

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0029a.7
(411 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
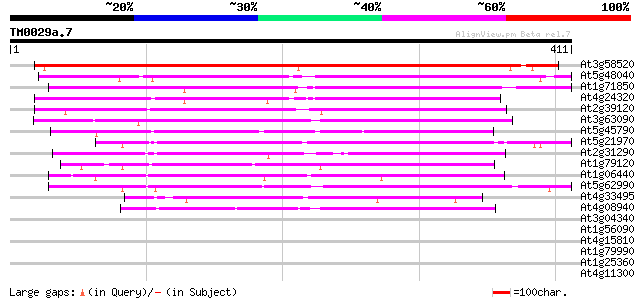
Score E
Sequences producing significant alignments: (bits) Value
At3g58520 unknown protein 547 e-156
At5g48040 putative protein 266 1e-71
At1g71850 hypothetical protein 209 3e-54
At4g24320 putative protein 196 3e-50
At2g39120 hypothetical protein 194 7e-50
At3g63090 unknown protein 164 6e-41
At5g45790 tyrosine-specific protein phosphatase-like protein 154 9e-38
At5g21970 unknown protein 150 9e-37
At2g31290 putative protein 143 2e-34
At1g79120 hypothetical protein 130 1e-30
At1g06440 unknown protein 124 9e-29
At5g62990 putative protein 120 1e-27
At4g33495 unknown protein 119 3e-27
At4g08940 unknown protein 102 5e-22
At3g04340 unknown protein 30 1.9
At1g56090 unknown protein 30 3.2
At4g15810 hypothetical protein 29 5.4
At1g79990 coatomer protein like complex, subunit beta 2 (beta pr... 29 5.4
At1g25360 29 5.4
At4g11300 unknown protein 28 7.0
>At3g58520 unknown protein
Length = 418
Score = 547 bits (1410), Expect = e-156
Identities = 275/396 (69%), Positives = 318/396 (79%), Gaps = 15/396 (3%)
Query: 19 QWRGIA---KVRLKWVKNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKL 75
QWRG+ KVRLKWVKN++LDH+ID ETDLKAA +LKDAI RS TGFLTAKS + WQKL
Sbjct: 21 QWRGMGAMTKVRLKWVKNKNLDHVIDTETDLKAACILKDAIKRSPTGFLTAKSVADWQKL 80
Query: 76 LGLTVPVLRFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERL 135
LGLTVPVLRF+RRYPTLFHEFPH R+ SLPCF+LTDTAL+L SQE+ +H HE DTVERL
Sbjct: 81 LGLTVPVLRFLRRYPTLFHEFPHARYASLPCFKLTDTALMLDSQEEIIHQSHEADTVERL 140
Query: 136 SKFLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWC 195
+ LMM +++TV L SL+ LK+D+GLPD++EKTL+ K+PD F FVKA NG ++L KW
Sbjct: 141 CRVLMMMRSKTVSLRSLHSLKFDLGLPDNYEKTLVMKYPDHFCFVKASNGNPCLKLVKWR 200
Query: 196 EEFAVSALQKSNERD---GGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKL 252
+EFA SALQK NE + G + YR+FKRG++ L FPM FPRGYGAQKKV+ WMDEF KL
Sbjct: 201 DEFAFSALQKRNEDNDVSGEESRYREFKRGQSTLTFPMSFPRGYGAQKKVKAWMDEFQKL 260
Query: 253 PYISPYADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRI 312
PYISPY D S IDP SDLMEKR V VLHELLSLT+HKKTKRNYLRS+R ELN+PHKFTR+
Sbjct: 261 PYISPYDDPSNIDPESDLMEKRAVAVLHELLSLTIHKKTKRNYLRSMRAELNIPHKFTRL 320
Query: 313 FTRYPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLLY--RGDGS 370
FTRYPGIFYLSLKCKTTTV L+EGY RGKLVDPHPL R RDKFYHVM+TG LY RG G
Sbjct: 321 FTRYPGIFYLSLKCKTTTVILKEGYRRGKLVDPHPLTRLRDKFYHVMRTGFLYRARGLGM 380
Query: 371 IPKPEGGALLL----DEIGDEGSEEEEVETSDEFSE 402
+ K E LLL D++ +EGSEEEE+ E E
Sbjct: 381 VSKEE---LLLDRPEDDLEEEGSEEEEIVEGSELEE 413
>At5g48040 putative protein
Length = 422
Score = 266 bits (681), Expect = 1e-71
Identities = 161/399 (40%), Positives = 224/399 (55%), Gaps = 27/399 (6%)
Query: 22 GIAKVRLKWVKNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLLGLT-- 79
G+ V+LKWVK+R LD ++ +E L+A L I+ S L + LGL
Sbjct: 31 GLVNVKLKWVKDRELDAVVVREKHLRAVCNLVSVISASPDLRLPIFKLLPHRGQLGLPQE 90
Query: 80 VPVLRFIRRYPTLFHEFPHPRWPS----LPCFRLTDTALLLHSQEQSLHNHHENDTVERL 135
+ + FIRRYP +F E H W S +PCF LT + L+ +E + +E D + RL
Sbjct: 91 LKLSAFIRRYPNIFVE--HCYWDSAGTSVPCFGLTRETIDLYYEEVDVSRVNERDVLVRL 148
Query: 136 SKFLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWC 195
K LM+T RT+ L S+ L+WD+GLP + +LI K PD F VK + + ++L W
Sbjct: 149 CKLLMLTCERTLSLHSIDHLRWDLGLPYDYRDSLITKHPDLFSLVKLSSDLDGLKLIHWD 208
Query: 196 EEFAVSALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYI 255
E AVS +Q D G+D + FP++F RG+G ++K W+ E+ +LPY
Sbjct: 209 EHLAVSQMQL--REDVGND---------ERMAFPVKFTRGFGLKRKSIEWLQEWQRLPYT 257
Query: 256 SPYADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTR 315
SPY D+S +DP +DL EKR VGV HELL LT+ KKT+R + +LR+ LP KFT++F R
Sbjct: 258 SPYVDASHLDPRTDLSEKRNVGVFHELLHLTIGKKTERKNVSNLRKPFALPQKFTKVFER 317
Query: 316 YPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLLYRGDGSIPKPE 375
+PGIFY+S+KC T TV LRE Y R L++ HPLV R+KF ++M G L R G K
Sbjct: 318 HPGIFYISMKCDTQTVILREAYDRRHLIEKHPLVEVREKFANMMNEGFLDRSRGLYQKSV 377
Query: 376 GGALLLDEIGDE---GSEEEEVETSDEFSEGEVSGSDSE 411
L + I S+EEE E +SG DS+
Sbjct: 378 EADLEKNNIKTSYPVWSDEEE-----ELDNNLISGYDSD 411
>At1g71850 hypothetical protein
Length = 470
Score = 209 bits (531), Expect = 3e-54
Identities = 129/390 (33%), Positives = 211/390 (54%), Gaps = 23/390 (5%)
Query: 29 KWVKNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLLGLTVPVLRFIRR 88
K+V++R LDH +++E +L+ +KD I + + + L + + + FIR
Sbjct: 30 KFVRDRGLDHAVEREKNLRPLLSIKDLIRSEPAKSVPISVITSQKDSLRVPLRPIEFIRS 89
Query: 89 YPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNH--HENDTVERLSKFLMMTKTRT 146
+P++F EF P LT L + EQ ++ ++ +RL K LM+ +
Sbjct: 90 FPSVFQEFLPGGIGIHPHISLTPEILNHDADEQLVYGSETYKQGLADRLLKLLMINRINK 149
Query: 147 VPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAP-NGVSA-VRLSKWCEEFAVSALQ 204
+PL L LKWD+GLP + +T++P+FPD F+ +K+ G S + L W E AVS L+
Sbjct: 150 IPLEILDLLKWDLGLPKDYVETMVPEFPDYFRVIKSKLRGCSGELELVCWSNEHAVSVLE 209
Query: 205 KSNE--RDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYISPYADSS 262
K R G ++ +G +A+ FPM+F G+ KK++ W+D++ KLPYISPY ++
Sbjct: 210 KKARTLRKG------EYTKG-SAIAFPMKFSNGFVVDKKMKKWIDDWQKLPYISPYENAL 262
Query: 263 KIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTRYPGIFYL 322
+ SD +K VLHE+++L + KK +++ + L E + L +F R+ +PGIFYL
Sbjct: 263 HLSATSDESDKWAAAVLHEIMNLFVSKKVEKDAILHLGEFMGLRSRFKRVLHNHPGIFYL 322
Query: 323 SLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLLYRGDGSIPKPEGGALLLD 382
S K +T TV LR+GY RG L++ + LV R+++ +M T + K
Sbjct: 323 SSKLRTHTVVLRDGYKRGMLIESNELVTSRNRYMKLMNT---------VKKDNKAVSSSS 373
Query: 383 EIGDEGSEEEEVETSDEFSEG-EVSGSDSE 411
+ D+G E EV +D +E ++SGSD E
Sbjct: 374 KKEDKGKVEGEVCDTDAKAENDDISGSDVE 403
>At4g24320 putative protein
Length = 395
Score = 196 bits (497), Expect = 3e-50
Identities = 123/349 (35%), Positives = 193/349 (55%), Gaps = 15/349 (4%)
Query: 19 QWRGIAKVRLKWVKNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLLGL 78
Q R R+KWV + LD + +E +LK LKD I S + L S S + L+ L
Sbjct: 14 QRRTFVNARVKWVSDHYLDEAVQREKNLKQVISLKDRIVSSPSKSLPLSSLSLLKPLVNL 73
Query: 79 TVPVLRFIRRYPTLFHEF-PHPRWPSLPCFRLTDTALLLHSQEQSLHNH--HENDTVERL 135
+ F ++YP++F F P P P RLT AL LH +E+++H N TV+RL
Sbjct: 74 HITAAAFFQKYPSVFTTFQPSPSHPLH--VRLTPQALTLHKEEETIHLSPPQRNVTVQRL 131
Query: 136 SKFLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVS-----AVR 190
+KFLM+T ++PL L ++D+GLP + +LI +P+ F+ + + ++ A+
Sbjct: 132 TKFLMLTGAGSLPLYVLDRFRFDLGLPRDYITSLIGDYPEYFEVTEIKDRLTGEKTLALT 191
Query: 191 LSKWCEEFAVSALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFH 250
+S VS +++ R+ DG R K+G + + M FP+GY K+V+ W++++
Sbjct: 192 ISSRRNNLPVSEMER---REATIDGSR-VKKG-LRIRYSMNFPKGYELHKRVKNWVEQWQ 246
Query: 251 KLPYISPYADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFT 310
LPYISPY ++ + SD EK V VLHELL L + KKT+ + + L E L +F
Sbjct: 247 NLPYISPYENAFHLGSYSDQAEKWAVAVLHELLFLLVSKKTETDNVICLGEYLGFGMRFK 306
Query: 311 RIFTRYPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVM 359
+ +PGIFY+S K +T TV LRE Y + L++ HPL+ R ++ ++M
Sbjct: 307 KALVHHPGIFYMSHKIRTQTVVLREAYHKVFLLEKHPLMGIRHQYIYLM 355
>At2g39120 hypothetical protein
Length = 387
Score = 194 bits (493), Expect = 7e-50
Identities = 119/351 (33%), Positives = 187/351 (52%), Gaps = 15/351 (4%)
Query: 19 QWRGIAKVRLKWVKNRSLDHI--IDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLL 76
Q R V +KW ++ D+I I + + LK+ LK+ I + + + S +
Sbjct: 20 QKRTYVDVYMKWKRDPYFDNIEHILRSSQLKSVVSLKNCIVQEPNRCIPISAISKKTRQF 79
Query: 77 GLTVPVLRFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLS 136
++ + F+R++P++F EF P + +LP FRLT A L QE+ ++ +D +RL
Sbjct: 80 DVSTKIAHFLRKFPSIFEEFVGPEY-NLPWFRLTPEATELDRQERVVYQTSADDLRDRLK 138
Query: 137 KFLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWCE 196
K ++M+K +PL + +KW +GLPD + + F+FV +GV + +
Sbjct: 139 KLILMSKDNVLPLSIVQGMKWYLGLPDDYLQFPDMNLDSSFRFVDMEDGVKGLAVDYNGG 198
Query: 197 EFAVSALQKSNERDGGDDGYRDFKRGKAALV---FPMRFPRGYGAQKKVRFWMDEFHKLP 253
+ +S LQK+ + +RG+ +L FP+ +G + K+ W+ EF KLP
Sbjct: 199 DKVLSVLQKNAMKK---------RRGEVSLEEIEFPLFPSKGCRLRVKIEDWLMEFQKLP 249
Query: 254 YISPYADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIF 313
Y+SPY D S +DP+SD+ EKRVVG LHELL L + +R L L++ LP K + F
Sbjct: 250 YVSPYDDYSCLDPSSDIAEKRVVGFLHELLCLFVEHSAERKKLLCLKKHFGLPQKVHKAF 309
Query: 314 TRYPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLL 364
R+P IFYLS+K KT T LRE Y V+ HP++ R K+ +MK L
Sbjct: 310 ERHPQIFYLSMKNKTCTAILREPYRDKASVETHPVLGVRKKYIQLMKNSEL 360
>At3g63090 unknown protein
Length = 404
Score = 164 bits (416), Expect = 6e-41
Identities = 110/356 (30%), Positives = 179/356 (49%), Gaps = 12/356 (3%)
Query: 18 LQWRGIAKVRLKWVKNRSLDHIIDKETDLK-AASLLKDAINRSSTGFLTAKSFSGWQKLL 76
+Q R I+ +++ W K+ LD I+++ K A ++K+ +N + + ++ L
Sbjct: 26 IQIRWISSLKVVWKKDTRLDEAIEQDKRYKLCARVVKEVLNEPGQ-VIPLRYLEKRRERL 84
Query: 77 GLTVPVLRFIRRYPTLF---HEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVE 133
LT F+ P+LF ++ P+ + R T +EQ +++ +E V
Sbjct: 85 RLTFKAKSFVEMNPSLFEIYYDRIKPKSDPVQFLRPTPRLRAFLDEEQRIYSENEPLLVS 144
Query: 134 RLSKFLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPN-GVSAVRLS 192
+L + LMM K + + L +K D G P+ F L+ K+P+ F+ P G S + L
Sbjct: 145 KLCQLLMMAKDKVISADKLVHVKRDFGFPNDFLMKLVQKYPNYFRLTGLPEEGKSFLELV 204
Query: 193 KWCEEFAVSALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKL 252
W +FA S ++ E + G R + ++ P G+ +K++R W ++ +
Sbjct: 205 SWNPDFAKSKIELRAEDETLKTGVRIRPN------YNVKLPSGFFLRKEMREWTRDWLEQ 258
Query: 253 PYISPYADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRI 312
YISPY D S +D S MEKR VGV+HELLSL++ K+ L +E + F+ +
Sbjct: 259 DYISPYEDVSHLDQASKEMEKRTVGVVHELLSLSLLKRVPVPTLGKFCDEFRFSNAFSSV 318
Query: 313 FTRYPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLLYRGD 368
FTR+ GIFYLSLK T LRE Y +LVD PL+ +DKF +++ G R D
Sbjct: 319 FTRHSGIFYLSLKGGIKTAVLREAYKDDELVDRDPLLAIKDKFLRLLEEGWQERKD 374
>At5g45790 tyrosine-specific protein phosphatase-like protein
Length = 423
Score = 154 bits (389), Expect = 9e-38
Identities = 95/329 (28%), Positives = 178/329 (53%), Gaps = 13/329 (3%)
Query: 31 VKNRSLDHIIDKET-DLKAASLLKDAINRSSTG----FLTAKSFSGWQKLLGLTVPVLRF 85
++NR+ DH +DK ++ +++ + S+ F++ + S W+ L+GL V V F
Sbjct: 49 LENRTRDHQLDKIIPQIRKLNIILEISKLMSSKKRGPFVSLQLMSRWKNLVGLNVNVGAF 108
Query: 86 IRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLSKFLMMTKTR 145
I +YP F F HP +L C ++T+ +L +E+++ E D V+R+ K L+++K
Sbjct: 109 IGKYPHAFEIFTHPFSKNL-CCKITEKFKVLIDEEENVVRECEVDAVKRVKKLLLLSKHG 167
Query: 146 TVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWCEEFAVSALQK 205
+ + +L ++ ++GLP+ F +++ K+ +F+ V + + + E V+ +++
Sbjct: 168 VLRVHALRLIRKELGLPEDFRDSILAKYSSEFRLVDLE---TLELVDRDDESLCVAKVEE 224
Query: 206 SNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYISPYADSSKID 265
E + + F+ A FP+ P G+ +K R + + ++PY+ PY D +I
Sbjct: 225 WREVEYREKWLSKFETNYA---FPIHLPTGFKIEKGFREELKNWQRVPYVKPY-DRKEIS 280
Query: 266 PNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTRYPGIFYLSLK 325
+ EKRVV V+HELLSLT+ K + L R++L + + ++PGIFY+S K
Sbjct: 281 RGLERFEKRVVAVIHELLSLTVEKMVEVERLAHFRKDLGIEVNVREVILKHPGIFYVSTK 340
Query: 326 CKTTTVTLREGYARGKLVDPHPLVRHRDK 354
+ T+ LRE Y++G L++P+P+ R K
Sbjct: 341 GSSQTLFLREAYSKGCLIEPNPIYNVRRK 369
>At5g21970 unknown protein
Length = 449
Score = 150 bits (380), Expect = 9e-37
Identities = 111/366 (30%), Positives = 177/366 (48%), Gaps = 27/366 (7%)
Query: 64 LTAKSFSGWQKLLGLTVP--VLRFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQ 121
+T +SF +++ + L P + FIR+ P LF + R L C LT+ L +
Sbjct: 89 MTVRSFEQYRRQINLPKPHKISDFIRKSPKLFELYKDQRGV-LWC-GLTEGGEDLLDEHD 146
Query: 122 SLHNHHENDTVERLSKFLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVK 181
L + + E +++ LMM+ + +PL + + D GLP F + FP F+ VK
Sbjct: 147 KLLEENGDKAAEHVTRCLMMSVDKKLPLDKIVHFRRDFGLPLDFRINWVYNFPQHFKVVK 206
Query: 182 APNGVSAVRLSKWCEEFAVSALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKK 241
+G + L W +A++ L+K D + K G +L FPM+FP Y +
Sbjct: 207 LGDGEEYLELVSWNPAWAITELEKKTLGITEDC---EHKPGMLSLAFPMKFPPSYKKMYR 263
Query: 242 VRFWMDEFHKLPYISPYADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLRE 301
R ++ F K Y+SPYAD+ ++ S +KR + V+HELLS T+ K+ ++L R
Sbjct: 264 YRGKIEHFQKRSYLSPYADARGLEAGSKEFDKRAIAVMHELLSFTLEKRLVTDHLTHFRR 323
Query: 302 ELNLPHKFTRIFTRYPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKT 361
E +P K RIF ++ GIFY+S + K +V L EGY +L++ PL+ ++K + T
Sbjct: 324 EFVMPQKLMRIFLKHCGIFYVSERGKRFSVFLTEGYEGPELIEKCPLILWKEKL--LKFT 381
Query: 362 GLLYRGDGSIPKPEGGALLLDE-------IGDE---------GSEEEEVETSDEFSEGEV 405
G YRG + L ++E GDE G ++ V DE GEV
Sbjct: 382 G--YRGRKRDIQTYSDTLDMEERELLESGSGDEDLSVGFEKDGDYDDVVTDDDEMDIGEV 439
Query: 406 SGSDSE 411
+ + E
Sbjct: 440 NDAYEE 445
>At2g31290 putative protein
Length = 415
Score = 143 bits (361), Expect = 2e-34
Identities = 103/334 (30%), Positives = 160/334 (47%), Gaps = 20/334 (5%)
Query: 32 KNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLLGLTVPVLRFIRRYPT 91
K+ L+ + + S LK+ I R + K K L L L ++++YP
Sbjct: 30 KDPDLESALSRNKRWIVNSRLKNIILRCPNQVASIKFLQKKFKTLDLQGKALNWLKKYPC 89
Query: 92 LFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLSKFLMMTKTRTVPLVS 151
FH + C RLT + L +E+ + + E +RL+K LM++ + + +V
Sbjct: 90 CFHVYLEN--DEYYC-RLTKPMMTLVEEEELVKDTQEPVLADRLAKLLMLSVNQRLNVVK 146
Query: 152 LYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSA--VRLSKWCEEFAVSALQKSNER 209
L K G PD + ++PK+ D F+ V S+ + L W E AVSA++ + ++
Sbjct: 147 LNEFKRSFGFPDDYVIRIVPKYSDVFRLVNYSGRKSSMEIELLLWKPELAVSAVEAAAKK 206
Query: 210 DGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYISPYADSSKIDPNSD 269
G + + + P + + + RF EF+ PYISPY D + S
Sbjct: 207 CGSEPSF--------SCSLPSTWTKPWE-----RFM--EFNAFPYISPYEDHGDLVEGSK 251
Query: 270 LMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTRYPGIFYLSLKCKTT 329
EKR VG++HELLSLT+ KK L + E LP K + ++PGIFY+ K +
Sbjct: 252 ESEKRSVGLVHELLSLTLWKKLSIVKLSHFKREFGLPEKLNGMLLKHPGIFYVGNKYQVH 311
Query: 330 TVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGL 363
TV LREGY +L+ PLV +DKF +M+ GL
Sbjct: 312 TVILREGYNGSELIHKDPLVVVKDKFGELMQQGL 345
>At1g79120 hypothetical protein
Length = 413
Score = 130 bits (328), Expect = 1e-30
Identities = 101/332 (30%), Positives = 154/332 (45%), Gaps = 20/332 (6%)
Query: 38 HIIDKETDL-KAASL---LKDAINRSSTGFLTAKSFSGWQKLLGLTVP--VLRFIRRYPT 91
H +DK DL K SL LK I G L + +K +G ++ I +YPT
Sbjct: 66 HDLDKAIDLNKKPSLILQLKSIIQAQKHGSLLLRDL---EKHVGFVHKWNLMAAIEKYPT 122
Query: 92 LFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLSKFLMMTKTRTVPLVS 151
+F+ + P P LT+ A + ++E E V L K LMM+ VPL
Sbjct: 123 IFYVGGGHKEP--PFVMLTEKAKKIAAEESEATESMEPILVNNLRKLLMMSVDCRVPLEK 180
Query: 152 LYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWCEEFAVSALQKSNERDG 211
+ ++ +GLP F+ TLIPK+ + F +K NG + L W A++A + R+G
Sbjct: 181 VEFIQSAMGLPQDFKSTLIPKYTEFFS-LKVINGKVNLVLENWDSSLAITAREDRLSREG 239
Query: 212 GDDGYRDFKRGKAAL--------VFPMRFPRGYGAQKKVRFWMDEFHKLPYISPYADSSK 263
+ D KR + F + FP G+ +++ K+ + SPY ++ +
Sbjct: 240 FSESVGDRKRVRITKDGNFLGRNAFKISFPPGFRPNASYLEEFEKWQKMEFPSPYLNARR 299
Query: 264 IDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTRYPGIFYLS 323
D KRVV VLHELLSLTM K+ L + E LP + ++ GIFY++
Sbjct: 300 FDAADPKARKRVVAVLHELLSLTMEKRVTCAQLDAFHSEYLLPSRLILCLIKHQGIFYIT 359
Query: 324 LKCKTTTVTLREGYARGKLVDPHPLVRHRDKF 355
K TV L++ YA L++ PL+ D+F
Sbjct: 360 NKGARGTVFLKDAYAGSNLIEKCPLLLFHDRF 391
>At1g06440 unknown protein
Length = 390
Score = 124 bits (311), Expect = 9e-29
Identities = 101/349 (28%), Positives = 165/349 (46%), Gaps = 19/349 (5%)
Query: 29 KWVKNRSLDHIIDKETDLKAASLLKDAINRSST--------GFLTAKSFSGWQKLLGLTV 80
++V +RS D + +K D K +LLK + T L+ + S + L L
Sbjct: 29 QYVASRSRDPVFEKLMD-KYKNLLKVIAIQDLTLANPTADPPSLSIEFLSRLSQKLHLNR 87
Query: 81 PVLRFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLSKFLM 140
F+R+YP +FH P + P RLTD A+ + QE + V+RL + L
Sbjct: 88 GAASFLRKYPHIFHVLYDPV-KAEPFCRLTDVAMEISRQEALAITATLSLVVDRLVRLLS 146
Query: 141 MTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNG----VSAVRLSKWCE 196
M+ ++++PL +++ + ++GLPD FE ++I K P F+ + V+ +
Sbjct: 147 MSISKSIPLRAVFKVWRELGLPDDFEDSVISKNPHLFKLSDGHESNTHILELVQEEEKRL 206
Query: 197 EFAVSALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYIS 256
EF + + +D D R + F +P G K + + E+ +LPY+
Sbjct: 207 EFEAAVEKWRVVECSKEDCSVD--RTEIQFSFKHSYPPGMRLSKTFKAKVKEWQRLPYVG 264
Query: 257 PYADS-SKIDPNSDLM--EKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIF 313
PY D K S +M EKR V + HE L+LT+ K + + R+ + +F
Sbjct: 265 PYEDMVGKKKSRSGVMGIEKRAVAIAHEFLNLTVEKMVEVEKISHFRKCFGIDLNIRDLF 324
Query: 314 TRYPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTG 362
+PG+FY+S K K TV LRE Y RG+L+DP+P+ R K ++ G
Sbjct: 325 LDHPGMFYVSTKGKRHTVFLREAYERGRLIDPNPVYDARRKLLDLVLLG 373
>At5g62990 putative protein
Length = 494
Score = 120 bits (301), Expect = 1e-27
Identities = 109/403 (27%), Positives = 182/403 (45%), Gaps = 34/403 (8%)
Query: 29 KWVKNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLLGLTVP--VLRFI 86
K V++ SLD + K+ ++ L + ++ + + L + P +L I
Sbjct: 58 KIVRSPSLDRHVVKQNRVRFVQKLNTLLLSKPKHYIPIEILYKCRSYLCIENPLAILSMI 117
Query: 87 RRYPTLFHEFPHPRWPSLP----------CFRLTDTALLLHSQEQSLHNHHENDTVERLS 136
RRYPT+F F P P LP C RLT A L QE +L + + +L
Sbjct: 118 RRYPTIFELFTTPT-PHLPMNATKPLSQLCVRLTSAASSLAMQELNLKSEISDKLATKLQ 176
Query: 137 KFLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWCE 196
K LM++ R + L L + D G P +F L +PD+F+ V G A+ L
Sbjct: 177 KLLMLSSHRRLLLSKLVHIAPDFGFPPNFRSRLCNDYPDKFKTVDTSYG-RALELVSSDP 235
Query: 197 EFAVSALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYIS 256
E A +R D FKR + RG +++ + ++ +F + P +
Sbjct: 236 ELANQMPSPEVDRGLIVDRPLKFKR--------LNLRRGLNLKRRHQGFLIKFRESPDVC 287
Query: 257 PYADSSKIDPNSDL-MEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTR 315
PY SS + + EKR V+ E+L LT+ K+T ++L R+E +LP+K + R
Sbjct: 288 PYKMSSDYLASESIEAEKRACAVVREVLGLTVEKRTLIDHLTHFRKEFSLPNKLRDLIVR 347
Query: 316 YPGIFYLSLKCKTTTVTLREGY-ARGKLVDPHPLVRHRDKFYHVMKTGLLYRGDGSIPKP 374
+P +FY+S+K +V L E Y G L+D + R++ +++ G R + +
Sbjct: 348 HPELFYVSIKGMRDSVFLVEAYNDNGDLLDKDERLVIRERLIDLIQEGKRIRRE----RR 403
Query: 375 EGGALLLDEIGDEGSEEEEV------ETSDEFSEGEVSGSDSE 411
GA L D+ +E +++E + DE+ +G + DSE
Sbjct: 404 RKGATLGDKSVEEKRKDDETIDDYDSDLDDEYEDGFENLFDSE 446
>At4g33495 unknown protein
Length = 354
Score = 119 bits (298), Expect = 3e-27
Identities = 88/275 (32%), Positives = 133/275 (48%), Gaps = 22/275 (8%)
Query: 85 FIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHE-----NDTVERLSKFL 139
F+ ++P +F + HP L C RLT AL +Q H H D V RL K +
Sbjct: 49 FLLKFPHVFEIYEHPVQRILYC-RLTRKAL-----DQIRHEHEAVLDQIPDAVTRLRKLV 102
Query: 140 MMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSA-VRLSKWCEEF 198
MM+ T + L + + + GLP+ FE ++I K P F+ + + + +
Sbjct: 103 MMSNTGRIRLEHVRIARTEFGLPEDFEYSVILKHPQFFRLIDGEETRDKYIEIVEKDPNL 162
Query: 199 AVSALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYISPY 258
++ A+++ E + G F + FP G+ K R + ++ +LPY SPY
Sbjct: 163 SICAIERVREIEYRTKG---IDAEDVRFSFVVNFPPGFKIGKYFRIAVWKWQRLPYWSPY 219
Query: 259 ADSSKIDPNS----DLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFT 314
D S D S + +EKR V +HELLSLT+ KK + R +NLP K
Sbjct: 220 EDISGYDLRSMEAQNRLEKRSVACIHELLSLTVEKKITLERIAHFRNVMNLPKKLKEFLL 279
Query: 315 RYPGIFYLSLK---CKTTTVTLREGYARGKLVDPH 346
++ GIFY+S + K TV LREGY RG+LV+P+
Sbjct: 280 QHQGIFYISTRGNYGKLHTVFLREGYKRGELVEPN 314
>At4g08940 unknown protein
Length = 395
Score = 102 bits (253), Expect = 5e-22
Identities = 78/276 (28%), Positives = 132/276 (47%), Gaps = 12/276 (4%)
Query: 82 VLRFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLSKFLMM 141
V +FI ++P +F + H F T+ L +E+ L E D V + K LMM
Sbjct: 119 VTKFIPKHPLIFQTYRHSDGKIWLGF--TEFMEDLLDEEKGLMESMELDRVNCVRKLLMM 176
Query: 142 TKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWCEEFAVS 201
TK + + L ++ + G+P+ F + + K+PD F+ V +G + L W AVS
Sbjct: 177 TKDKRILLSKIHHTRLLFGIPEDF-RDRVAKYPDYFRVVTGGDGNRVLELVNWDTNLAVS 235
Query: 202 ALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYISPYADS 261
L++ D D R FK FP++ + ++K ++ + P +SPY+D
Sbjct: 236 ELERLFMIDE-DKAKRVFK-------FPVKHGKELELEEKDTRKLNLLNTFPLVSPYSDG 287
Query: 262 SKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTRYPGIFY 321
K+D S EK VG++HE L+LT+ K+ +++ ++E +L + ++ + FY
Sbjct: 288 WKLDVWSLEAEKYRVGIVHEFLNLTLEKRASIHHIVEFKDEFSLTRQTYQMLKKQSMSFY 347
Query: 322 LSLKCKTTTVTLREGY-ARGKLVDPHPLVRHRDKFY 356
L+ TV L++ Y G LV P V +K Y
Sbjct: 348 LAGTEMNWTVFLKDAYDENGVLVSKDPQVVFNEKLY 383
>At3g04340 unknown protein
Length = 1320
Score = 30.4 bits (67), Expect = 1.9
Identities = 22/81 (27%), Positives = 41/81 (50%), Gaps = 6/81 (7%)
Query: 258 YADSSKIDPNSDLMEKRVVGVLHELLSL-----TMHKKTKRNYLRSLREELN-LPHKFTR 311
Y D ++ DL+ KR ++ E LSL + +K R + L E ++ + ++ +
Sbjct: 221 YRDMRRLRKERDLLMKRADKIVDEALSLKKQSEKLLRKGAREKMEKLEESVDIMESEYNK 280
Query: 312 IFTRYPGIFYLSLKCKTTTVT 332
I+ R I + LK +TTT++
Sbjct: 281 IWERIDEIDDIILKKETTTLS 301
>At1g56090 unknown protein
Length = 272
Score = 29.6 bits (65), Expect = 3.2
Identities = 23/69 (33%), Positives = 37/69 (53%), Gaps = 6/69 (8%)
Query: 339 RGKLVDPH-PLVRHRDKFYHVMKTGLLYRGDGSIPKPEGGALL----LDEIGDEGSEEEE 393
+G++V P P VR ++ V +G +IPKP+G + L + + D+ SEEE+
Sbjct: 187 KGEVVAPKTPEVREQNS-KEVPMSGKQSNAWQAIPKPKGHSTLDYARWNTVEDDSSEEED 245
Query: 394 VETSDEFSE 402
E SD+ E
Sbjct: 246 DEDSDDSDE 254
>At4g15810 hypothetical protein
Length = 526
Score = 28.9 bits (63), Expect = 5.4
Identities = 17/42 (40%), Positives = 23/42 (54%), Gaps = 11/42 (26%)
Query: 379 LLLDEIGDEGSE-----------EEEVETSDEFSEGEVSGSD 409
LL +E+G +GSE +E+ E SDE E EVSG +
Sbjct: 92 LLGEELGGDGSEFQVGVGFIDHLQEDGENSDEVCESEVSGDE 133
>At1g79990 coatomer protein like complex, subunit beta 2 (beta
prime)
Length = 920
Score = 28.9 bits (63), Expect = 5.4
Identities = 14/35 (40%), Positives = 21/35 (60%), Gaps = 3/35 (8%)
Query: 380 LLDEIGDE---GSEEEEVETSDEFSEGEVSGSDSE 411
+LDE+G+E G EEEE + +E S+G + E
Sbjct: 855 VLDEVGEEGEDGEEEEEEDRQEESSDGRQQNVEEE 889
>At1g25360
Length = 790
Score = 28.9 bits (63), Expect = 5.4
Identities = 21/81 (25%), Positives = 34/81 (41%), Gaps = 12/81 (14%)
Query: 289 KKTKRNYLRSLREELNLPHKFTRIFTRYPGIFYLSLKCKTTTVTLREGYARGKLVDPHPL 348
K ++ NY R L +E++ P K R +S C + +TL G + + P+
Sbjct: 61 KSSELNYARQLFDEISEPDKIAR-------TTMVSGYCASGDITLARG-----VFEKAPV 108
Query: 349 VRHRDKFYHVMKTGLLYRGDG 369
Y+ M TG + DG
Sbjct: 109 CMRDTVMYNAMITGFSHNNDG 129
>At4g11300 unknown protein
Length = 371
Score = 28.5 bits (62), Expect = 7.0
Identities = 18/55 (32%), Positives = 29/55 (52%), Gaps = 3/55 (5%)
Query: 184 NGVSAVRLSKWCEEFAVSALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGA 238
NGV +VR + C E AV+AL+++ DG R KR +L+ + + G+
Sbjct: 120 NGVDSVRQIQRCAEIAVTALKQTPLSDG---SVRRAKRALTSLLAALNADKNSGS 171
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.138 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,822,089
Number of Sequences: 26719
Number of extensions: 446041
Number of successful extensions: 1595
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 1542
Number of HSP's gapped (non-prelim): 27
length of query: 411
length of database: 11,318,596
effective HSP length: 102
effective length of query: 309
effective length of database: 8,593,258
effective search space: 2655316722
effective search space used: 2655316722
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0029a.7


