADIALDNRE 951
++++ + + A ++A E
Sbjct: 267 NEKMSENEKVEAALKSSAGELAAVQEE 293
>At1g12150 hypothetical protein
Length = 548
Score = 46.6 bits (109), Expect = 7e-05
Identities = 50/194 (25%), Positives = 88/194 (44%), Gaps = 33/194 (17%)
Query: 749 EKFNAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELE 808
E +++ E++K E+ + + AA ++ + L + + A++ + I E+E
Sbjct: 362 EALKQESLKLEQMKTEAIEARNEAANMNRKIESLKKETEAAMIAAEEAEKRLELVIREVE 421
Query: 809 DQLALMKEEADELD--ASLQACKKEKEQAEKDLI------------ARGEALIAKESELA 854
+ + ++ +E+ + Q KK+ E++ I GE A E +LA
Sbjct: 422 EAKSAEEKVREEMKMISQKQESKKQDEESSGSKIKITIQEFESLKRGAGETEAAIEKKLA 481
Query: 855 VLCAELELVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEI 914
+ AELE + K AE + K +EA ++A EE+K+ATE LA K AE
Sbjct: 482 TIAAELEEINKRRAEADNK-----------LEANLKAI-EEMKQATE-----LAQKSAES 524
Query: 915 ASQLAKIKSLEDEL 928
A A + +E EL
Sbjct: 525 AE--AAKRMVESEL 536
Score = 32.3 bits (72), Expect = 1.4
Identities = 41/207 (19%), Positives = 89/207 (42%), Gaps = 18/207 (8%)
Query: 749 EKFNAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELE 808
E++ + VE + K + K +++ ++ D AT A A + + K+ EL
Sbjct: 152 EQYISTTVELDAAKQQLNKIRQS---FDSAMDFKAT-ALNQAAEAQRALQVNSAKVNELS 207
Query: 809 DQLALMKEEADELDASLQACKKEKEQA----EKDLIARGEALIAKESELAVLCAELELVK 864
+++ MK+ +L L A + +E A EKD + +E+E +L +++
Sbjct: 208 KEISDMKDAIHQL--KLAAAQNLQEHANIVKEKDDLRECYRTAVEEAEKKLL-----VLR 260
Query: 865 KALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSL 924
K E E + + +L + + ++ EE+KKA E+ + E+ +++
Sbjct: 261 K---EYEPELSRTLEAKLLETTSEIEVLREEMKKAHESEMNTVKIITNELNEATMRLQEA 317
Query: 925 EDELATEKAKAIEAREQAADIALDNRE 951
D+ + ++ R + D+ + E
Sbjct: 318 ADDECSLRSLVNSLRMELEDLRREREE 344
>At4g37450 arabinogalactan protein AGP18
Length = 209
Score = 46.2 bits (108), Expect = 9e-05
Identities = 37/138 (26%), Positives = 55/138 (39%), Gaps = 17/138 (12%)
Query: 554 SNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAG 613
S+P PTT +AP + A T PT PA TP +A A++ V + S + S
Sbjct: 26 SSPTKSPTTPSAP---TTSPTKSPAVTSPTTAPAKTP-TASASSPVESPKSPAPVSES-- 79
Query: 614 VNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPGAA 673
+ TP+ E P +PP P P D+ P + P +
Sbjct: 80 -------SPPPTPVPESSPPVPAPMVSSPVSSPPVPAPVADSPPAPVAAPVADVPAPAPS 132
Query: 674 ----TTSEAAQIEQAPAP 687
TT ++ + + APAP
Sbjct: 133 KHKKTTKKSKKHQAAPAP 150
Score = 35.8 bits (81), Expect = 0.13
Identities = 33/131 (25%), Positives = 48/131 (36%), Gaps = 10/131 (7%)
Query: 551 AGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVP-ASTPASAGATAAVAAESSVGATA 609
A S+P P + A + + ++ P P S+P S+ A A+S A
Sbjct: 61 ASASSPVESPKSPAPVSESSPPPTPVPESSPPVPAPMVSSPVSSPPVPAPVADSPPAPVA 120
Query: 610 ASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPP-----PTRDAGSMPSPPHQ 664
A A + + +K P P PP+PP P DA S P P
Sbjct: 121 APVADVPAPAPSKHKKTTKKSKKHQAAPAPAPELLGPPAPPTESPGPNSDAFS-PGPSAD 179
Query: 665 GEKSCPGAATT 675
+ GAA+T
Sbjct: 180 DQS---GAAST 187
>At4g12770 auxilin-like protein
Length = 909
Score = 46.2 bits (108), Expect = 9e-05
Identities = 76/331 (22%), Positives = 141/331 (41%), Gaps = 70/331 (21%)
Query: 637 PKSPPHQEAPPSPPPTR------DAGSMPSPPHQGEKSCPGAATTSE--AAQIEQAPAPE 688
P S P PP P PTR + S+P+ + G A+ + A+Q+++
Sbjct: 366 PTSAPPPTRPPPPRPTRPIKKKVNEPSIPTSAYHSHVPSSGRASVNSPTASQMDELDDFS 425
Query: 689 VG-SSSYYNMLPNAIEPSEFLLAGLNRDVIEKEVLSRG-LNDTKEETLACLLRAGCIFAH 746
+G + + N P +PS +G + DV S + D + +A F H
Sbjct: 426 IGRNQTAANGYP---DPS----SGEDSDVFSTAAASAAAMKDAMD-------KAEAKFRH 471
Query: 747 TFEKFNAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGE 806
E+ E E LKA ++ + ++ R +L
Sbjct: 472 AKERR-----EKESLKASRSREGDHTENYDSRERELR----------------------- 503
Query: 807 LEDQLALMKEEAD---ELDASLQACKKEKEQAEKDLIARGEALIAKES---------ELA 854
E Q+ L +E A+ E++ + ++E+E+ +K + E L+A+++ E A
Sbjct: 504 -EKQVRLDRERAEREAEMEKTQAREREEREREQKRIERERERLLARQAVERATREARERA 562
Query: 855 VLCAELELVKKALAE--QEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDA 912
A ++ + A+ + ++ AE A+ ++ EA +A + +KA + AEA +A
Sbjct: 563 ATEAHAKVQRAAVGKVTDARERAERAAVQRAHAEARERAAAGAREKAEKAAAEARERANA 622
Query: 913 EIASQLAKIKSLEDELATEKAKAIEAREQAA 943
E+ + AK+++ + A E+A A EAR +AA
Sbjct: 623 EVREKEAKVRA--ERAAVERA-AAEARGRAA 650
Score = 30.0 bits (66), Expect = 6.9
Identities = 49/237 (20%), Positives = 83/237 (34%), Gaps = 24/237 (10%)
Query: 504 KARERRMARFSPKFNTEGDAKKRGGAENQAESTKAP--KRRRLTKTSSGAGTSNPGAQPT 561
+ RER +AR + + T +A++R E A+ +A K + + A A+
Sbjct: 540 RERERLLARQAVERATR-EARERAATEAHAKVQRAAVGKVTDARERAERAAVQRAHAEAR 598
Query: 562 TAAAPKGKNVAEASIAAATE-PTAVPASTPASAGATAAVAAESSVGATAASAGVNATKAA 620
AA + AE + A A E A A A A ++ A +A K
Sbjct: 599 ERAAAGAREKAEKAAAEARERANAEVREKEAKVRAERAAVERAAAEARGRAAAQAKAKQQ 658
Query: 621 ASSDTPIGEKEKENETPKSPPHQEAPPSPP------------PTRDAGSMPSPPHQGEKS 668
++ + P S P Q P P +R + +PS P + +
Sbjct: 659 QENNNDLDSFFNSVSRPSSVPRQRTNPPDPFQDSWNKGGSFESSRPSSRVPSGPTENLRK 718
Query: 669 CPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFLLAGLNRDVIEKEVLSRG 725
A + + S + LPN + + L+ L RD + ++RG
Sbjct: 719 ASSATNIVD-------DLSSIFGGSAISELPNLV-GFKMLMEKLKRDDVPGWNVTRG 767
>At3g19060 hypothetical protein
Length = 1647
Score = 46.2 bits (108), Expect = 9e-05
Identities = 52/201 (25%), Positives = 90/201 (43%), Gaps = 27/201 (13%)
Query: 757 EAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKE 816
E LK E K + A E R+ + A K YAD E E+++ L++
Sbjct: 1388 EVLNLKEEFGKMKSEAKEMEARYIEAQQIAESRKTYAD-----------EREEEVKLLEG 1436
Query: 817 EADELDASLQACKKE----KEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEK 872
+EL+ ++ + + K++AE+ + R E E EL + ++E + A E ++
Sbjct: 1437 SVEELEYTINVLENKVNVVKDEAERQRLQREEL----EMELHTIRQQMESARNADEEMKR 1492
Query: 873 KSAE---SLALAKSDMEAVMQATSE---EIKKATETHAEALATKDAEIASQLAKIKSLED 926
E LA AK +EA+ + T++ EI + +E +E +A+ + + K K L
Sbjct: 1493 ILDEKHMDLAQAKKHIEALERNTADQKTEITQLSEHISELNLHAEAQASEYMHKFKEL-- 1550
Query: 927 ELATEKAKAIEAREQAADIAL 947
E E+ K QA D +L
Sbjct: 1551 EAMAEQVKPEIHVSQAIDSSL 1571
Score = 33.9 bits (76), Expect = 0.48
Identities = 41/190 (21%), Positives = 84/190 (43%), Gaps = 25/190 (13%)
Query: 791 VYADKMIGTAGIKIGELED------QLALMKEEADELDASLQA-------CKKEKEQAEK 837
V+ + + T +K ELED L + +E+ E+ +L+ C+++ K
Sbjct: 1261 VHVEALEKTLALKTFELEDAVSHAQMLEVRLQESKEITRNLEVDTEKARKCQEKLSAENK 1320
Query: 838 DLIARGEALIAKESELAVLCAELELVK-KALAEQEKKSAESLALAKSDMEAVMQATSEEI 896
D+ A E L+A++ L E E+++ K ++E + +L A + + T ++
Sbjct: 1321 DIRAEAEDLLAEKCSL-----EEEMIQTKKVSESMEMELFNLRNALGQLNDTVAFTQRKL 1375
Query: 897 KKATETHAEALATKDAEIASQLAKIKSLEDELATEKAKAIEARE--QAADIALDNRERGF 954
A + + L + + + K+KS E+ +A+ IEA++ ++ D RE
Sbjct: 1376 NDAID-ERDNLQDEVLNLKEEFGKMKSEAKEM---EARYIEAQQIAESRKTYADEREEEV 1431
Query: 955 YLAKDQAQHL 964
L + + L
Sbjct: 1432 KLLEGSVEEL 1441
Score = 33.5 bits (75), Expect = 0.63
Identities = 43/177 (24%), Positives = 74/177 (41%), Gaps = 16/177 (9%)
Query: 756 VEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMK 815
VE LK E + + + + LA Q M K+ LE ++A MK
Sbjct: 604 VEMANLKGEKEELKASEKRSLSNLNDLAAQICNLNTVMSNMEEQYEHKMETLEHEIAKMK 663
Query: 816 EEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAV--LCAELELVKKALAEQEKK 873
EAD+ K+ E+A+ + +E+++ V L E ++ L +Q+K+
Sbjct: 664 IEADQEYVENLCILKKFEEAQGTI---------READITVNELVIANEKMRFDLEKQKKR 714
Query: 874 SAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELAT 930
+ K+ +E + + S +K+ E LA + S L I +L +ELAT
Sbjct: 715 GISLVGEKKALVEKLQELESINVKE-----NEKLAYLEKLFESSLMGIGNLVEELAT 766
>At1g79280 hypothetical protein
Length = 2111
Score = 46.2 bits (108), Expect = 9e-05
Identities = 50/207 (24%), Positives = 86/207 (41%), Gaps = 13/207 (6%)
Query: 749 EKFNAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELE 808
EK A + AE K + Q + EK T KD +K + A ++
Sbjct: 1389 EKLKAKDAHAEDCKKVLLEKQNKISLLEKEL----TNCKKDLSEREKRLDDAQQAQATMQ 1444
Query: 809 DQLALMKEEADE---LDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKK 865
+ K+E ++ + +L K++ E+ EKD +++ +AK+ E A A
Sbjct: 1445 SEFNKQKQELEKNKKIHYTLNMTKRKYEK-EKDELSKQNQSLAKQLEEAKEEAGKRTTTD 1503
Query: 866 ALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLE 925
A+ EQ K E ++ + +E++K TE L KD E+ + ++ KS+E
Sbjct: 1504 AVVEQSVKEREEKEKRIQILDKYVHQLKDEVRKKTED----LKKKDEELTKERSERKSVE 1559
Query: 926 DELATEKAKAIEAREQAADIALDNRER 952
E+ K I+ + D L ER
Sbjct: 1560 KEVGDSLTK-IKKEKTKVDEELAKLER 1585
Score = 46.2 bits (108), Expect = 9e-05
Identities = 48/212 (22%), Positives = 91/212 (42%), Gaps = 34/212 (16%)
Query: 746 HTFEKFNAANVEAERLKAESAKHQEAAAAWE-------KRFDKLATQAGKDKVYADKMIG 798
H FEK A++ + ES + + K +KL + K D++
Sbjct: 1308 HNFEKCQEMREVAQKARMESENFENLLKTKQTELDLCMKEMEKLRMETDLHKKRVDELRE 1367
Query: 799 T-AGIKIGE---LEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELA 854
T I I + L+D++ ++E+ DA + CKK L+ K+++++
Sbjct: 1368 TYRNIDIADYNRLKDEVRQLEEKLKAKDAHAEDCKK--------------VLLEKQNKIS 1413
Query: 855 VLCAELELVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEI 914
+L EL KK L+E+EK+ + A++ M++ +E++K + H TK
Sbjct: 1414 LLEKELTNCKKDLSEREKR-LDDAQQAQATMQSEFNKQKQELEKNKKIHYTLNMTK---- 1468
Query: 915 ASQLAKIKSLEDELATEKAKAIEAREQAADIA 946
K + +DEL+ + + E+A + A
Sbjct: 1469 ----RKYEKEKDELSKQNQSLAKQLEEAKEEA 1496
Score = 38.1 bits (87), Expect = 0.026
Identities = 46/175 (26%), Positives = 80/175 (45%), Gaps = 17/175 (9%)
Query: 800 AGIKIGE-LEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCA 858
AG +IG + +L KEE ++L +++ K Q + IA+ K+ E A
Sbjct: 955 AGFRIGSAMSIELRTAKEEIEKLRGEVESSKSHMLQYKS--IAQVNETALKQMESAHENF 1012
Query: 859 ELELVKKALAEQEKKSAESLALAK--SDMEAVMQATSEEIKKATETHAEALATKDAEIAS 916
LE K+ Q AE ++L + S++E SE++ A +AL + AEIAS
Sbjct: 1013 RLEAEKR----QRSLEAELVSLRERVSELENDCIQKSEQLATAAAGKEDALLSASAEIAS 1068
Query: 917 -------QLAKIKSLEDELATEKAKAIEAREQAADIALDNRERGFYLAKDQAQHL 964
+ ++I+++ +++T K +E + +A N ER L + Q L
Sbjct: 1069 LREENLVKKSQIEAMNIQMSTLK-NDLETEHEKWRVAQRNYERQVILLSETIQEL 1122
Score = 35.8 bits (81), Expect = 0.13
Identities = 37/157 (23%), Positives = 69/157 (43%), Gaps = 10/157 (6%)
Query: 800 AGIKIGELEDQLALMKEEADELDASLQACKKEKEQA----EKDLIARGEALIAKESELAV 855
A + ++ + + E D + A A EQ E+ ++ + + ES+ A
Sbjct: 18 ASVVAERADEYIRKIYAELDSVRAKADAASITAEQTCSLLEQKYLSLSQDFSSLESQNAK 77
Query: 856 LCAELE--LVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAE 913
L ++ + L + A ++ +K ++ K M E+ K+ E L KDAE
Sbjct: 78 LQSDFDDRLAELAQSQAQKHQLHLQSIEKDGEVERMSTEMSELHKSKRQLMELLEQKDAE 137
Query: 914 IASQLAKIKSLEDELA--TEKAKAIEAR--EQAADIA 946
I+ + + IKS D++ T+ + EAR E A++A
Sbjct: 138 ISEKNSTIKSYLDKIVKLTDTSSEKEARLAEATAELA 174
Score = 30.8 bits (68), Expect = 4.1
Identities = 44/195 (22%), Positives = 91/195 (46%), Gaps = 25/195 (12%)
Query: 749 EKFNAANVEAERLKAESAKHQEAAAAWEKRFDKLA-TQAGKDKVYADKMIGTAGIKIGEL 807
+K+ + + + L++++AK Q + ++ R +LA +QA K +++ + E
Sbjct: 59 QKYLSLSQDFSSLESQNAKLQ---SDFDDRLAELAQSQAQKHQLHLQSI---------EK 106
Query: 808 EDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKAL 867
+ ++ M E EL K K Q + L + + K S + ++ +
Sbjct: 107 DGEVERMSTEMSELH-------KSKRQLMELLEQKDAEISEKNSTIKSYLDKIVKLTDTS 159
Query: 868 AEQEKKSAESLA-LAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLED 926
+E+E + AE+ A LA+S +A+ S+E K+ TE HA+ L + A+++
Sbjct: 160 SEKEARLAEATAELARS--QAMCSRLSQE-KELTERHAKWLDEELTAKVDSYAELRRRHS 216
Query: 927 ELATE-KAKAIEARE 940
+L +E AK ++ +
Sbjct: 217 DLESEMSAKLVDVEK 231
>At1g77580 unknown protein
Length = 779
Score = 46.2 bits (108), Expect = 9e-05
Identities = 65/276 (23%), Positives = 112/276 (40%), Gaps = 33/276 (11%)
Query: 657 SMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFLLAGLNRDV 716
++P P KS P + T E + A E+ +L + I+ E L L
Sbjct: 313 ALPETPDGNGKSGPESVTEEVVVPSENSLASEI------EVLTSRIKELEEKLEKLEA-- 364
Query: 717 IEKEVLSRGLNDTKEETLACLLRAGCIFAHTFE---KFNAANVEAERLKAESAKHQEAAA 773
EK L + +EE + + + + + T E K E E LK+E ++E A
Sbjct: 365 -EKHELENEVKCNREEAVVHIENSEVLTSRTKELEEKLEKLEAEKEELKSEVKCNREKAV 423
Query: 774 AWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEADELDASLQACKKEK- 832
+ + + A+ + T+ K ELE+QL ++ E EL++ ++ C +E+
Sbjct: 424 VHVE-----------NSLAAEIEVLTSRTK--ELEEQLEKLEAEKVELESEVK-CNREEA 469
Query: 833 -EQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKSAESLALAKSDMEAVMQA 891
Q E L E L + +L +LE+ K L + K + E + + ++EA+
Sbjct: 470 VAQVENSLATEIEVLTCRIKQLEEKLEKLEVEKDELKSEVKCNREVESTLRFELEAIACE 529
Query: 892 TSEEIKKATETHAEALATKDAEIASQLAKIKSLEDE 927
K E E L + AE+ IK +E
Sbjct: 530 -----KMELENKLEKLEVEKAELQISFDIIKDKYEE 560
Score = 38.5 bits (88), Expect = 0.020
Identities = 50/223 (22%), Positives = 96/223 (42%), Gaps = 43/223 (19%)
Query: 729 TKEETLACLLRAGCIFAHTFEKFNAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGK 788
T+ E L C ++ EK VE + LK+E ++E + + +A
Sbjct: 479 TEIEVLTCRIK------QLEEKLEKLEVEKDELKSEVKCNREVESTLRFELEAIA----- 527
Query: 789 DKVYADKMIGTAGIKIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIA 848
+KM ELE++L ++ E EL S K + E+++ L
Sbjct: 528 ----CEKM---------ELENKLEKLEVEKAELQISFDIIKDKYEESQV-------CLQE 567
Query: 849 KESELAVLCAELELVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALA 908
E++L + E++LV + AE E ++ A AK+ A +++ E+++K E A
Sbjct: 568 IETKLGEIQTEMKLVNELKAEVESQTIAMEADAKTK-SAKIESLEEDMRK------ERFA 620
Query: 909 TKDAEIASQLAKIKSLEDELATEKAKAIEAREQAADIALDNRE 951
+ K ++LE+E++ K +I++ + I ++ E
Sbjct: 621 FDELR-----RKCEALEEEISLHKENSIKSENKEPKIKQEDIE 658
Score = 32.3 bits (72), Expect = 1.4
Identities = 27/131 (20%), Positives = 52/131 (39%)
Query: 807 LEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKA 866
LE+Q+ +++EL ++ KE +L+AR E L + E + E K
Sbjct: 168 LENQIFETATKSEELSQMAESVAKENVMLRHELLARCEELEIRTIERDLSTQAAETASKQ 227
Query: 867 LAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLED 926
+ KK A+ A + + S ++T++H++ D + A +E
Sbjct: 228 QLDSIKKVAKLEAECRKFRMLAKSSASFNDHRSTDSHSDGGERMDVSCSDSWASSTLIEK 287
Query: 927 ELATEKAKAIE 937
+ +IE
Sbjct: 288 RSLQGTSSSIE 298
>At5g38560 putative protein
Length = 681
Score = 45.4 bits (106), Expect = 2e-04
Identities = 41/161 (25%), Positives = 60/161 (36%), Gaps = 12/161 (7%)
Query: 544 LTKTSSGAGTSNPG---AQPTTAAAPKGKNVAEAS-------IAAATEPTAVPASTPASA 593
L+ SS + T+ P QPTT +AP + ++++ P V + P+S+
Sbjct: 10 LSPPSSNSSTTAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVVSSPPPSSS 69
Query: 594 GATAAVAAESSVGATAASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSP--PP 651
+ S A+S A+ TP +T PP +A PSP P
Sbjct: 70 PPPSPPVITSPPPTVASSPPPPVVIASPPPSTPATTPPAPPQTVSPPPPPDASPSPPAPT 129
Query: 652 TRDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSS 692
T + PSP GE P T S P P +S
Sbjct: 130 TTNPPPKPSPSPPGETPSPPGETPSPPKPSPSTPTPTTTTS 170
Score = 40.8 bits (94), Expect = 0.004
Identities = 32/143 (22%), Positives = 54/143 (37%), Gaps = 9/143 (6%)
Query: 548 SSGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGA 607
SS +S+P P +P ++A++ P V AS P S AT A +V
Sbjct: 62 SSPPPSSSPPPSPPVITSPP------PTVASSPPPPVVIASPPPSTPATTPPAPPQTVSP 115
Query: 608 TAA-SAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPS--PPHQ 664
A + ++ P ETP P +PP P P+ + + PP
Sbjct: 116 PPPPDASPSPPAPTTTNPPPKPSPSPPGETPSPPGETPSPPKPSPSTPTPTTTTSPPPPP 175
Query: 665 GEKSCPGAATTSEAAQIEQAPAP 687
+ P ++ ++ + + P P
Sbjct: 176 ATSASPPSSNPTDPSTLAPPPTP 198
Score = 31.2 bits (69), Expect = 3.1
Identities = 28/130 (21%), Positives = 42/130 (31%), Gaps = 9/130 (6%)
Query: 582 PTAVPASTPASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEKEKENETPKSPP 641
P + + +++ TA ++ +A V + S P+ SPP
Sbjct: 6 PLPILSPPSSNSSTTAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVVSSPP 65
Query: 642 HQEAPP-------SPPPTRDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSSY 694
+PP SPPPT S P PP P T+ A + P +S
Sbjct: 66 PSSSPPPSPPVITSPPPT--VASSPPPPVVIASPPPSTPATTPPAPPQTVSPPPPPDASP 123
Query: 695 YNMLPNAIEP 704
P P
Sbjct: 124 SPPAPTTTNP 133
Score = 30.0 bits (66), Expect = 6.9
Identities = 22/98 (22%), Positives = 34/98 (34%), Gaps = 1/98 (1%)
Query: 554 SNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAG 613
+ P A P T + P + A S A T P +P+ G T + E+ + +
Sbjct: 104 TTPPAPPQTVSPPPPPD-ASPSPPAPTTTNPPPKPSPSPPGETPSPPGETPSPPKPSPST 162
Query: 614 VNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPP 651
T + P + P P PP+P P
Sbjct: 163 PTPTTTTSPPPPPATSASPPSSNPTDPSTLAPPPTPLP 200
>At5g24710 unknown protein
Length = 1003
Score = 45.4 bits (106), Expect = 2e-04
Identities = 52/197 (26%), Positives = 78/197 (39%), Gaps = 29/197 (14%)
Query: 515 PKFNTEGDAKKRG-------GAENQAESTKAPKRRRLTKTSSGAGTSNPGA--------- 558
PK + DA ++ G + S APK+ LT+ ++ + P A
Sbjct: 785 PKLTSLSDASRKPPIEILPPGMSSIFASITAPKKPLLTQKTAQPEVAKPLALEEPTKPLA 844
Query: 559 --QPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAGVNA 616
P ++ AP+ ++ E + AA + A + A TAAVA E+ TAA A V+
Sbjct: 845 IEAPPSSEAPQTESAPETAAAAESPAPETAAVAESPAPGTAAVA-EAPASETAA-APVDG 902
Query: 617 TKAAASSDTPIGEKE----KENETPKSPPHQEAPPSPPPTRDAGSMP-----SPPHQGEK 667
S+ P EKE +E P S P+ E S T + P +PP
Sbjct: 903 PVTETVSEPPPVEKEETSLEEKSDPSSTPNTETATSTENTSQTTTTPESVTTAPPEPITT 962
Query: 668 SCPGAATTSEAAQIEQA 684
+ P TT+ E A
Sbjct: 963 APPETVTTTAVKPTENA 979
>At3g28830 hypothetical protein
Length = 536
Score = 45.4 bits (106), Expect = 2e-04
Identities = 64/341 (18%), Positives = 132/341 (37%), Gaps = 26/341 (7%)
Query: 547 TSSGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVG 606
+S + T + + + + K K E+S + ++ +V + S+G +AA ++ S G
Sbjct: 198 SSESSSTKSGSVSSSGSVSTKSK---ESSSSGSSASGSVATKSKESSGGSAATKSKESSG 254
Query: 607 ATAASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPPHQGE 666
+AA+ ++ +A++ G + +PK+ P A + S QG
Sbjct: 255 GSAATKSKESSGGSATTGKTSGSP---SGSPKASPSGSVSGKSSSKGSASAQGSASAQGS 311
Query: 667 KSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFLLAGLNRDVIEKEVLSRGL 726
S G+A+ +A ++ + + S ++ S+ + V +
Sbjct: 312 ASAQGSASAQGSASAQRRESGAMAMSKSRETKTSSQRQSKSSSESSSSSTTTTTV-KQVE 370
Query: 727 NDTKEETLACLLRAGCIFAHTFEKFNAANVEAERLKAESAKHQEAAAAWEKRFDKLATQA 786
++T +E ++ +++ EK AA E ++ ES K A+A + ++
Sbjct: 371 SETSKEVMSFIMQ--------LEKKYAAKAEL-KVFFESLKSSMQASA------SVGSKT 415
Query: 787 GKDKVYADK----MIGTAGIKIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIAR 842
KD V A K + A + + A MK D + C K+ + +++
Sbjct: 416 AKDYVSASKAATGKLSEAMASVSSKNVKSAKMKSNLDTSKDEMLKCVKQIQDINGKMVSG 475
Query: 843 GEALIAKESELAVLCAELELVKKALAEQEKKSAESLALAKS 883
++SEL + E V E S+ S + + S
Sbjct: 476 KTVSSTQQSELKQTITKWEKVTTQFVETAASSSSSSSSSSS 516
Score = 37.0 bits (84), Expect = 0.057
Identities = 34/178 (19%), Positives = 65/178 (36%), Gaps = 20/178 (11%)
Query: 516 KFNTEGDAKKRGGAENQAESTKAPKRRRLTKTSSGAGTSNPGAQPTTAAAPKGKNVAEAS 575
K ++ + G +++ + +K SSG + + + +A GK S
Sbjct: 220 KESSSSGSSASGSVATKSKESSGGSAATKSKESSGGSAATKSKESSGGSATTGKTSGSPS 279
Query: 576 IAAATEPTAVPASTPASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIG--EKEKE 633
+ P+ + +S G+ +A + S+ G+ +A +A +A++ G K
Sbjct: 280 GSPKASPSGSVSGKSSSKGSASAQGSASAQGSASAQGSASAQGSASAQRRESGAMAMSKS 339
Query: 634 NETPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGS 691
ET S Q S +S + TT+ Q+E + EV S
Sbjct: 340 RETKTSSQRQSKSSS------------------ESSSSSTTTTTVKQVESETSKEVMS 379
>At1g63300 hypothetical protein
Length = 1029
Score = 45.4 bits (106), Expect = 2e-04
Identities = 47/222 (21%), Positives = 102/222 (45%), Gaps = 21/222 (9%)
Query: 743 IFAHTFEKFNAANVEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDK---VYADKMIGT 799
+ A+ ++ E E LK K+Q++ ++ + L K K + A+ +
Sbjct: 722 VTANLNQEIKILKEEIENLK----KNQDSLMLQAEQAENLRVDLEKTKKSVMEAEASLQR 777
Query: 800 AGIKIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAE 859
+K ELE +++LM++E++ L A LQ K K++ E A+ ++EL + ++
Sbjct: 778 ENMKKIELESKISLMRKESESLAAELQVIKLAKDEKE-------TAISLLQTELETVRSQ 830
Query: 860 LELVKKALAEQE---KKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIAS 916
+ +K +L+E + +K + +A KS+++ + + KK E+ T +
Sbjct: 831 CDDLKHSLSENDLEMEKHKKQVAHVKSELKKKEETMANLEKKLKESRTAITKTAQRNNIN 890
Query: 917 QLAKI----KSLEDELATEKAKAIEAREQAADIALDNRERGF 954
+ + + S E + +K K +E + + + AL++ F
Sbjct: 891 KGSPVGAHGGSKEVAVMKDKIKLLEGQIKLKETALESSSNMF 932
Score = 35.8 bits (81), Expect = 0.13
Identities = 45/228 (19%), Positives = 95/228 (40%), Gaps = 41/228 (17%)
Query: 759 ERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEA 818
E +K + + + A +E + +L+ + ++M+ K E+++Q ++
Sbjct: 664 EMIKDANDELRANQAEYEAKLHELSEKLSFKTSQMERMLENLDEKSNEIDNQKRHEEDVT 723
Query: 819 DELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQE------- 871
L+ ++ K+E E +K+ ++L+ + + L +LE KK++ E E
Sbjct: 724 ANLNQEIKILKEEIENLKKNQ----DSLMLQAEQAENLRVDLEKTKKSVMEAEASLQREN 779
Query: 872 -------------KKSAESLA-------LAKSDME---AVMQATSEEIKKATETHAEALA 908
+K +ESLA LAK + E +++Q E ++ + +L+
Sbjct: 780 MKKIELESKISLMRKESESLAAELQVIKLAKDEKETAISLLQTELETVRSQCDDLKHSLS 839
Query: 909 TKDAE-------IASQLAKIKSLEDELATEKAKAIEAREQAADIALDN 949
D E +A +++K E+ +A + K E+R A N
Sbjct: 840 ENDLEMEKHKKQVAHVKSELKKKEETMANLEKKLKESRTAITKTAQRN 887
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.129 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,208,764
Number of Sequences: 26719
Number of extensions: 1077465
Number of successful extensions: 9039
Number of sequences better than 10.0: 457
Number of HSP's better than 10.0 without gapping: 105
Number of HSP's successfully gapped in prelim test: 361
Number of HSP's that attempted gapping in prelim test: 6868
Number of HSP's gapped (non-prelim): 1513
length of query: 1016
length of database: 11,318,596
effective HSP length: 109
effective length of query: 907
effective length of database: 8,406,225
effective search space: 7624446075
effective search space used: 7624446075
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0029b.2


