

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0028b.5
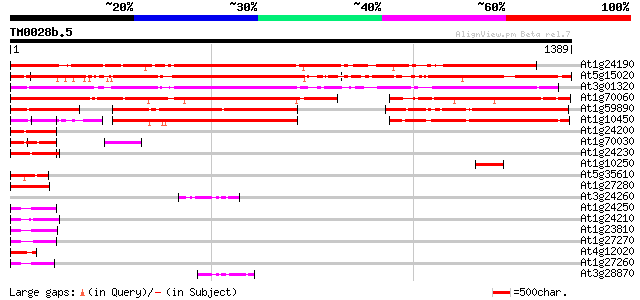
(1389 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g24190 hypothetical protein 1109 0.0
At5g15020 transcriptional regulatory - like protein 1075 0.0
At3g01320 unknown protein 1035 0.0
At1g70060 hypothetical protein 857 0.0
At1g59890 hypothetical protein 464 e-130
At1g10450 unknown protein 442 e-124
At1g24200 hypothetical protein 144 3e-34
At1g70030 unknown protein 127 3e-29
At1g24230 hypothetical protein 103 5e-22
At1g10250 unknown protein 102 2e-21
At5g35610 putative protein 90 8e-18
At1g27280 hypothetical protein 88 4e-17
At3g24260 hypothetical protein 70 1e-11
At1g24250 hypothetical protein 69 2e-11
At1g24210 unknown protein 67 5e-11
At1g23810 hypothetical protein 66 1e-10
At1g27270 hypothetical protein 65 4e-10
At4g12020 putative disease resistance protein 64 6e-10
At1g27260 hypothetical protein 63 1e-09
At3g28870 hypothetical protein 60 9e-09
>At1g24190 hypothetical protein
Length = 1263
Score = 1109 bits (2869), Expect = 0.0
Identities = 676/1348 (50%), Positives = 853/1348 (63%), Gaps = 176/1348 (13%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNK 60
+DTAGVI RVKELF+GH++LILGFNTFLPKG+EITL ED QPP KK VEFEEAI+FVNK
Sbjct: 48 VDTAGVITRVKELFKGHQELILGFNTFLPKGFEITLQPEDGQPPLKKRVEFEEAISFVNK 107
Query: 61 IKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTHFLPDASAA 120
IK RF+GDD VYKSFLDILNMYR+++K I EVY EVA+LF+ H+DLL EFTHFLPD SA
Sbjct: 108 IKTRFQGDDRVYKSFLDILNMYRRDSKSITEVYQEVAILFRDHSDLLVEFTHFLPDTSAT 167
Query: 121 ASSYYMSARNSVLRDRRSAMPTVRQAHVVKRERTTVSHCDRDPSVDRPDPDHDRGLLRAE 180
AS +P+V+ + RER VS D+ + P PDHD G +
Sbjct: 168 AS-----------------IPSVKTS---VRERG-VSLADKKDRIITPHPDHDYGTEHID 206
Query: 181 KEQRRRVEKE-KDH-REDGKRDWERDGRDHEHDGGQYRERLSHKRKSDRKAEDFGAEPLL 238
+++ R ++KE K+H R K + RD RD E + + L+ K+K + +D +
Sbjct: 207 QDRERPIKKENKEHMRGTNKENEHRDARDFEPHSKK-EQFLNKKQKLHIRGDD--PAEIS 263
Query: 239 DADEFFGTRPMSSTCDDKKSLKTKYPEEFAFCDKVKEKLRNPDGYQEFLKCLHIYSKEII 298
+ + G P SST D+K ++K+ Y ++ A D+VKEKL N YQEFL+CL+++SKEII
Sbjct: 264 NQSKLSGAVPSSSTYDEKGAMKS-YSQDLAIVDRVKEKL-NASEYQEFLRCLNLFSKEII 321
Query: 299 TQQELQSLVGDLLGKYPDLMEEFNEFLVQAEKNDG------------GFLAGVMNKKSLW 346
++ ELQSLVG+L+G YPDLM+ F EFLVQ EKN+ G L+G++ KKSLW
Sbjct: 322 SRPELQSLVGNLIGVYPDLMDSFIEFLVQCEKNEKRQICNLLNLLAEGLLSGILTKKSLW 381
Query: 347 IEGPGLKPMKAEDRDRDRDRDRYRDDGMKERDRECRERDRSTVVSNKEVSGLKIKDKHFL 406
EG +P D DRD++ RDDG+++RD E +ER K + LK +
Sbjct: 382 SEGKYPQP----SLDNDRDQEHKRDDGLRDRDHE-KER------LEKAAANLK-----WA 425
Query: 407 KPINELDLSNCDRCTPSYRLLPKNYPIPIASYKSKIGAKVLNDHWVSVTSGSEDYSFKHM 466
KPI+ELDLSNC++CTPSYRLLPKNYPI IAS K++IG VLNDHWVSVTSGSEDYSF HM
Sbjct: 426 KPISELDLSNCEQCTPSYRLLPKNYPISIASQKTEIGKLVLNDHWVSVTSGSEDYSFSHM 485
Query: 467 RKNQYEESLFRCEDDRFELDMLLESVNLTTKRVEELFEKINNNVIKGDRPVRIDEHLTAL 526
RKNQYEESLF+CEDDRFELDMLLESVN TTK VEEL KIN+N +K + P+R+++HLTAL
Sbjct: 486 RKNQYEESLFKCEDDRFELDMLLESVNSTTKHVEELLTKINSNELKTNSPIRVEDHLTAL 545
Query: 527 NLRCIERLYGDHGLDVMEVLRKNAPLALPVILTRLKQKQDEWARCRADFSKVWAEIYAKN 586
NLRCIERLYGDHGLDVM+VL+KN LALPVILTRLKQKQ+EWARCR+DF KVWAEIYAKN
Sbjct: 546 NLRCIERLYGDHGLDVMDVLKKNVSLALPVILTRLKQKQEEWARCRSDFDKVWAEIYAKN 605
Query: 587 YHKSLDHRSFYFKQQDTKRLSTKALLAEVKEISEKKHKEDDVLLAIAAGSRQPIIPNLEF 646
Y+KSLDHRSFYFKQQD+K+LS KALLAE+KEI+EKK +EDD LLA AAG+R I P+LEF
Sbjct: 606 YYKSLDHRSFYFKQQDSKKLSMKALLAEIKEITEKK-REDDSLLAFAAGNRLSISPDLEF 664
Query: 647 HYPDLDIHEDLYQLVKYSCGELCTAEQLDKVMKVWTTFLEPMLCVPSRAQGAEDTEDVVK 706
YPD D+HEDLYQL+KYSC E+C+ EQLDKVMK+WTTF+E + VPSR QGAED EDVVK
Sbjct: 665 DYPDHDLHEDLYQLIKYSCAEMCSTEQLDKVMKIWTTFVEQIFGVPSRPQGAEDQEDVVK 724
Query: 707 AKNTEDVVNTENNSVKSG--------IVSVAESGDSK-AWQLNGDSGVREDRCLDSDHTV 757
+ N N ++ S +G SVA+S SK + + N S + + + D
Sbjct: 725 SMNQ----NVKSGSSSAGESEGSPHNYASVADSRRSKSSRKANEHSQLGQTSNSERDGAA 780
Query: 758 YKTETAGNNT-QHGKTNIIGFTPDELSGVNKQNQSSERLVNANVSPASG-MEQSNGRTKI 815
+T A T QH K T DE +KQ S ER ++ G ++QSNG + I
Sbjct: 781 GRTSDALCETAQHEKMLKNVVTSDE-KPESKQAVSIERAHDSTALAVDGLLDQSNGGSSI 839
Query: 816 DNTSGHTATPSGNGNTSVEGGLELPSSEPILFQTRKFPSNYSIFSLMLDILINFGLHLHE 875
+ +GH N V G EL + + N + L+ G+ +
Sbjct: 840 VHMTGH----CNNNLKPVTCGTEL--------ELKMNDGNGPKLEVGNKKLLTNGIAVEI 887
Query: 876 CILQRILFVKHPSYGWLMILVVDISSRFQIRQGGDSTRPGTF--TNGATTGGTEVHRY-- 931
Q + R+ G+ + G F N A T+ +
Sbjct: 888 TSDQEMAGTSKVE-----------------REEGELSPNGDFEEDNFAVYAKTDFETFSK 930
Query: 932 -QEQSIRHFKSER-EEGELS-----PNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQS 984
+ + + +R EGE S D E D A S D+ ++ + GD+S +S
Sbjct: 931 ANDSTGNNISGDRSREGEPSCLETRAENDAEGDENAA--RSSEDSRNEYENGDVSGT-ES 987
Query: 985 RHGEQVCCEAKGENDVDADDEGEESPHRSSDDSENASENVDVSGSESADGE-ECSREEHE 1043
GE E+D+D +++GE ++GE EC + H+
Sbjct: 988 GGGED------PEDDLDNNNKGE------------------------SEGEAECMADAHD 1017
Query: 1044 DREHDNKAESEGEAEGMVDAHDVEGDGMSLTISERFLQTVKPLAKYVPA--ALHEKD--- 1098
AE G A L +S RFL VKPL KYVP+ ALH+KD
Sbjct: 1018 -------AEENGSA---------------LPVSARFLLHVKPLVKYVPSAIALHDKDKDS 1055
Query: 1099 -RNSQVFYGSDSFYVLFRLHQTLYERIQSAKFNSSSAEKKWKASHDTGSTDQYDRFMNAL 1157
+NSQVFYG+DSFYVLFRLH+ LYERI SAK NSSS E KW+ S+ TD Y RFM AL
Sbjct: 1056 LKNSQVFYGNDSFYVLFRLHRILYERILSAKVNSSSPEGKWRTSNTKNPTDSYARFMTAL 1115
Query: 1158 YSLLDGSSDNTKFEDDCRAIIGTQSYLLFTLDKLIYKLVKQLQAIASEEMDNKLLQLYAY 1217
Y+LLDG+SDN KFEDDCRAIIGTQSY+LFTLDKLI+K +K LQ + ++EMDNKLLQLY Y
Sbjct: 1116 YNLLDGTSDNAKFEDDCRAIIGTQSYILFTLDKLIHKFIKHLQVVVADEMDNKLLQLYFY 1175
Query: 1218 EKSRKLGKFIDIVYHENACVILHEENIYRIEYS-PGPKKLSIQLMDCGHDKPEVTAVSVD 1276
EKSR+ D VY++N V+L +ENIYRIE P KLSIQLM G DKP+VT+VS+D
Sbjct: 1176 EKSRRPETIFDAVYYDNTRVLLPDENIYRIECRLSTPAKLSIQLMCNGLDKPDVTSVSID 1235
Query: 1277 PKFSTYLHNDFLSVVPDKKEKSRLFLKR 1304
P F+ YLHNDFLS+ P+ +E R++L R
Sbjct: 1236 PTFAAYLHNDFLSIQPNAREDRRIYLNR 1263
>At5g15020 transcriptional regulatory - like protein
Length = 1343
Score = 1075 bits (2781), Expect = 0.0
Identities = 661/1416 (46%), Positives = 872/1416 (60%), Gaps = 189/1416 (13%)
Query: 2 DTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNKI 61
DT+GVI+RVKELF+GH +LI GFNTFLPKG+EITL +D + P+KK VEFEEAI+FVNKI
Sbjct: 87 DTSGVISRVKELFKGHNNLIFGFNTFLPKGFEITL--DDVEAPSKKTVEFEEAISFVNKI 144
Query: 62 KARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTHFLPDASAA- 120
K RF+ ++ VYKSFL+ILNMYRK+NK I EVY+EV+ LF+ H+DLL+EFT FLPD+ A
Sbjct: 145 KTRFQHNELVYKSFLEILNMYRKDNKDITEVYNEVSTLFEDHSDLLEEFTRFLPDSLAPH 204
Query: 121 ASSYYMSARNSVLRDRRSAMPTVRQAHVVK---RERTTVSHCDRDPSVDRPDPDHDRGLL 177
+ + ++ DR S P VR+ + K RERT S DRD SVDR D + D+ ++
Sbjct: 205 TEAQLLRSQAQRYDDRGSGPPLVRRMFMEKDRRRERTVASRGDRDHSVDRSDLNDDKSMV 264
Query: 178 RAEKEQRRRVEKEKDHREDGKRDWERDGRDHEHDGGQYRERLSHKRKSDRKAEDFGAEPL 237
+ ++QR+RV+K D+RE RD E DG + E D Q+ S KRKS R+ E F A
Sbjct: 265 KMHRDQRKRVDK--DNRERRSRDLE-DG-EAEQDNLQH---FSEKRKSSRRMEGFEAY-- 315
Query: 238 LDADEFFGTRPMSSTCDDKKSLKTKYPEEFAFCDKVKEKLRNPDGYQEFLKCLHIYSKEI 297
+ P S + +K +LK+ Y + F FC+KVKE+L + D YQ FLKCL+++S I
Sbjct: 316 --------SGPASHS--EKNNLKSMYNQAFLFCEKVKERLCSQDDYQAFLKCLNMFSNGI 365
Query: 298 ITQQELQSLVGDLLGKYPDLMEEFNEFLVQAEKNDG-GFLAGVMNKKSLWIEGPGLKPMK 356
I +++LQ+LV D+LGK+PDLM+EFN+F + E DG LAGVM+KKSL E + +K
Sbjct: 366 IQRKDLQNLVSDVLGKFPDLMDEFNQFFERCESIDGFQHLAGVMSKKSLGSEENLSRSVK 425
Query: 357 AEDRDRDRDRDRYRDDGMKERDRECRERDRSTVVSNKEVSGLKIKDKHFLKPINELDLSN 416
E++DR+ RD + KE++R KDK+ K I ELDLS+
Sbjct: 426 GEEKDREHKRDV---EAAKEKERS--------------------KDKYMGKSIQELDLSD 462
Query: 417 CDRCTPSYRLLPKNYPIPIASYKSKIGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLF 476
C+RCTPSYRLLP +YPIP ++ K GA VLNDHWVSVTSGSEDYSFKHMR+NQYEESLF
Sbjct: 463 CERCTPSYRLLPPDYPIPSVRHRQKSGAAVLNDHWVSVTSGSEDYSFKHMRRNQYEESLF 522
Query: 477 RCEDDRFELDMLLESVNLTTKRVEELFEKINNNVIKGDRPVRIDEHLTALNLRCIERLYG 536
RCEDDRFELDMLLESV K EEL I + I + RI++H TALNLRCIERLYG
Sbjct: 523 RCEDDRFELDMLLESVGSAAKSAEELLNIIIDKKISFEGSFRIEDHFTALNLRCIERLYG 582
Query: 537 DHGLDVMEVLRKNAPLALPVILTRLKQKQDEWARCRADFSKVWAEIYAKNYHKSLDHRSF 596
DHGLDV +++RKN ALPVILTRLKQKQDEW +CR F+ VWA++YAKN++KSLDHRSF
Sbjct: 583 DHGLDVTDLIRKNPAAALPVILTRLKQKQDEWTKCREGFNVVWADVYAKNHYKSLDHRSF 642
Query: 597 YFKQQDTKRLSTKALLAEVKEISEKKHKEDDVLLAIAAGSRQPIIPNLEFHYPDLDIHED 656
YFKQQD+K LS KAL++EVK++ EK KEDDV+L+I+AG RQPIIP+LE+ Y D IHED
Sbjct: 643 YFKQQDSKNLSAKALVSEVKDLKEKSQKEDDVVLSISAGYRQPIIPHLEYDYLDRAIHED 702
Query: 657 LYQLVKYSCGELC-TAEQLDKVMKVWTTFLEPMLCVPSRAQGAEDTEDVVKAKNTEDVVN 715
L++LV++SC E+C T EQ KV+K+W FLE ML V RA+G++ EDVV+ ++ +
Sbjct: 703 LFKLVQFSCEEICSTKEQTGKVLKLWANFLELMLDVAPRAKGSDSVEDVVETQHQRAFTS 762
Query: 716 TENNSVKSGIVSVAESGDSKAWQLNGDSGVREDRCLDSDHTVYKTETAGNNTQHGKTNII 775
E N I V+ + NGD + ++HG+T ++
Sbjct: 763 GEANESSDAISLVSRQ---LKFATNGD-----------------VHASSGVSKHGETGLL 802
Query: 776 GFTPDELSGVNKQNQSSERLVNANVSPASGMEQSNGRTKIDNTSGHTATPSGNGNTSVEG 835
+ SG K+N L N +V+ + Q + GNG G
Sbjct: 803 N---RDSSG--KENLKDGDLANKDVATCAEKPQKDQEI-------------GNGAAKRSG 844
Query: 836 GLELPSSEPILFQTRKFPSNYSIFSLMLDILINFGLHLHECILQRILFVKHPSYGWLMIL 895
++ E + + FPS + + + G R + K PS
Sbjct: 845 DVD----ERVATSSSSFPSGVENNNGKVGSRDSSG--------SRGILSK-PSEA----- 886
Query: 896 VVDISSRFQIRQGGDSTRPGTFTNGATTGGTEVHRYQEQSIRHFKSEREEGELSPNGDME 955
+D Q QG D R NG + ++ + ++S K E+EEGELSP GD
Sbjct: 887 -IDKVDSIQHTQGVDIGRIIVLGNGLQSDTSKANSNYDESGGPSKIEKEEGELSPVGD-S 944
Query: 956 EDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGENDVDADDEGEESPHRSSD 1015
EDNF VYE+ L A K + H EA+GEND DADDE D
Sbjct: 945 EDNFVVYEDRELKATAK-----------TEHS----VEAEGENDEDADDE-------DGD 982
Query: 1016 DSENASENVDVSGSESADGEECSREEH---EDREHDN---KAESEGEAEGMVDAHDVEGD 1069
D+ A E D SG+ES G+ECS++++ E+ EHD KAESEGEAEGM ++H +E
Sbjct: 983 DASEAGE--DASGTESI-GDECSQDDNGVEEEGEHDEIDGKAESEGEAEGM-ESHLIEDK 1038
Query: 1070 GMSLTISERFLQTVKPLAKYVPAA--LHEKDRNSQVFYGSDSFYVLFRLHQT-------- 1119
G+ SER L +VKPL+K++ AA + EK ++S+VFYG+D FYVLFRLH+
Sbjct: 1039 GL-FPSSERVLLSVKPLSKHIAAAALVDEKKKDSRVFYGNDDFYVLFRLHRVSAIDSYDL 1097
Query: 1120 ----LYERIQSAKFNSSSAEKKWKASHDTGSTDQYDRFMNALYSLLDGSSDNTKFEDDCR 1175
LYERI SAK S +E K + + DT S D Y RFMNAL+SLL+GS++N+KFED+CR
Sbjct: 1098 LSHILYERILSAKTYCSGSEMKLRNTKDTCSPDPYARFMNALFSLLNGSAENSKFEDECR 1157
Query: 1176 AIIGTQSYLLFTLDKLIYKLVKQLQAIASEEMDNKLLQLYAYEKSRKLGKFIDIVYHENA 1235
AIIG QSY+LFTL+KLIYKLVKQLQA+ +++MDNKLLQLY YE SR+ G+
Sbjct: 1158 AIIGNQSYVLFTLEKLIYKLVKQLQAVVADDMDNKLLQLYEYENSRRPGR---------- 1207
Query: 1236 CVILHEENIYRIEYSPGPKKLSIQLMDCGHDKPEVTAVSVDPKFSTYLHNDFLSVVPDKK 1295
S P +LSIQLMD +KP+ AVS++P F++YL N+FLS KK
Sbjct: 1208 --------------SSSPSRLSIQLMDNIIEKPDAYAVSMEPTFTSYLQNEFLSNSSGKK 1253
Query: 1296 EKSRLFLKRNKHRYAGSSEF--SSQAMEGLQVFNGLECKIACSSSKVSYVLDTEDVLYRK 1353
E + L+RN Y G + + +AMEG+QV NGLECK++CSS K+SYVLDTED +RK
Sbjct: 1254 ELQDIVLQRNMRGYNGLDDLAVACKAMEGVQVINGLECKMSCSSYKISYVLDTEDFFHRK 1313
Query: 1354 KSKRRILHPTSSCNEPAKSSNIRSSRALRFCKLFSS 1389
K +++ + + + +R RF K S+
Sbjct: 1314 KKQKK--------SNNLSLAKLSQNRIARFHKFLSA 1341
Score = 557 bits (1435), Expect = e-158
Identities = 350/856 (40%), Positives = 478/856 (54%), Gaps = 118/856 (13%)
Query: 52 EEAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFT 111
++A+ ++ ++K F+ Y FL+++ ++ + + V V LF+GH +L+ F
Sbjct: 51 DDALTYLKEVKEMFQDQRDKYDMFLEVMKDFKAQKTDTSGVISRVKELFKGHNNLIFGFN 110
Query: 112 HFLP----------DASAAASSYYMSARNSVLRDR----------RSAMPTVRQAHVVKR 151
FLP +A + + + A + V + + +S + + +
Sbjct: 111 TFLPKGFEITLDDVEAPSKKTVEFEEAISFVNKIKTRFQHNELVYKSFLEILNMYRKDNK 170
Query: 152 ERTTV---------SHCDRDPSVDRPDPD----HDRGLLRAEKEQR-----------RRV 187
+ T V H D R PD H L + QR RR+
Sbjct: 171 DITEVYNEVSTLFEDHSDLLEEFTRFLPDSLAPHTEAQLLRSQAQRYDDRGSGPPLVRRM 230
Query: 188 EKEKDHRED------GKRDWERDGRDHEHDGGQYRERLSHKRKSDRKAEDFGAEPLLDAD 241
EKD R + G RD D D D + +++ D+ + + L D +
Sbjct: 231 FMEKDRRRERTVASRGDRDHSVDRSDLNDDKSMVKMHRDQRKRVDKDNRERRSRDLEDGE 290
Query: 242 ------EFFGTRPMSS-------------TCDDKKSLKTKYPEEFAFCDKVKEKLRNPDG 282
+ F + SS + +K +LK+ Y + F FC+KVKE+L + D
Sbjct: 291 AEQDNLQHFSEKRKSSRRMEGFEAYSGPASHSEKNNLKSMYNQAFLFCEKVKERLCSQDD 350
Query: 283 YQEFLKCLHIYSKEIITQQELQSLVGDLLGKYPDLMEEFNEFLVQAEKNDG-GFLAGVMN 341
YQ FLKCL+++S II +++LQ+LV D+LGK+PDLM+EFN+F + E DG LAGVM+
Sbjct: 351 YQAFLKCLNMFSNGIIQRKDLQNLVSDVLGKFPDLMDEFNQFFERCESIDGFQHLAGVMS 410
Query: 342 KKSLWIEGPGLKPMKAEDRDRDRDRDRYRDDGMKERDRECRERDRSTVVSNKEVSGLKIK 401
KKSL E + +K E++DR+ RD + KE++R K
Sbjct: 411 KKSLGSEENLSRSVKGEEKDREHKRD---VEAAKEKERS--------------------K 447
Query: 402 DKHFLKPINELDLSNCDRCTPSYRLLPKNYPIPIASYKSKIGAKVLNDHWVSVTSGSEDY 461
DK+ K I ELDLS+C+RCTPSYRLLP +YPIP ++ K GA VLNDHWVSVTSGSEDY
Sbjct: 448 DKYMGKSIQELDLSDCERCTPSYRLLPPDYPIPSVRHRQKSGAAVLNDHWVSVTSGSEDY 507
Query: 462 SFKHMRKNQYEESLFRCEDDRFELDMLLESVNLTTKRVEELFEKINNNVIKGDRPVRIDE 521
SFKHMR+NQYEESLFRCEDDRFELDMLLESV K EEL I + I + RI++
Sbjct: 508 SFKHMRRNQYEESLFRCEDDRFELDMLLESVGSAAKSAEELLNIIIDKKISFEGSFRIED 567
Query: 522 HLTALNLRCIERLYGDHGLDVMEVLRKNAPLALPVILTRLKQKQDEWARCRADFSKVWAE 581
H TALNLRCIERLYGDHGLDV +++RKN ALPVILTRLKQKQDEW +CR F+ VWA+
Sbjct: 568 HFTALNLRCIERLYGDHGLDVTDLIRKNPAAALPVILTRLKQKQDEWTKCREGFNVVWAD 627
Query: 582 IYAKNYHKSLDHRSFYFKQQDTKRLSTKALLAEVKEISEKKHKEDDVLLAIAAGSRQPII 641
+YAKN++KSLDHRSFYFKQQD+K LS KAL++EVK++ EK KEDDV+L+I+AG RQPII
Sbjct: 628 VYAKNHYKSLDHRSFYFKQQDSKNLSAKALVSEVKDLKEKSQKEDDVVLSISAGYRQPII 687
Query: 642 PNLEFHYPDLDIHEDLYQLVKYSCGELC-TAEQLDKVMKVWTTFLEPMLCVPSRAQGAED 700
P+LE+ Y D IHEDL++LV++SC E+C T EQ KV+K+W FLE ML V RA+G++
Sbjct: 688 PHLEYDYLDRAIHEDLFKLVQFSCEEICSTKEQTGKVLKLWANFLELMLDVAPRAKGSDS 747
Query: 701 TEDVVKAKNTEDVVNTENNSVKSGIVSV-------------AESGDSKAWQ---LNGDSG 744
EDVV+ ++ + E N I V A SG SK + LN DS
Sbjct: 748 VEDVVETQHQRAFTSGEANESSDAISLVSRQLKFATNGDVHASSGVSKHGETGLLNRDSS 807
Query: 745 VREDRCLDSDHTVYKTETAGNNTQHGKTNIIGFTPDELSGVNKQNQSSERLVNANVSPAS 804
+E+ D D T Q K IG + SG ER+ ++ S S
Sbjct: 808 GKEN-LKDGDLANKDVATCAEKPQ--KDQEIGNGAAKRSG-----DVDERVATSSSSFPS 859
Query: 805 GMEQSNGRTKIDNTSG 820
G+E +NG+ ++SG
Sbjct: 860 GVENNNGKVGSRDSSG 875
>At3g01320 unknown protein
Length = 1324
Score = 1035 bits (2675), Expect = 0.0
Identities = 630/1369 (46%), Positives = 819/1369 (59%), Gaps = 176/1369 (12%)
Query: 2 DTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNKI 61
DT GVIARVKELF+GH +LI GFNTFLPKGYEITL ED+ P KK VEFE+AINFVNKI
Sbjct: 92 DTGGVIARVKELFKGHNNLIYGFNTFLPKGYEITLIEEDDALP-KKTVEFEQAINFVNKI 150
Query: 62 KARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTHFLPDASAAA 121
K RF+ D+HVYKSFL+ILNMYRKENK I EVY+EV++LFQGH DLL++FT FLP AS +
Sbjct: 151 KMRFKHDEHVYKSFLEILNMYRKENKEIKEVYNEVSILFQGHLDLLEQFTRFLP-ASLPS 209
Query: 122 SSYYMSARNSVLR--DRRSAMPTVRQAHVVKRERTTVSHCDR-DPSVDRPDPDHDRGLLR 178
S +R+ + DR S P + Q V K R + R D SV+R D + D+ +++
Sbjct: 210 HSAAQHSRSQAQQYSDRGSDPPLLHQMQVEKERRRERAVALRGDYSVERYDLNDDKTMVK 269
Query: 179 AEKEQRRRVEKEKDHREDGKRDWERDGRDHEHDGGQYRERLSHKRKSDRKAEDFGAEPLL 238
++EQR+R++KE R D G+D+ H KRKS R+AE A
Sbjct: 270 IQREQRKRLDKENRARRGRDLDDREAGQDNLH-------HFPEKRKSSRRAEALEA---- 318
Query: 239 DADEFFGTRPMSSTCDDKKSLKTKYPEEFAFCDKVKEKLRNPDGYQEFLKCLHIYSKEII 298
+ G S++ +K +LK+ Y + F FC+KVK++L + D YQ FLKCL+I+S II
Sbjct: 319 ----YSG----SASHSEKDNLKSMYKQAFVFCEKVKDRLCSQDDYQTFLKCLNIFSNGII 370
Query: 299 TQQELQSLVGDLLGKYPDLMEEFNEFLVQAEKNDG----GF--LAGVMNKKSLWIEGPGL 352
+++LQ+LV DLLGK+PDLM+EFN+F + E G GF LAGVM+KK E
Sbjct: 371 QRKDLQNLVSDLLGKFPDLMDEFNQFFERCESITGTEIHGFQRLAGVMSKKLFSSEEQLS 430
Query: 353 KPMKAEDRDRDRDRDRYRDDGMKERDRECRERDRSTVVSNKEVSGLKIKDKHFLKPINEL 412
+PMK E+++ + + + +KE + +C+ KE G K I EL
Sbjct: 431 RPMKVEEKESEHKPEL---EAVKETE-QCK----------KEYMG---------KSIQEL 467
Query: 413 DLSNCDRCTPSYRLLPKNYPIPIASYKSKIGAKVLNDHWVSVTSGSEDYSFKHMRKNQYE 472
DLS+C+ CTPSYRLLP +YPIPIAS +S++GA+VLNDHWVSVTSGSEDYSFKHMR+NQYE
Sbjct: 468 DLSDCECCTPSYRLLPADYPIPIASQRSELGAEVLNDHWVSVTSGSEDYSFKHMRRNQYE 527
Query: 473 ESLFRCEDDRFELDMLLESVNLTTKRVEELFEKINNNVIKGDRPVRIDEHLTALNLRCIE 532
ESLFRCEDDRFELDMLLESV+ + E L I I RI++H TALNLRCIE
Sbjct: 528 ESLFRCEDDRFELDMLLESVSSAARSAESLLNIITEKKISFSGSFRIEDHFTALNLRCIE 587
Query: 533 RLYGDHGLDVMEVLRKNAPLALPVILTRLKQKQDEWARCRADFSKVWAEIYAKNYHKSLD 592
RLYGDHGLDV+++L KN ALPVILTRLKQKQ EW +CR DF KVWA +YAKN++KSLD
Sbjct: 588 RLYGDHGLDVIDILNKNPATALPVILTRLKQKQGEWKKCRDDFDKVWANVYAKNHYKSLD 647
Query: 593 HRSFYFKQQDTKRLSTKALLAEVKEISEKKHKEDDVLLAIAAGSRQPIIPNLEFHYPDLD 652
HRSFYFKQQD+K LS K+LLAE+KE+ EK +DDVLL+I+AG RQPI PNLE+ Y +
Sbjct: 648 HRSFYFKQQDSKNLSAKSLLAEIKELKEKSQNDDDVLLSISAGYRQPINPNLEYEYLNRA 707
Query: 653 IHEDLYQLVKYSCGELC-TAEQLDKVMKVWTTFLEPMLCVPSRAQGAEDTEDVVKAKNTE 711
IHED++++V++SC ELC T EQL KV+++W FLE +L VP RA+G + EDVV T
Sbjct: 708 IHEDMFKVVQFSCEELCSTKEQLSKVLRLWENFLEAVLGVPPRAKGTDLVEDVVINPKTL 767
Query: 712 DVVNTENNSVKSGIVSVAESGDSKAWQLNGDSGVREDRCLDSDHTVYKTETAGNNTQHGK 771
DV +++ +G +V+ GD+ K ++A N ++
Sbjct: 768 DV---NHSTSPNGEAAVSSGGDTARLASR------------------KLKSAANGDENSS 806
Query: 772 TNIIGFTPDELSGVNKQNQSSERLVNANVSPASGMEQSNGRTKIDNTSGHTATPSGNGNT 831
+ G + +NK + E L + ++ G+ S + + + +G+ A
Sbjct: 807 S---GTFKHGIGLLNKDSTGKENLEDVEIANRDGVACSAVKPQKEQETGNEA-------- 855
Query: 832 SVEGGLELPSSEPILFQTRKFPSNYSIFSLMLDILINFGLHLHECILQRILFVKHPSYGW 891
E +PI P + S + + I I G + C++
Sbjct: 856 ------EKRFGKPI-------PMDISERAAISSISIPSGAENNHCVV------------- 889
Query: 892 LMILVVDISSRFQIRQGGDSTRPGTFTNGATTGGTEVHRYQEQSIRHFKSEREEGELSPN 951
G PG N G + E + +K + P
Sbjct: 890 -----------------GKEVLPGPSRNEKEEGELSPNGDFEDNFGVYKDHGVKSTSKPE 932
Query: 952 GDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGENDVDADDEGEESPH 1011
E + A E D + S+ + G VC + D D E E H
Sbjct: 933 NSAEAEVEADAEVENEDDADDVDSENASEASGTESGGDVCSQ-------DEDREEENGEH 985
Query: 1012 RSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEGEAEGMVDAHDVEGDGM 1071
E D KAESEGEAEGM D H +EG+
Sbjct: 986 ---------------------------------DEIDGKAESEGEAEGM-DPHLLEGESE 1011
Query: 1072 SLTISERFLQTVKPLAKYVPAAL-HEKDRNSQVFYGSDSFYVLFRLHQTLYERIQSAKFN 1130
L SER L +V+PL+K+V A L E+ ++ QVFYG+D FYVLFRLHQ LYERI AK N
Sbjct: 1012 LLPQSERVLLSVRPLSKHVAAVLCDERTKDLQVFYGNDDFYVLFRLHQILYERILYAKRN 1071
Query: 1131 SSSAEKKWKASHDTGSTDQYDRFMNALYSLLDGSSDNTKFEDDCRAIIGTQSYLLFTLDK 1190
S E K K DT + D Y RFM LY LLDGS++NTKFED+CRAIIG QSY+LFTLDK
Sbjct: 1072 CSGGELKSKNLKDTNAGDPYARFMRVLYGLLDGSAENTKFEDECRAIIGNQSYVLFTLDK 1131
Query: 1191 LIYKLVKQLQAIASEEMDNKLLQLYAYEKSRKLGKFIDIVYHENACVILHEENIYRIEYS 1250
LIY+LVKQLQAI ++EMDNKLLQLY YEKSRK G+ ID VY+EN V++HEENIYR+E S
Sbjct: 1132 LIYRLVKQLQAIVADEMDNKLLQLYEYEKSRKPGRVIDSVYYENVRVLVHEENIYRLECS 1191
Query: 1251 PGPKKLSIQLMDCGHDKPEVTAVSVDPKFSTYLHNDFLSVVPDKKEKSR-LFLKRNKHRY 1309
P +LSIQLMD +KPE AVS+DP F++Y+ + LSV KKE+ + L+RN
Sbjct: 1192 SLPSRLSIQLMDNIIEKPEAYAVSMDPTFASYMQTELLSVSSGKKEEGHDIVLQRN---L 1248
Query: 1310 AGSSEFSSQAMEGLQVFNGLECKIACSSSKVSYVLDTEDVLYRKKSKRR 1358
G + +AMEG++V NGLECK++CSS K++YVLDTED +RKK K++
Sbjct: 1249 TGLYDL-CKAMEGVEVVNGLECKMSCSSYKIAYVLDTEDYFHRKKKKKK 1296
>At1g70060 hypothetical protein
Length = 1386
Score = 857 bits (2215), Expect = 0.0
Identities = 483/857 (56%), Positives = 594/857 (68%), Gaps = 75/857 (8%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPA-KKPVEFEEAINFVN 59
+DT GVI RVKELF+G+R+LILGFNTFLPKG+EITL ED+QP A KKPVEFEEAI+FVN
Sbjct: 48 VDTTGVILRVKELFKGNRELILGFNTFLPKGFEITLRPEDDQPAAPKKPVEFEEAISFVN 107
Query: 60 KIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTHFLPDASA 119
KIK RF+GDD VYKSFLDILNMYRKENK I EVYHEVA+LF+ H DLL EFTHFLPD SA
Sbjct: 108 KIKTRFQGDDRVYKSFLDILNMYRKENKSITEVYHEVAILFRDHHDLLGEFTHFLPDTSA 167
Query: 120 AASSYYMSARNSVLRDRR-SAMPTVRQAHVVKRERTTVSHCDRDPSVDRPDPDHDRGLLR 178
AS+ + +RDR ++PT+RQ + K++R SH +R + D DH+R LL+
Sbjct: 168 TASTN--DSVKVPVRDRGIKSLPTMRQIDLDKKDRIITSHPNRALKTENMDVDHERSLLK 225
Query: 179 AEKEQRRRVEKEKDHREDGKRDWERDGRDHEHDGGQYRERLSHKRKSDRKAEDFGAE--- 235
KE+ RR++K+ D +D R +D R +HD ++E + +K + +D AE
Sbjct: 226 DSKEEVRRIDKKNDFMDDRDR---KDYRGLDHDS--HKEHFFNSKKKLIRKDDDSAEMSD 280
Query: 236 PLLDADEFFGTRPMSSTCDDKKSL---KTKYPEEFAFCDKVKEKLRNPDGYQEFLKCLHI 292
+ D+F G P SST D+K + + +E AF D+VK KL D QEFL+CL++
Sbjct: 281 QAREGDKFSGAIPSSSTYDEKGFIIDFLESHSQELAFVDRVKAKLDTADN-QEFLRCLNL 339
Query: 293 YSKEIITQQELQSLVGDLLGKYPDLMEEFNEFLVQAEKNDGGFLAGVMNK---------- 342
YSKEII+Q ELQSLV DL+G YPDLM+ F FL Q +KNDG L+G+++K
Sbjct: 340 YSKEIISQPELQSLVSDLIGVYPDLMDAFKVFLAQCDKNDG-LLSGIVSKSKSSTFYNIL 398
Query: 343 ------KSLWIEGPGLKPMKAEDRDRDRDRDRYRDDGMKERDRECRERDRSTVVSNKEVS 396
+SLW EG +P K+ D+D DR+R++ + +ERDRE +ER S K
Sbjct: 399 LTYLFGQSLWSEGKCPQPTKSLDKDTDREREKI--ERYRERDRE-KERLEKVAASQK--- 452
Query: 397 GLKIKDKHFLKPINELDLSNCDRCTPSYRLLPKN-----------YPIPIASYKSKIGAK 445
+ KPI+ELDLSNC++CTPSYR LPKN YPIPIAS K +IG++
Sbjct: 453 --------WAKPISELDLSNCEQCTPSYRRLPKNLNVHTYFVLLQYPIPIASQKMEIGSQ 504
Query: 446 VLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESVNLTTKRVEELFEK 505
VLNDHWVSVTSGSEDYSFKHMRKNQYEESLF+CEDDRFELDMLLESV T RVEEL K
Sbjct: 505 VLNDHWVSVTSGSEDYSFKHMRKNQYEESLFKCEDDRFELDMLLESVISATNRVEELLAK 564
Query: 506 INNNVIKGDRPVRIDEHLTALNLRCIERLYGDHGLDVMEVLRKNAPLALPVILTRLKQKQ 565
IN+N +K D P+ I++HLTALNLRCIERLY DHGLDV+++L+KNA LALPVILTRLKQKQ
Sbjct: 565 INSNELKTDTPICIEDHLTALNLRCIERLYSDHGLDVLDLLKKNAYLALPVILTRLKQKQ 624
Query: 566 DEWARCRADFSKVWAEIYAKNYHKSLDHRSFYFKQQDTKRLSTKALLAEVKEISEKKHKE 625
+EWARCR +F+KVWA+IY KNYH+SLDHRSFYFKQQD+K LSTKALLAE+KEISEKK E
Sbjct: 625 EEWARCRTEFNKVWADIYTKNYHRSLDHRSFYFKQQDSKNLSTKALLAEIKEISEKKRGE 684
Query: 626 DDVLLAIAAGSRQPIIPNLEFHYPDLDIHEDLYQLVKYSCGELCTAEQLDKVMKVWTTFL 685
DD LLA+AAG+R+ I N+ F YPD D+HEDLYQL+KYSCGE+C+ EQLDKVMKVWT FL
Sbjct: 685 DDALLALAAGNRRTISSNMSFDYPDPDLHEDLYQLIKYSCGEMCSTEQLDKVMKVWTEFL 744
Query: 686 EPMLCVPSRAQGAEDTEDVVKAKN-----TEDVVNTENNSVKSGIVSVAESGDSKA-WQL 739
EP+ VPSR QGAED ED VK+ N ED V+ +N + S+A S S ++
Sbjct: 745 EPIFGVPSRPQGAEDREDAVKSTNHDREDQEDAVSPQNGA------SIANSMRSNGPRKV 798
Query: 740 NGDSGVREDRCLDSDHTVYKTETA---GNNTQHGKTNIIGFTPDELSGVNKQNQSSERLV 796
N + VR+ LD D T KT A +NTQ+ K TPDE KQ S ER
Sbjct: 799 NESNQVRQASELDKDVTSSKTSDALLSCDNTQNDKMPKNLTTPDE-RAETKQAVSIERAH 857
Query: 797 NANVSPASG-MEQSNGR 812
N+N P G + Q NG+
Sbjct: 858 NSNALPLDGLLPQRNGK 874
Score = 463 bits (1192), Expect = e-130
Identities = 265/473 (56%), Positives = 323/473 (68%), Gaps = 55/473 (11%)
Query: 940 KSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGEND 999
K EREEGELSP GD EEDN+AV+ + ++A K K END
Sbjct: 943 KVEREEGELSPTGDFEEDNYAVHGENDMEALSKSK----------------------END 980
Query: 1000 VDADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEGEAE- 1058
ADD S RSSD S N S N DVSG++S DGE+C RE+ D NK ESEGEAE
Sbjct: 981 ATADDA---SAPRSSDGSGNTSHNGDVSGTDSGDGEDCYREDDIDH---NKVESEGEAEE 1034
Query: 1059 GMVDAHD-VEGDGMSLTISERFLQTVKPLAKYVPAALHEKD-----RNSQVFYGSDSFYV 1112
GM D HD EGD L+IS + L VKPLAKYVP AL++KD +NSQVFYG+DSFYV
Sbjct: 1035 GMSDGHDDTEGDMPVLSISVKNLLHVKPLAKYVPPALYDKDNDDSRKNSQVFYGNDSFYV 1094
Query: 1113 LFRLHQTLYERIQSAKFNSSSAEKKWKASHDTGSTDQYDRFMNALYSLLDGSSDNTKFED 1172
LFRLHQ LY+RI SAK NSSS ++KWK S+ T D Y R M+ALY+LLDG+SDN+KFED
Sbjct: 1095 LFRLHQILYDRILSAKINSSSPDRKWKTSNPTNPADSYARIMDALYNLLDGTSDNSKFED 1154
Query: 1173 DCRAIIGTQSYLLFTLDKLIYKLVK-------------QLQAIASEEMDNKLLQLYAYEK 1219
DCRAIIGTQSY+LFTLDKLIYKL+K QLQA+A++EMDNKL QLYAYEK
Sbjct: 1155 DCRAIIGTQSYVLFTLDKLIYKLIKHSSHMIQLTNVILQLQAVAADEMDNKLQQLYAYEK 1214
Query: 1220 SRKLGKFIDIVYHENACVILHEENIYRIEYSPG-PKKLSIQLMDCGHDKPEVTAVSVDPK 1278
SRK KF+D VY+ENA V+L +E+IYRIE P KLSIQL+D GHDKP+VT++S+DP
Sbjct: 1215 SRKPEKFLDAVYYENALVLLPDEDIYRIECEQSTPSKLSIQLLDYGHDKPDVTSISMDPT 1274
Query: 1279 FSTYLHNDFLSVVPDKKEKSRLFLKRNKHRYAGSSEFSSQAMEGLQVFNGLECKIACSSS 1338
F+ YLHN FLS P+ KE R++LKRNK + G E + + +++ NGLECKI CSSS
Sbjct: 1275 FAAYLHNVFLSYQPNAKENPRIYLKRNKRKNGGDDELCT--TDEVKIINGLECKITCSSS 1332
Query: 1339 KVSYVLDTEDVLYRKKSKRRILHPTS---SCNEPAKSSNIRSSRALRFCKLFS 1388
KVSYVLDTEDVL+R K +R++L+ + + + S IR R R+ KL +
Sbjct: 1333 KVSYVLDTEDVLHRAK-RRKLLNQSGLPLAHDSVCSGSLIRQRRTQRYQKLLT 1384
>At1g59890 hypothetical protein
Length = 1125
Score = 464 bits (1193), Expect = e-130
Identities = 246/468 (52%), Positives = 316/468 (66%), Gaps = 21/468 (4%)
Query: 254 DDKKSLKTKYPEEFAFCDKVKEKLRNPD-GYQEFLKCLHIYSKEIITQQELQSLVGDLLG 312
D+K + F +++K + D Y++FL L++Y KE + E+ V L
Sbjct: 118 DEKPKKPVDFQVAIEFVNRIKARFGGDDRAYKKFLDILNMYRKETKSINEVYQEVTLLFQ 177
Query: 313 KYPDLMEEFNEFLVQ----AEKNDGGFLAGVM--NKKSLWIEGPGLKPMKAEDRDRDRDR 366
+ DL+ EF FL ND F + ++ S + PG+ P E + +
Sbjct: 178 DHEDLLGEFVHFLPDFRGSVSVNDPLFQRNTIPRDRNSTF---PGMHPKHFEKKIK---- 230
Query: 367 DRYRDDGMKERDRECRERDRSTVVSNKEVSGLK--IKDKHFLKPINELDLSNCDRCTPSY 424
R R D E + + D + V + ++ K + H K INELDL++C +CTPSY
Sbjct: 231 -RSRHDEYTELSDQREDGDENLVAYSADIGSQKNLLSTNHMAKAINELDLTDCAQCTPSY 289
Query: 425 RLLPKNYPIPIASYKSKIGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFE 484
R LP +YPI I SY++ +G KVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFE
Sbjct: 290 RRLPDDYPIQIPSYRNSLGEKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFE 349
Query: 485 LDMLLESVNLTTKRVEELFEKINNNVIKGDRPVRIDEHLTALNLRCIERLYGDHGLDVME 544
LDMLLESV+ KRVE L EKINNN I + P+ I EHL+ +CIERLYGD+GLDVM+
Sbjct: 350 LDMLLESVSAAIKRVESLLEKINNNTISIETPICIREHLSG---KCIERLYGDYGLDVMD 406
Query: 545 VLRKNAPLALPVILTRLKQKQDEWARCRADFSKVWAEIYAKNYHKSLDHRSFYFKQQDTK 604
L+KN+ +ALPVILTRLKQKQ+EWARCRADF KVWAE+YAKN+HKSLDHRSFYFKQQD+K
Sbjct: 407 FLKKNSHIALPVILTRLKQKQEEWARCRADFRKVWAEVYAKNHHKSLDHRSFYFKQQDSK 466
Query: 605 RLSTKALLAEVKEISEKKHKEDDVLLAIAAGSRQPIIPNLEFHYPDLDIHEDLYQLVKYS 664
LSTK L+AE+K+ISE+KHKE D+L AIA G++ P++EF Y D +H DLY+L+KY
Sbjct: 467 NLSTKGLVAEIKDISERKHKE-DLLRAIAVGTKPSFTPDVEFIYTDTKVHTDLYKLIKYY 525
Query: 665 CGELCTAEQLDKVMKVWTTFLEPMLCVPSRAQGAEDTEDVVKAKNTED 712
C E+C EQ DKVMK+W TFLEPM VPSR++ E +DV K ++ ++
Sbjct: 526 CEEICATEQSDKVMKLWVTFLEPMFGVPSRSETIETMKDVAKIEDNQE 573
Score = 415 bits (1067), Expect = e-116
Identities = 244/465 (52%), Positives = 310/465 (66%), Gaps = 27/465 (5%)
Query: 930 RYQEQSIRHFKSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQ 989
R + S+ K EREEGELSP E++NF VY+ +GL+ K ++S R ++
Sbjct: 665 RVSDVSMGGHKVEREEGELSPTESCEQENFEVYKENGLEPVQKLPDNEISNT--DREPKE 722
Query: 990 VCC--EAKGENDVDADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREH 1047
C EA ++ +D+ + + S+ ENAS+ + VS S+ G+ S EEH+
Sbjct: 723 GACGTEAVTRSNALPEDDDNKITQKLSEGDENASKFI-VSASKFG-GQVSSDEEHKG--- 777
Query: 1048 DNKAESEGEAEGMVDAHDVEGDGMSLTISERFLQTVKPLAKYVPAALHEKD----RNSQV 1103
AESE EA GMV++++ E DG T SER+LQ VKPLAK+VP L + +S+V
Sbjct: 778 ---AESENEAGGMVNSNEGE-DGSFFTFSERYLQPVKPLAKHVPGTLQASECDTRNDSRV 833
Query: 1104 FYGSDSFYVLFRLHQTLYERIQSAKFNSSSAEKKWKASHDTGSTDQYDRFMNALYSLLDG 1163
FYG+DS YVLFRLHQ LYERIQSAK +S E+KWKA D+ STD Y RFM ALY+LLDG
Sbjct: 834 FYGNDSLYVLFRLHQMLYERIQSAKIHS---ERKWKAP-DSTSTDSYTRFMEALYNLLDG 889
Query: 1164 SSDNTKFEDDCRAIIGTQSYLLFTLDKLIYKLVKQLQAIASEEMDNKLLQLYAYEKSRKL 1223
SSDNTKFED+CRAIIG QSY+LFTLDKL+ K VK L A+A++E D KLLQLYAYE RK
Sbjct: 890 SSDNTKFEDECRAIIGAQSYVLFTLDKLVQKFVKHLHAVAADETDTKLLQLYAYENYRKP 949
Query: 1224 GKFIDIVYHENACVILHEENIYRIEYSPGPKKLSIQLMDCGHDKPEVTAVSVDPKFSTYL 1283
G+F DIVYHENA +LH++NIYRIEYS +L IQLM+ +D+PEVTAV+V+P F+ YL
Sbjct: 950 GRFFDIVYHENARALLHDQNIYRIEYSSAQTRLGIQLMNSWNDQPEVTAVTVEPGFANYL 1009
Query: 1284 HNDFLSVVPDKKEKSRLFLKRNKHRYAGSSEFS---SQAMEGLQVFNGLECKIACSSSKV 1340
NDFLS V D +EK LFLKRNK + +G E S S+A+EGL + N +ECKIACSS KV
Sbjct: 1010 QNDFLSFVSD-EEKPGLFLKRNKAKLSGPGEESLGMSRALEGLNIINEVECKIACSSFKV 1068
Query: 1341 SYVLDTEDVLYRKKSKRRILHPTSSCNEPAKSSN--IRSSRALRF 1383
Y T D+LYR+K K+ L+PT N S+ R R RF
Sbjct: 1069 KYEPHTADLLYRRKQKKATLNPTGPENVKTSDSSELSRKKRISRF 1113
Score = 186 bits (471), Expect = 1e-46
Identities = 99/174 (56%), Positives = 121/174 (68%), Gaps = 3/174 (1%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNK 60
+DT GVIARVK+LF+G+ DL+LGFNTFLPKGY+ITL EDE+P KKPV+F+ AI FVN+
Sbjct: 79 VDTNGVIARVKDLFKGYDDLLLGFNTFLPKGYKITLQPEDEKP--KKPVDFQVAIEFVNR 136
Query: 61 IKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTHFLPDASAA 120
IKARF GDD YK FLDILNMYRKE K INEVY EV +LFQ H DLL EF HFLPD +
Sbjct: 137 IKARFGGDDRAYKKFLDILNMYRKETKSINEVYQEVTLLFQDHEDLLGEFVHFLPDFRGS 196
Query: 121 AS-SYYMSARNSVLRDRRSAMPTVRQAHVVKRERTTVSHCDRDPSVDRPDPDHD 173
S + + RN++ RDR S P + H K+ + + + S R D D +
Sbjct: 197 VSVNDPLFQRNTIPRDRNSTFPGMHPKHFEKKIKRSRHDEYTELSDQREDGDEN 250
Score = 40.0 bits (92), Expect = 0.009
Identities = 19/63 (30%), Positives = 35/63 (55%)
Query: 53 EAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTH 112
+A+ ++ +K F+ + Y++FL ++ ++ + N V V LF+G+ DLL F
Sbjct: 45 DALTYLKAVKDMFQDNKEKYETFLGVMKDFKAQRVDTNGVIARVKDLFKGYDDLLLGFNT 104
Query: 113 FLP 115
FLP
Sbjct: 105 FLP 107
>At1g10450 unknown protein
Length = 1173
Score = 442 bits (1136), Expect = e-124
Identities = 241/490 (49%), Positives = 313/490 (63%), Gaps = 31/490 (6%)
Query: 254 DDKKSLKTKYPEEFAFCDKVKEKLRNPD-GYQEFLKCLHIYSKEIITQQELQSLVGDLLG 312
++K ++ + + F K+K + + + Y+ FL L++Y KE + E+ V L
Sbjct: 157 EEKPKIRVDFKDAIGFVTKIKTRFGDDEHAYKRFLDILNLYRKEKKSISEVYEEVTMLFK 216
Query: 313 KYPDLMEEFNEFLVQAEKNDGGFLAGVMNKK-----SLWIEGPGLKPMKAEDRDRDRDRD 367
+ DL+ EF FL ++ V K ++ + + K ED D+
Sbjct: 217 GHEDLLMEFVNFLPNCPESAPSTKNAVPRHKGTATTAMHSDKKRKQRCKLEDYSGHSDQR 276
Query: 368 RYRDDGMKE---------------RDRECRE---------RDRSTVVSNKEVSGLKIKDK 403
D+ + RD E RE ++S ++++ K K
Sbjct: 277 EDGDENLVTCSADSPVGEGQPGYFRDYENREDTETDTADRTEKSAASGSQDIGNHKSTTK 336
Query: 404 HFLKPINELDLSNCDRCTPSYRLLPKNYPIPIASYKSKIGAKVLNDHWVSVTSGSEDYSF 463
+ PINELDLS C +CTPSYRLLPK+Y + I SY++ +G K LNDH VSVTSGSEDYSF
Sbjct: 337 YVGTPINELDLSECTQCTPSYRLLPKDYAVEIPSYRNTLGKKTLNDHLVSVTSGSEDYSF 396
Query: 464 KHMRKNQYEESLFRCEDDRFELDMLLESVNLTTKRVEELFEKINNNVIKGDRPVRIDEHL 523
HMRKNQYEESLFRCEDDR+E+DMLL SV+ K+VE L EK+NNN I D + I++HL
Sbjct: 397 SHMRKNQYEESLFRCEDDRYEMDMLLGSVSSAIKQVEILLEKMNNNTISVDSTICIEKHL 456
Query: 524 TALNLRCIERLYGDHGLDVMEVLRKNAPLALPVILTRLKQKQDEWARCRADFSKVWAEIY 583
+A+NLRCIERLYGD+GLDVM++L+KN ALPVILTRLKQKQ+EWARC +DF KVWAE+Y
Sbjct: 457 SAMNLRCIERLYGDNGLDVMDLLKKNMHSALPVILTRLKQKQEEWARCHSDFQKVWAEVY 516
Query: 584 AKNYHKSLDHRSFYFKQQDTKRLSTKALLAEVKEISEKKHKEDDVLLAIAAGSRQPIIPN 643
AKN+HKSLDHRSFYFKQQD+K LSTK L+AEVK+ISEKKH+E D+L AIA P+
Sbjct: 517 AKNHHKSLDHRSFYFKQQDSKNLSTKCLVAEVKDISEKKHQE-DLLQAIAVRVMPLFTPD 575
Query: 644 LEFHYPDLDIHEDLYQLVKYSCGELCTAEQLDKVMKVWTTFLEPMLCVPSRAQGAEDTED 703
LEF+Y D IHEDLY L+KY C E+C EQ DKVMK+W TFLEP+ + SR+Q ED
Sbjct: 576 LEFNYCDTQIHEDLYLLIKYYCEEICATEQSDKVMKLWITFLEPIFGILSRSQDNLALED 635
Query: 704 VVKAKNTEDV 713
V K KN ++
Sbjct: 636 VSKLKNNREL 645
Score = 360 bits (923), Expect = 4e-99
Identities = 211/453 (46%), Positives = 282/453 (61%), Gaps = 30/453 (6%)
Query: 940 KSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGEND 999
K E EEGELSP E+ NF V + K ++ +S +Q C + +N
Sbjct: 740 KHELEEGELSPTASREQSNFEVNGQNAFKPLQK-----VTDNVRSNKDKQSCDKKGAKNK 794
Query: 1000 VDADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEGEAEG 1059
A+D+ +E+ H+ S++++ ASE + VSG++ + C HE+ G G
Sbjct: 795 TRAEDDKQENCHKLSENNKTASEML-VSGTKVS----C----HEENNRVMNCNGRGSVAG 845
Query: 1060 MVDAHDVEGDGMSLTISERFLQTVKPLAKYVPAALHEKD----RNSQVFYGSDSFYVLFR 1115
+ A+ +G+ S SERFLQTVKP+AK++ L + +SQVFYG+DS+YVLFR
Sbjct: 846 EM-ANGNQGEDGSFAFSERFLQTVKPVAKHLSWPLQASETCSQNDSQVFYGNDSYYVLFR 904
Query: 1116 LHQTLYERIQSAKFNSSSAEKKWKASHDTGSTDQYDRFMNALYSLLDGSSDNTKFEDDCR 1175
LHQ LYERIQ+AK +S EKKWKA+ +T + D Y RFM+ALY+LLDGS DNTKFED+CR
Sbjct: 905 LHQMLYERIQTAKKHS---EKKWKAADNT-TPDSYPRFMDALYNLLDGSIDNTKFEDECR 960
Query: 1176 AIIGTQSYLLFTLDKLIYKLVKQLQAIASEEMDNKLLQLYAYEKSRKLGKFIDIVYHENA 1235
AI G QSY+LFTLDKL+ K VK L ++AS+E D KLLQL+AYE RK GKF D+VYHENA
Sbjct: 961 AIFGAQSYVLFTLDKLVQKFVKHLHSVASDETDTKLLQLHAYENYRKPGKFFDLVYHENA 1020
Query: 1236 CVILHEENIYRIEYSPGPKKLSIQLMDCGHDKPEVTAVSVDPKFSTYLHNDFLSVVPDKK 1295
C +LHE NIYRI YS +LSIQLM+ G+++ EV V+++P F+ YL N L V D +
Sbjct: 1021 CALLHEANIYRIRYSSEGTRLSIQLMNSGNNQLEVMGVAMEPAFADYLQNKCLKSVND-E 1079
Query: 1296 EKSRLFLKRNKHRYAGSSEFSSQ--AMEGLQVFNGLECKIACSSSKVSYVLDTEDVLYRK 1353
E LFL RNK ++ E AME L + N +EC++ACSSSKV YV +T D+LYR
Sbjct: 1080 ENHGLFLNRNKKKFTSLDESRGMPVAMERLNIINEMECRMACSSSKVKYVANTSDLLYRS 1139
Query: 1354 KSKRRILHPTSSCNEPAKSSNIRSSRALRFCKL 1386
K + P S +E K I + C+L
Sbjct: 1140 KQGK----PNSRVSEILKQRRISRFHIMLNCRL 1168
Score = 158 bits (399), Expect = 2e-38
Identities = 97/233 (41%), Positives = 132/233 (56%), Gaps = 41/233 (17%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNK 60
IDT GVI R+K LF+G+RDL+LGFNTFLPKGY+ITL E+E+P + V+F++AI FV K
Sbjct: 118 IDTNGVIERIKVLFKGYRDLLLGFNTFLPKGYKITLLPEEEKPKIR--VDFKDAIGFVTK 175
Query: 61 IKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTHFLPDASAA 120
IK RF D+H YK FLDILN+YRKE K I+EVY EV +LF+GH DLL EF +FLP+ +
Sbjct: 176 IKTRFGDDEHAYKRFLDILNLYRKEKKSISEVYEEVTMLFKGHEDLLMEFVNFLPNCPES 235
Query: 121 ASSYYMSARNSVLRDRRSAMPTVRQAHVVKRERTTVSHCDRDPSVDRPDPDHDRGLLRAE 180
A S +N+V R + +A TT H D+ +
Sbjct: 236 AP----STKNAVPRHKGTA--------------TTAMHSDK------------------K 259
Query: 181 KEQRRRVEK---EKDHREDGKRDWERDGRDHEHDGGQYRERLSHKRKSDRKAE 230
++QR ++E D REDG + D GQ ++ + D + +
Sbjct: 260 RKQRCKLEDYSGHSDQREDGDENLVTCSADSPVGEGQPGYFRDYENREDTETD 312
Score = 43.9 bits (102), Expect = 6e-04
Identities = 20/63 (31%), Positives = 36/63 (56%)
Query: 53 EAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTH 112
+A+ ++ +K F + Y+SFL+++ ++ + N V + VLF+G+ DLL F
Sbjct: 84 DALTYLKAVKDIFHDNKEKYESFLELMKEFKAQTIDTNGVIERIKVLFKGYRDLLLGFNT 143
Query: 113 FLP 115
FLP
Sbjct: 144 FLP 146
>At1g24200 hypothetical protein
Length = 196
Score = 144 bits (364), Expect = 3e-34
Identities = 74/115 (64%), Positives = 85/115 (73%), Gaps = 3/115 (2%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNK 60
ID +GVI R+KEL + + L+LGFN FLP GY IT EQP KKPVE EAI+F+NK
Sbjct: 48 IDISGVIIRMKELLKEQQGLLLGFNAFLPNGYMIT---HHEQPSQKKPVELGEAISFINK 104
Query: 61 IKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTHFLP 115
IK RF+GDD VYKS LDILNMYRK+ K I VY EVA+LF H +LL EFTHFLP
Sbjct: 105 IKTRFQGDDRVYKSVLDILNMYRKDRKPITAVYREVAILFLDHNNLLVEFTHFLP 159
Score = 36.6 bits (83), Expect = 0.10
Identities = 22/72 (30%), Positives = 36/72 (49%), Gaps = 1/72 (1%)
Query: 45 AKKPVEFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHA 104
A KP +A+ ++ +KA+F+G Y FL I+ Y+ + I+ V + L +
Sbjct: 7 AHKPTT-NDALKYLRAVKAKFQGQREKYDEFLQIMIDYKTQRIDISGVIIRMKELLKEQQ 65
Query: 105 DLLDEFTHFLPD 116
LL F FLP+
Sbjct: 66 GLLLGFNAFLPN 77
>At1g70030 unknown protein
Length = 160
Score = 127 bits (320), Expect = 3e-29
Identities = 64/113 (56%), Positives = 84/113 (73%), Gaps = 2/113 (1%)
Query: 3 TAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNKIK 62
T VI++VKELF+G +L+LGFNTFLP G+EITL +DE K F+EA FVNK+K
Sbjct: 47 TCDVISKVKELFKGQPELLLGFNTFLPTGFEITLS-DDELTSNSKFAHFDEAYEFVNKVK 105
Query: 63 ARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTHFLP 115
RF+ +D V+ SFL++L ++KENK + E+Y EVA+LFQGH DLL+EF FLP
Sbjct: 106 TRFQNND-VFNSFLEVLKTHKKENKSVAELYQEVAILFQGHRDLLEEFHLFLP 157
Score = 45.1 bits (105), Expect = 3e-04
Identities = 22/71 (30%), Positives = 41/71 (56%)
Query: 45 AKKPVEFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHA 104
A K + +A++F++ +K +++ + +Y SFL I+ +R + +V +V LF+G
Sbjct: 3 ANKKLTTTDALDFLHLVKTKYQDNREIYDSFLTIMKDFRGQRAKTCDVISKVKELFKGQP 62
Query: 105 DLLDEFTHFLP 115
+LL F FLP
Sbjct: 63 ELLLGFNTFLP 73
Score = 45.1 bits (105), Expect = 3e-04
Identities = 31/94 (32%), Positives = 48/94 (50%), Gaps = 3/94 (3%)
Query: 235 EPLLDADEFFGTRPMSSTCDDKKSLKTKYP---EEFAFCDKVKEKLRNPDGYQEFLKCLH 291
E LL + F T + DD+ + +K+ E + F +KVK + +N D + FL+ L
Sbjct: 63 ELLLGFNTFLPTGFEITLSDDELTSNSKFAHFDEAYEFVNKVKTRFQNNDVFNSFLEVLK 122
Query: 292 IYSKEIITQQELQSLVGDLLGKYPDLMEEFNEFL 325
+ KE + EL V L + DL+EEF+ FL
Sbjct: 123 THKKENKSVAELYQEVAILFQGHRDLLEEFHLFL 156
>At1g24230 hypothetical protein
Length = 245
Score = 103 bits (258), Expect = 5e-22
Identities = 54/124 (43%), Positives = 79/124 (63%), Gaps = 5/124 (4%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNK 60
+D +AR+ EL + HR+L+LGF+ L G T PLE E P K + NF++K
Sbjct: 120 VDAPSAVARMTELMKDHRNLVLGFSVLLSTGDTKTTPLEAE-PDNNKRIR---VANFISK 175
Query: 61 IKARFEGDD-HVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTHFLPDASA 119
+KARF+G+D HVY+SFL+IL MY++ NK +N++Y EV L QGH DL+ EF++ +
Sbjct: 176 LKARFQGNDGHVYESFLEILTMYQQGNKSVNDLYQEVVALLQGHEDLVMEFSNVFKRTTG 235
Query: 120 AASS 123
+ S
Sbjct: 236 PSGS 239
Score = 46.2 bits (108), Expect = 1e-04
Identities = 30/114 (26%), Positives = 50/114 (43%), Gaps = 15/114 (13%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNK 60
+D A V+ R++EL + H +L+L FN FL PA+ F + +++
Sbjct: 49 VDLATVVPRMRELLKDHVNLLLRFNAFL---------------PAEAKETFHDVRSYIYS 93
Query: 61 IKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTHFL 114
+K F + Y FL+ILN Y + L + H +L+ F+ L
Sbjct: 94 LKESFRDEPAKYAQFLEILNDYSARRVDAPSAVARMTELMKDHRNLVLGFSVLL 147
Score = 36.6 bits (83), Expect = 0.10
Identities = 21/65 (32%), Positives = 32/65 (48%), Gaps = 1/65 (1%)
Query: 52 EEAINFVNKIKARFEGDDHV-YKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEF 110
+EA +++N +K F D Y+ FLDI+ R + V + L + H +LL F
Sbjct: 13 DEATSYINAVKEAFGADQPAKYREFLDIMLDLRANRVDLATVVPRMRELLKDHVNLLLRF 72
Query: 111 THFLP 115
FLP
Sbjct: 73 NAFLP 77
Score = 31.2 bits (69), Expect = 4.3
Identities = 22/79 (27%), Positives = 39/79 (48%), Gaps = 7/79 (8%)
Query: 246 TRPMSSTCDDKKSLKTKYPEEFAFCDKVKEKLRNPDG--YQEFLKCLHIYSKEIITQQEL 303
T P+ + D+ K ++ F K+K + + DG Y+ FL+ L +Y + + +L
Sbjct: 154 TTPLEAEPDNNKRIRVAN-----FISKLKARFQGNDGHVYESFLEILTMYQQGNKSVNDL 208
Query: 304 QSLVGDLLGKYPDLMEEFN 322
V LL + DL+ EF+
Sbjct: 209 YQEVVALLQGHEDLVMEFS 227
>At1g10250 unknown protein
Length = 77
Score = 102 bits (253), Expect = 2e-21
Identities = 51/71 (71%), Positives = 59/71 (82%), Gaps = 1/71 (1%)
Query: 1154 MNALYSLLDGS-SDNTKFEDDCRAIIGTQSYLLFTLDKLIYKLVKQLQAIASEEMDNKLL 1212
M+ALY+LLDGS DNTKFED+CRAI G QSY+LFTLDKL+ K VK L A+AS+E D KLL
Sbjct: 1 MDALYNLLDGSIDDNTKFEDECRAIFGAQSYVLFTLDKLVQKFVKHLHAVASDETDTKLL 60
Query: 1213 QLYAYEKSRKL 1223
QL+AYE RKL
Sbjct: 61 QLHAYENYRKL 71
>At5g35610 putative protein
Length = 155
Score = 90.1 bits (222), Expect = 8e-18
Identities = 51/107 (47%), Positives = 71/107 (65%), Gaps = 15/107 (14%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEI---------TLPLEDEQPPAKK--PV 49
ID +GV + VKELF+ ++ I GFNTFLPKG+EI + LE EQ P KK +
Sbjct: 44 IDPSGVKSVVKELFKEDQEPISGFNTFLPKGFEIKPECDQNGFKIKLECEQTPPKKYVDI 103
Query: 50 EFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEV 96
E+ EA++FV K+K DD +YKSF+ I++MY+K+NK ++EV EV
Sbjct: 104 EYSEALDFVRKVK----DDDRIYKSFVTIMDMYKKKNKSLDEVCREV 146
>At1g27280 hypothetical protein
Length = 225
Score = 87.8 bits (216), Expect = 4e-17
Identities = 45/100 (45%), Positives = 66/100 (66%), Gaps = 1/100 (1%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNK 60
ID AG I RV+EL + H++L++ N FLP + L L+ EQ A + + +F+ K
Sbjct: 115 IDVAGGITRVEELLKAHKNLLVRLNAFLPPEAQRILHLKIEQRAASDINKRKRVASFIGK 174
Query: 61 IKARFEGDD-HVYKSFLDILNMYRKENKCINEVYHEVAVL 99
+K RF+GDD HVY+SFL+IL MY++ NK +N++Y EV L
Sbjct: 175 LKERFQGDDRHVYESFLEILTMYQEGNKSVNDLYQEVGFL 214
>At3g24260 hypothetical protein
Length = 374
Score = 69.7 bits (169), Expect = 1e-11
Identities = 50/151 (33%), Positives = 73/151 (48%), Gaps = 27/151 (17%)
Query: 418 DRCTPSYRLLPKNYPIPIASYKSKIGAKVLNDHWVSVTSGSEDYSFKHMRK-NQYEESLF 476
+R TP+Y+L+PK P++S VLN+ WV Y + + E+ ++
Sbjct: 212 ERATPNYKLIPKEEQTPVSS-------TVLNNTWVV-----NSYDIQAQKNLTDIEKDMY 259
Query: 477 RCEDDRFELDMLLESVNLTTKRVEELFEKINNNVIKGDRPVRIDEHLTALNLRCIERLYG 536
ED FELDML+ + K EE VI G+R ++ L RC E LY
Sbjct: 260 NWEDQMFELDMLVGFLTSAAKNAEE--------VINGERDLK---DLGGKFYRCAENLY- 307
Query: 537 DHGLDVMEVLRKNAPLALPVILTRLKQKQDE 567
G D++E++++N LP IL RL QK E
Sbjct: 308 --GRDMLEIVKENHQRVLPAILNRLNQKLRE 336
>At1g24250 hypothetical protein
Length = 252
Score = 68.6 bits (166), Expect = 2e-11
Identities = 43/117 (36%), Positives = 57/117 (47%), Gaps = 24/117 (20%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNK 60
+D A VIARV+EL + H +L+ GF FL +F K
Sbjct: 154 VDAACVIARVEELMKDHLNLLFGFCVFL-----------------------SATTSFTTK 190
Query: 61 IKARFEGD-DHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTHFLPD 116
+KARF+GD V S L I+ MY + NK ++ Y EV L QGH DL+ E + L D
Sbjct: 191 LKARFQGDGSQVVDSVLQIMRMYGEGNKSKHDAYQEVVALVQGHDDLVMELSQILTD 247
Score = 40.8 bits (94), Expect = 0.005
Identities = 31/126 (24%), Positives = 52/126 (40%), Gaps = 19/126 (15%)
Query: 8 ARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQ------------PP-------AKKP 48
AR+++L + H L LG N LP Y++T+P E + PP +
Sbjct: 56 ARMQDLLKDHPSLCLGLNVLLPPEYQLTIPPEASEEFHKVVGRSVPVPPKVVGRSLPRPE 115
Query: 49 VEFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLD 108
++A +++ +K F + Y L +L ++ V V L + H +LL
Sbjct: 116 PTIDDATSYLIAVKEAFHDEPAKYGEMLKLLKDFKARRVDAACVIARVEELMKDHLNLLF 175
Query: 109 EFTHFL 114
F FL
Sbjct: 176 GFCVFL 181
>At1g24210 unknown protein
Length = 155
Score = 67.4 bits (163), Expect = 5e-11
Identities = 40/124 (32%), Positives = 63/124 (50%), Gaps = 22/124 (17%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNK 60
+D VIARV+EL + H+DL+LGF+ FLP PV E+ F+NK
Sbjct: 48 VDKYSVIARVEELMKDHQDLLLGFSVFLP------------------PVSVED---FINK 86
Query: 61 IKARFEG-DDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTHFLPDASA 119
+K RF+ D HV + ++ M+++ + EV EV + H DL+++F F
Sbjct: 87 LKTRFQSLDTHVVGAIRGLMKMFKEGKMSVKEVQEEVIDVLFYHEDLIEDFLRFFTKNPV 146
Query: 120 AASS 123
+ +S
Sbjct: 147 STAS 150
>At1g23810 hypothetical protein
Length = 241
Score = 66.2 bits (160), Expect = 1e-10
Identities = 40/119 (33%), Positives = 57/119 (47%), Gaps = 24/119 (20%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNK 60
++ A VIARV+EL + H +L+ GF FL +F K
Sbjct: 143 VNAASVIARVEELMKDHSNLLFGFCVFL-----------------------SATTSFTTK 179
Query: 61 IKARFEGD-DHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTHFLPDAS 118
+KA+F+GD V S L I+ MY + NK ++ Y E+ L QGH DL+ E + D S
Sbjct: 180 LKAKFQGDGSQVVDSVLQIMRMYGEGNKSKHDAYQEIVALVQGHDDLVMELSQIFTDPS 238
>At1g27270 hypothetical protein
Length = 241
Score = 64.7 bits (156), Expect = 4e-10
Identities = 44/117 (37%), Positives = 58/117 (48%), Gaps = 24/117 (20%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNK 60
I+ A VIAR++EL + H +L+LGF FL P ++ F+ K
Sbjct: 147 INAASVIARMEELLKDHLNLLLGFCVFLS--------------PTRR---------FITK 183
Query: 61 IKARFEGD-DHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTHFLPD 116
+KARF GD V S L IL M+ + NK +E EV L QGH DLL E + D
Sbjct: 184 LKARFLGDGSQVVDSVLQILRMHSEGNKSKDEASQEVRALIQGHEDLLMELSEIFSD 240
>At4g12020 putative disease resistance protein
Length = 1895
Score = 63.9 bits (154), Expect = 6e-10
Identities = 31/66 (46%), Positives = 47/66 (70%), Gaps = 11/66 (16%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNK 60
+DT+GVIAR+K+LF+GH DL+LGFNT+L K Y+IT+ ED+ P I+F++K
Sbjct: 341 VDTSGVIARLKDLFKGHDDLLLGFNTYLSKEYQITILPEDDFP-----------IDFLDK 389
Query: 61 IKARFE 66
++ +E
Sbjct: 390 VEGPYE 395
Score = 39.7 bits (91), Expect = 0.012
Identities = 21/62 (33%), Positives = 36/62 (57%), Gaps = 1/62 (1%)
Query: 53 EAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTH 112
+A+N++ +K +FE D Y +FL++LN + + + V + LF+GH DLL F
Sbjct: 308 DALNYLKAVKDKFE-DSEKYDTFLEVLNDCKHQGVDTSGVIARLKDLFKGHDDLLLGFNT 366
Query: 113 FL 114
+L
Sbjct: 367 YL 368
>At1g27260 hypothetical protein
Length = 222
Score = 62.8 bits (151), Expect = 1e-09
Identities = 39/110 (35%), Positives = 54/110 (48%), Gaps = 24/110 (21%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNK 60
+D A IA V+EL + H+ L+ GF+ FL + F+ K
Sbjct: 127 VDAASFIASVEELMKDHQTLLNGFSVFL-----------------------SAEMKFIRK 163
Query: 61 IKARFEGD-DHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDE 109
+KA+F+GD HV S L IL MY + NK +E + EV L Q H DL+ E
Sbjct: 164 LKAKFQGDGSHVADSVLQILRMYSEGNKSKSEAFQEVVPLVQDHEDLVME 213
>At3g28870 hypothetical protein
Length = 355
Score = 60.1 bits (144), Expect = 9e-09
Identities = 42/142 (29%), Positives = 66/142 (45%), Gaps = 17/142 (11%)
Query: 465 HMRKNQYEESLFRCEDDRFELDMLLESVNLTTKRVEELFEKINNNVIKGDRPVRIDEHLT 524
H EE +++ ED+ FE+DML+ + + +E VIKG+ ++ L
Sbjct: 142 HKNLTDIEEDMYKWEDEMFEVDMLMRVLTSAVESAKE--------VIKGEMELK---DLG 190
Query: 525 ALNLRCIERLYGDHGLDVMEVLRKNAPLALPVILTRLKQKQDEWARCRADFSKVWAEIYA 584
A RC+E LYG+ D+ E + ++ ALP+IL+RLKQK R +W +
Sbjct: 191 AKFYRCVEMLYGE---DMFETVTEDHQRALPMILSRLKQKLRHVTTARERLKPLWKQTIE 247
Query: 585 KNYHKSLDHRSFYFKQQDTKRL 606
K S + R +D L
Sbjct: 248 K---LSTNQRGSTISSEDNNNL 266
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.133 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,296,960
Number of Sequences: 26719
Number of extensions: 1590955
Number of successful extensions: 7422
Number of sequences better than 10.0: 277
Number of HSP's better than 10.0 without gapping: 96
Number of HSP's successfully gapped in prelim test: 190
Number of HSP's that attempted gapping in prelim test: 5410
Number of HSP's gapped (non-prelim): 1372
length of query: 1389
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1277
effective length of database: 8,326,068
effective search space: 10632388836
effective search space used: 10632388836
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0028b.5


