

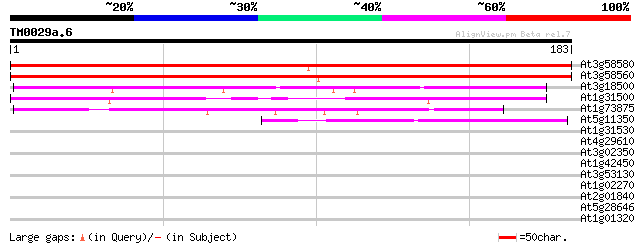
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0029a.6
(183 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g58580 putative protein 308 1e-84
At3g58560 unknown protein 296 5e-81
At3g18500 unknown protein 74 6e-14
At1g31500 unknown protein 58 2e-09
At1g73875 putative protein 52 2e-07
At5g11350 unknown protein 50 5e-07
At1g31530 putative protein 37 0.005
At4g29610 cytidine deaminase 6 (CDA6) 28 2.8
At3g02350 unknown protein 28 2.8
At1g42450 mudrA-like protein 28 2.8
At3g53130 Cytochrom P450 -like protein 28 3.6
At1g02270 unknown protein 28 3.6
At2g01840 putative non-LTR retroelement reverse transcriptase 27 4.7
At5g28646 putative protein 27 8.0
At1g01320 unknown protein 27 8.0
>At3g58580 putative protein
Length = 605
Score = 308 bits (789), Expect = 1e-84
Identities = 152/185 (82%), Positives = 163/185 (87%), Gaps = 2/185 (1%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKV 60
ANTHVNV QDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFN+ PGSAPH LL MGKV
Sbjct: 420 ANTHVNVQQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNTLPGSAPHTLLVMGKV 479
Query: 61 DPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFAR--TIGLGYEQHKRRLDSATNEPLF 118
DP HPDLA+DPL+ILRPH+KL HQLPLVSAYSSF R +GLG EQH+RR+D TNEPLF
Sbjct: 480 DPMHPDLAVDPLNILRPHTKLTHQLPLVSAYSSFVRKGIMGLGLEQHRRRIDLNTNEPLF 539
Query: 119 TNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCC 178
TN TRDFIG DYIFYTAD+L+VESLLELLDE+ LRKDTALPSPEWSS+HIALLAEFRC
Sbjct: 540 TNCTRDFIGTHDYIFYTADTLMVESLLELLDEDGLRKDTALPSPEWSSNHIALLAEFRCT 599
Query: 179 KNRSR 183
R
Sbjct: 600 PRTRR 604
>At3g58560 unknown protein
Length = 602
Score = 296 bits (757), Expect = 5e-81
Identities = 149/184 (80%), Positives = 159/184 (85%), Gaps = 1/184 (0%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKV 60
ANTHVNV +LKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFN+ P SAPH LLA+GKV
Sbjct: 415 ANTHVNVPHELKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNTVPASAPHTLLAVGKV 474
Query: 61 DPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIG-LGYEQHKRRLDSATNEPLFT 119
DP HPDL +DPL ILRPHSKL HQLPLVSAYS FA+ G + EQ +RRLD A++EPLFT
Sbjct: 475 DPLHPDLMVDPLGILRPHSKLTHQLPLVSAYSQFAKMGGNVITEQQRRRLDPASSEPLFT 534
Query: 120 NVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCK 179
N TRDFIG LDYIFYTAD+L VESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRC
Sbjct: 535 NCTRDFIGTLDYIFYTADTLTVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCMP 594
Query: 180 NRSR 183
R
Sbjct: 595 RARR 598
>At3g18500 unknown protein
Length = 251
Score = 73.6 bits (179), Expect = 6e-14
Identities = 59/200 (29%), Positives = 94/200 (46%), Gaps = 28/200 (14%)
Query: 2 NTHVNVHQDLKDVKLWQVHTLLKGLEKIAAS-ADIPMLVCGDFNSNPGSAPHALLAMGKV 60
N HV + + DVKL QV +L ++ DIP+++CGDFNS P S + LA ++
Sbjct: 48 NIHVLYNPNQGDVKLGQVRSLCSKAHLLSKKWGDIPIVLCGDFNSTPKSPLYNFLASSEL 107
Query: 61 DPSHPDLA-------IDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQ-----HKRR 108
+ D P +L SK + + S+++ + G E H +
Sbjct: 108 NVMEHDKKELSGQKNCRPTKVLETGSKSSNTITF-SSWTKEEIRVATGQENSYWAAHPLK 166
Query: 109 LDS-------------ATNEPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRK 155
L+S + EPL T+ F+G +DY++Y +D L+ +L+ L + L K
Sbjct: 167 LNSSYASVKGSANTRDSVGEPLATSYHSKFLGTVDYLWY-SDGLLPARVLDTLPIDVLCK 225
Query: 156 DTALPSPEWSSDHIALLAEF 175
LP E SDH+AL++EF
Sbjct: 226 TKGLPCQELGSDHLALVSEF 245
>At1g31500 unknown protein
Length = 335
Score = 58.2 bits (139), Expect = 2e-09
Identities = 54/182 (29%), Positives = 76/182 (41%), Gaps = 37/182 (20%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAA------SADIPMLVCGDFNSNPGSAPHAL 54
ANTH+ +L DVKL Q LL L + +L+ GDFNS PG ++
Sbjct: 179 ANTHLYWDPELADVKLAQAKYLLSRLAQFKTLISDEFECTPSLLLAGDFNSIPGDMVYSY 238
Query: 55 LAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSATN 114
L G P+ +I + + PL S Y +
Sbjct: 239 LVSGNAKPTE--------TIEEEEAPV----PLSSVY------------------EVTRG 268
Query: 115 EPLFTNVTRDFIGALDYIFYT-ADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLA 173
EP FTN T F LDYIF + +D + S+L+L + +S LP+ SDH+ + A
Sbjct: 269 EPKFTNCTPGFTNTLDYIFISPSDFIKPVSILQLPEPDSPDVVGFLPNHHHPSDHLPIGA 328
Query: 174 EF 175
EF
Sbjct: 329 EF 330
>At1g73875 putative protein
Length = 201
Score = 52.0 bits (123), Expect = 2e-07
Identities = 50/181 (27%), Positives = 79/181 (43%), Gaps = 28/181 (15%)
Query: 2 NTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKVD 61
N HV + D+KL Q + L + +IP+ + GD NS P SA + +A +D
Sbjct: 22 NIHVLFNPKRGDIKLGQAYKLSQEW------GNIPVAIAGDLNSTPQSAIYDFIASADLD 75
Query: 62 PS-HPDLAIDPLSILRPHSKLVHQL--PLVSAYSSFARTIGLG-----YEQHKRRLDSA- 112
H I + + P + L++ +S + G + QH+ +L+SA
Sbjct: 76 TQLHDRRQISGQTEVEPKESASASISGSLLNEWSQEELQLATGGQETTHVQHQLKLNSAY 135
Query: 113 ------------TNEPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALP 160
EPL T F+G +DYI++T + LV +LE L + LR+ LP
Sbjct: 136 SGVPGTYRTRDQRGEPLATTYHSRFLGTVDYIWHTKE-LVPVRVLETLPADVLRRTGGLP 194
Query: 161 S 161
S
Sbjct: 195 S 195
>At5g11350 unknown protein
Length = 754
Score = 50.4 bits (119), Expect = 5e-07
Identities = 28/100 (28%), Positives = 51/100 (51%), Gaps = 10/100 (10%)
Query: 83 HQLPLVSAYSSFARTIGLGYEQHKRRLDSATNEPLFTNVTRDFIGALDYIFYTADSLVVE 142
H L L S YS + + EP+ T+ R F+G +DYI + ++ L
Sbjct: 661 HALELKSTYSEV---------EGQANTRDENGEPVVTSYHRCFMGTVDYI-WRSEGLQTV 710
Query: 143 SLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKNRS 182
+L + +++++ P+P+W SDHIAL++E C +++
Sbjct: 711 RVLAPIPKQAMQWTPGFPTPKWGSDHIALVSELAFCSSKT 750
Score = 38.5 bits (88), Expect = 0.002
Identities = 27/73 (36%), Positives = 38/73 (51%), Gaps = 3/73 (4%)
Query: 2 NTHVNVHQDLKDVKLWQVHTLLKGLEKIAAS-ADIPMLVCGDFNSNPGSAPHALLAMGKV 60
N HV + D KL QV TLL ++ D P+++CGDFN P S + ++ K+
Sbjct: 331 NIHVLFNPKRGDFKLGQVRTLLDKAHAVSKLWDDAPIVLCGDFNCTPKSPLYNFISDRKL 390
Query: 61 DPSHPDLAIDPLS 73
D S LA D +S
Sbjct: 391 DLS--GLARDKVS 401
>At1g31530 putative protein
Length = 283
Score = 37.4 bits (85), Expect = 0.005
Identities = 25/66 (37%), Positives = 35/66 (52%), Gaps = 4/66 (6%)
Query: 115 EPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAE 174
EP FTN F LDY+FYT ++ S ++LLD S + LP+ SDH+ + E
Sbjct: 221 EPRFTNNVPGFAETLDYMFYTHSEII--SPVKLLD--SPDEVDFLPNEIHPSDHLPIGVE 276
Query: 175 FRCCKN 180
F +N
Sbjct: 277 FEINRN 282
>At4g29610 cytidine deaminase 6 (CDA6)
Length = 293
Score = 28.1 bits (61), Expect = 2.8
Identities = 15/37 (40%), Positives = 20/37 (53%)
Query: 10 DLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSN 46
++KDVK+ Q T L+KIA D +L C N N
Sbjct: 257 EMKDVKVSQEATARTFLDKIAPKCDFKVLHCYKTNKN 293
>At3g02350 unknown protein
Length = 561
Score = 28.1 bits (61), Expect = 2.8
Identities = 23/78 (29%), Positives = 37/78 (46%), Gaps = 13/78 (16%)
Query: 52 HALLAMGKVDPSHPDLAI--DPLSILRP--HSKLVHQLPLVSAYSSFARTIGLGYEQHKR 107
H L +MG LA+ DPL H + + L LV+AY+++AR K
Sbjct: 57 HTLPSMGNAYMQRTFLALQSDPLKTRLDLIHKQAIDHLTLVNAYAAYAR---------KL 107
Query: 108 RLDSATNEPLFTNVTRDF 125
+LD++ LF ++ +F
Sbjct: 108 KLDASKQLKLFEDLAINF 125
>At1g42450 mudrA-like protein
Length = 601
Score = 28.1 bits (61), Expect = 2.8
Identities = 14/36 (38%), Positives = 22/36 (60%), Gaps = 2/36 (5%)
Query: 38 LVCGDFNSNPGSAPHALLAMGKVDPSHPDLAIDPLS 73
+V G +SNP + L +G VDP+ PD+++ LS
Sbjct: 154 MVIGHTSSNPEESIR--LTLGSVDPARPDVSVVALS 187
>At3g53130 Cytochrom P450 -like protein
Length = 539
Score = 27.7 bits (60), Expect = 3.6
Identities = 22/55 (40%), Positives = 28/55 (50%), Gaps = 5/55 (9%)
Query: 111 SATN-EPLFTNVTRDFIGA--LDYIF--YTADSLVVESLLELLDEESLRKDTALP 160
SA N E F+ +T D IG +Y F T DS V+E++ L E LR LP
Sbjct: 208 SAVNMEAKFSQMTLDVIGLSLFNYNFDSLTTDSPVIEAVYTALKEAELRSTDLLP 262
>At1g02270 unknown protein
Length = 484
Score = 27.7 bits (60), Expect = 3.6
Identities = 17/50 (34%), Positives = 28/50 (56%), Gaps = 5/50 (10%)
Query: 2 NTHVNVHQD--LKDVKLWQVHTLLKGLEKIAASADI---PMLVCGDFNSN 46
NTH+ D L V+L QV+ +L+ LE + P+++CGD+N +
Sbjct: 215 NTHLLFPHDSSLSIVRLHQVYKILEYLEAYQKENKLNHMPIILCGDWNGS 264
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 27.3 bits (59), Expect = 4.7
Identities = 10/25 (40%), Positives = 17/25 (68%)
Query: 20 HTLLKGLEKIAASADIPMLVCGDFN 44
H L + L++++A P ++CGDFN
Sbjct: 476 HHLWEKLQRVSAHRSGPWMMCGDFN 500
>At5g28646 putative protein
Length = 293
Score = 26.6 bits (57), Expect = 8.0
Identities = 16/69 (23%), Positives = 28/69 (40%)
Query: 100 LGYEQHKRRLDSATNEPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTAL 159
+ Y +R S NEP F+ +DF DS+ ++D+ L++D +
Sbjct: 176 VAYVDRERLQHSINNEPRFSKEAKDFFQWRKSCEEEIDSVNKNKTWFIVDKTKLKEDVCI 235
Query: 160 PSPEWSSDH 168
E +H
Sbjct: 236 DQYEIFEEH 244
>At1g01320 unknown protein
Length = 1047
Score = 26.6 bits (57), Expect = 8.0
Identities = 22/81 (27%), Positives = 36/81 (44%), Gaps = 10/81 (12%)
Query: 44 NSNPGSAPHALLAMGKVDPSHPDLAIDPLS-ILRPHSKLVHQLPLVSAYSSFARTIGLGY 102
NSNP ++ +P HPD + D + RP S + + +S IG Y
Sbjct: 476 NSNPITSSDV-----STEPQHPDGSEDGWQPVQRPRSAGSYGRRMKQRRAS----IGKVY 526
Query: 103 EQHKRRLDSATNEPLFTNVTR 123
K+ +++ + PLF N T+
Sbjct: 527 TYQKKNVEADIDNPLFQNATQ 547
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,148,879
Number of Sequences: 26719
Number of extensions: 164342
Number of successful extensions: 395
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 374
Number of HSP's gapped (non-prelim): 16
length of query: 183
length of database: 11,318,596
effective HSP length: 93
effective length of query: 90
effective length of database: 8,833,729
effective search space: 795035610
effective search space used: 795035610
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0029a.6


