

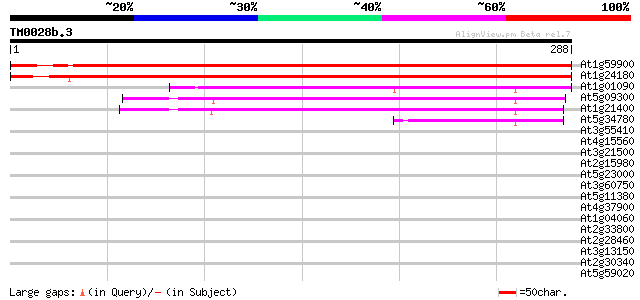
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0028b.3
(288 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g59900 pyruvate dehydrogenase E1 alpha subunit, putative 477 e-135
At1g24180 pyruvate dehydrogenase E1 alpha subunit 475 e-134
At1g01090 pyruvate dehydrogenase E1 alpha subunit 178 3e-45
At5g09300 branched-chain alpha keto-acid dehydrogenase E1 alpha ... 102 2e-22
At1g21400 branched-chain alpha keto-acid dehydrogenase E1 alpha ... 99 3e-21
At5g34780 3-methyl-2-oxobutanoate dehydrogenase-like protein 50 1e-06
At3g55410 2-oxoglutarate dehydrogenase, E1 subunit - like protein 35 0.036
At4g15560 1-D-deoxyxylulose 5-phosphate synthase, putative 35 0.061
At3g21500 1-D-deoxyxylulose 5-phosphate synthase, putative 31 0.68
At2g15980 F7H1.1 31 0.68
At5g23000 myb like transcription factor (MYB37) 30 1.5
At3g60750 transketolase - like protein 30 1.5
At5g11380 1-D-deoxyxylulose 5-phosphate synthase - like protein 29 2.6
At4g37900 putative protein 29 2.6
At1g04060 hypothetical protein 29 2.6
At2g33800 30S ribosomal protein S5 29 3.4
At2g28460 unknown protein 29 3.4
At3g13150 hypothetical protein 28 4.4
At2g30340 unknown protein 28 4.4
At5g59020 putative protein 28 7.5
>At1g59900 pyruvate dehydrogenase E1 alpha subunit, putative
Length = 389
Score = 477 bits (1228), Expect = e-135
Identities = 236/288 (81%), Positives = 258/288 (88%), Gaps = 10/288 (3%)
Query: 1 MALSRLASSSSSSSSTVGSKFLKPLTAAFSLRRSISSDSNPITIETSVPFTAHNCDPPSR 60
MALSRL+S S+ + +P +AAFS R IS+D+ PITIETS+PFTAH CDPPSR
Sbjct: 1 MALSRLSSRSNIIT--------RPFSAAFS--RLISTDTTPITIETSLPFTAHLCDPPSR 50
Query: 61 SVETSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKD 120
SVE+S+ EL FFR MA MRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEA ITKKD
Sbjct: 51 SVESSSQELLDFFRTMALMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAAITKKD 110
Query: 121 AIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQ 180
AIITAYRDHC FLGRGG+L E+FSELMGR GCSKGKGGSMHFY+KE++FYGGHGIVGAQ
Sbjct: 111 AIITAYRDHCIFLGRGGSLHEVFSELMGRQAGCSKGKGGSMHFYKKESSFYGGHGIVGAQ 170
Query: 181 VPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMG 240
VPLGCG+AFAQKY+K+EAVTFALYGDGAANQGQLFEALNI+ALWDLPAILVCENNHYGMG
Sbjct: 171 VPLGCGIAFAQKYNKEEAVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYGMG 230
Query: 241 TAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGP 288
TAEWRAAKSP+YYKRGDYVPGLKVDGMD AVKQACKFAK+HAL+ GP
Sbjct: 231 TAEWRAAKSPSYYKRGDYVPGLKVDGMDAFAVKQACKFAKQHALEKGP 278
>At1g24180 pyruvate dehydrogenase E1 alpha subunit
Length = 393
Score = 475 bits (1223), Expect = e-134
Identities = 231/290 (79%), Positives = 259/290 (88%), Gaps = 10/290 (3%)
Query: 1 MALSRLASSSSSSSSTVGSKFLKPLTAAF--SLRRSISSDSNPITIETSVPFTAHNCDPP 58
MALSRL+S S++ FLKP A S+RR +S+DS+PITIET+VPFT+H C+ P
Sbjct: 1 MALSRLSSRSNT--------FLKPAITALPSSIRRHVSTDSSPITIETAVPFTSHLCESP 52
Query: 59 SRSVETSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITK 118
SRSVETS+ E+ +FFR MA MRRMEIAADSLYKAKLIRGFCHLYDGQEA+A+GMEA ITK
Sbjct: 53 SRSVETSSEEILAFFRDMARMRRMEIAADSLYKAKLIRGFCHLYDGQEALAVGMEAAITK 112
Query: 119 KDAIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVG 178
KDAIIT+YRDHCTF+GRGG L++ FSELMGR GCS GKGGSMHFY+K+ +FYGGHGIVG
Sbjct: 113 KDAIITSYRDHCTFIGRGGKLVDAFSELMGRKTGCSHGKGGSMHFYKKDASFYGGHGIVG 172
Query: 179 AQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYG 238
AQ+PLGCG+AFAQKY+KDEAVTFALYGDGAANQGQLFEALNI+ALWDLPAILVCENNHYG
Sbjct: 173 AQIPLGCGLAFAQKYNKDEAVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYG 232
Query: 239 MGTAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGP 288
MGTA WR+AKSPAY+KRGDYVPGLKVDGMD LAVKQACKFAKEHALKNGP
Sbjct: 233 MGTATWRSAKSPAYFKRGDYVPGLKVDGMDALAVKQACKFAKEHALKNGP 282
>At1g01090 pyruvate dehydrogenase E1 alpha subunit
Length = 428
Score = 178 bits (451), Expect = 3e-45
Identities = 92/215 (42%), Positives = 130/215 (59%), Gaps = 10/215 (4%)
Query: 83 EIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCTFLGRGGTLLEI 142
++ A Y+ K+ GF HLY+GQEAV+ G +TK D++++ YRDH L +G + +
Sbjct: 99 DMCAQMYYRGKMF-GFVHLYNGQEAVSTGFIKLLTKSDSVVSTYRDHVHALSKGVSARAV 157
Query: 143 FSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQKYSKD------ 196
SEL G+V GC +G+GGSMH + KE N GG +G +P+ G AF+ KY ++
Sbjct: 158 MSELFGKVTGCCRGQGGSMHMFSKEHNMLGGFAFIGEGIPVATGAAFSSKYRREVLKQDC 217
Query: 197 EAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSPAYYKRG 256
+ VT A +GDG N GQ FE LN+AAL+ LP I V ENN + +G + RA P +K+G
Sbjct: 218 DDVTVAFFGDGTCNNGQFFECLNMAALYKLPIIFVVENNLWAIGMSHLRATSDPEIWKKG 277
Query: 257 DY--VPGLKVDGMDVLAVKQACKFAKEHALK-NGP 288
+PG+ VDGMDVL V++ K A A + GP
Sbjct: 278 PAFGMPGVHVDGMDVLKVREVAKEAVTRARRGEGP 312
>At5g09300 branched-chain alpha keto-acid dehydrogenase E1 alpha
subunit-like protein
Length = 472
Score = 102 bits (254), Expect = 2e-22
Identities = 64/232 (27%), Positives = 109/232 (46%), Gaps = 9/232 (3%)
Query: 59 SRSVETSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLYD---GQEAVAIGMEAG 115
S+ V+ S + M T++ M D+++ +G Y G+EA+ I A
Sbjct: 120 SQFVQVSEEVAVKIYSDMVTLQIM----DNIFYEAQRQGRLSFYATAIGEEAINIASAAA 175
Query: 116 ITKKDAIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHG 175
+T +D I YR+ L RG TL E ++ G KG+ +H+ + N++
Sbjct: 176 LTPQDVIFPQYREPGVLLWRGFTLQEFANQCFGNKSDYGKGRQMPVHYGSNKLNYFTVSA 235
Query: 176 IVGAQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENN 235
+ Q+P G A++ K K +A +GDG ++G ALNIAA+ + P + +C NN
Sbjct: 236 TIATQLPNAVGAAYSLKMDKKDACAVTYFGDGGTSEGDFHAALNIAAVMEAPVLFICRNN 295
Query: 236 HYGMGTAEWRAAKSPAYYKRGDY--VPGLKVDGMDVLAVKQACKFAKEHALK 285
+ + T +S +G + ++VDG D LA+ A A+E A++
Sbjct: 296 GWAISTPTSDQFRSDGVVVKGRAYGIRSIRVDGNDALAMYSAVHTAREMAIR 347
>At1g21400 branched-chain alpha keto-acid dehydrogenase E1 alpha
subunit
Length = 472
Score = 99.0 bits (245), Expect = 3e-21
Identities = 64/233 (27%), Positives = 105/233 (44%), Gaps = 9/233 (3%)
Query: 57 PPSRSVETSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLY---DGQEAVAIGME 113
P S + S + QMAT++ M D ++ +G Y G+EA+ I
Sbjct: 118 PDSDFIPVSEKLAVRMYEQMATLQVM----DHIFYEAQRQGRISFYLTSVGEEAINIASA 173
Query: 114 AGITKKDAIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGG 173
A ++ D ++ YR+ L RG TL E ++ G KG+ +H+ N++
Sbjct: 174 AALSPDDVVLPQYREPGVLLWRGFTLEEFANQCFGNKADYGKGRQMPIHYGSNRLNYFTI 233
Query: 174 HGIVGAQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCE 233
+ Q+P GV ++ K K A T GDG ++G LN AA+ + P + +C
Sbjct: 234 SSPIATQLPQAAGVGYSLKMDKKNACTVTFIGDGGTSEGDFHAGLNFAAVMEAPVVFICR 293
Query: 234 NNHYGMGTAEWRAAKSPAYYKRGDY--VPGLKVDGMDVLAVKQACKFAKEHAL 284
NN + + T +S +G + ++VDG D LAV A + A+E A+
Sbjct: 294 NNGWAISTHISEQFRSDGIVVKGQAYGIRSIRVDGNDALAVYSAVRSAREMAV 346
>At5g34780 3-methyl-2-oxobutanoate dehydrogenase-like protein
Length = 365
Score = 50.4 bits (119), Expect = 1e-06
Identities = 30/89 (33%), Positives = 45/89 (49%), Gaps = 4/89 (4%)
Query: 198 AVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSPAYYKRGD 257
AVTF GDG ++G LN AA+ + P + +C NN + + T +S +G
Sbjct: 30 AVTFI--GDGGTSEGDFHAGLNFAAVMEAPVVFICRNNGWAISTHISEQFRSDGIVVKGQ 87
Query: 258 Y--VPGLKVDGMDVLAVKQACKFAKEHAL 284
+ ++VDG D LAV A A+E A+
Sbjct: 88 AYGIRSIRVDGNDALAVYSAVCSAREMAV 116
>At3g55410 2-oxoglutarate dehydrogenase, E1 subunit - like protein
Length = 1017
Score = 35.4 bits (80), Expect = 0.036
Identities = 33/102 (32%), Positives = 50/102 (48%), Gaps = 12/102 (11%)
Query: 191 QKYSKD----EAVTFALYGDGA-ANQGQLFEALNIAALWDLPA---ILVCENNHYGMGTA 242
Q YS D + + ++GDG+ A QG ++E L+++AL + I + NN T
Sbjct: 380 QYYSNDLDRTKNLGILIHGDGSFAGQGVVYETLHLSALPNYTTGGTIHIVVNNQVAF-TT 438
Query: 243 EWRAAKSPAY---YKRGDYVPGLKVDGMDVLAVKQACKFAKE 281
+ RA +S Y + P V+G DV AV AC+ A E
Sbjct: 439 DPRAGRSSQYCTDVAKALSAPIFHVNGDDVEAVVHACELAAE 480
>At4g15560 1-D-deoxyxylulose 5-phosphate synthase, putative
Length = 717
Score = 34.7 bits (78), Expect = 0.061
Identities = 19/63 (30%), Positives = 28/63 (44%)
Query: 173 GHGIVGAQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVC 232
G G + G G+A + A+ GDGA GQ +EA+N A D I++
Sbjct: 185 GTGHSSTTISAGLGMAVGRDLKGKNNNVVAVIGDGAMTAGQAYEAMNNAGYLDSDMIVIL 244
Query: 233 ENN 235
+N
Sbjct: 245 NDN 247
>At3g21500 1-D-deoxyxylulose 5-phosphate synthase, putative
Length = 628
Score = 31.2 bits (69), Expect = 0.68
Identities = 26/98 (26%), Positives = 43/98 (43%), Gaps = 5/98 (5%)
Query: 141 EIFSELMGRVDGCSKGKGGSMHFYRKET---NFYGGHGIVGAQVPLGCGVAFAQKYSKDE 197
+I + G++ + G S + R+E+ +F GH LG V K +
Sbjct: 130 KILTGRRGKMKTIRQTNGLSGYTKRRESEHDSFGTGHSSTTLSAGLGMAVGRDLKGMNNS 189
Query: 198 AVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENN 235
V+ + GDGA GQ +EA+N A I++ +N
Sbjct: 190 VVS--VIGDGAMTAGQAYEAMNNAGYLHSNMIVILNDN 225
>At2g15980 F7H1.1
Length = 498
Score = 31.2 bits (69), Expect = 0.68
Identities = 36/111 (32%), Positives = 45/111 (40%), Gaps = 24/111 (21%)
Query: 79 MRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAY---RDHCTFL-- 133
M+R AD L L+ G C DGQ V EA KDA+ A +C L
Sbjct: 377 MKRKGFEADGLTIEALVEGLCDDRDGQRVV----EAADIVKDAVREAMFYPSRNCYELLV 432
Query: 134 ------GRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVG 178
G+ L I +E++G KG S YR F G+GIVG
Sbjct: 433 KRLCEDGKMDRALNIQAEMVG------KGFKPSQETYRA---FIDGYGIVG 474
>At5g23000 myb like transcription factor (MYB37)
Length = 329
Score = 30.0 bits (66), Expect = 1.5
Identities = 42/182 (23%), Positives = 71/182 (38%), Gaps = 33/182 (18%)
Query: 6 LASSSSSSSSTVGSKFLKPLTAAFSLRRSISSDSNPITIETSV--PFTAHNCDPPSRSVE 63
L+SSS SSSS + S +L P++ + + ++P TI +S P+ + P++S+
Sbjct: 118 LSSSSDSSSSAMASPYLNPIS------QDVKRPTSPTTIPSSSYNPYAENPNQYPTKSLI 171
Query: 64 TSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAII 123
+S + +Q+ + D LY G +
Sbjct: 172 SSINGFEAGDKQIISYINPNYPQD-------------LYLSDSNNNTSNANGFLLNHNMC 218
Query: 124 TAYRDHCTFLGR-GGTLLEIF---SELM------GRVDGCSKGKGGSMHFYRKETNFYGG 173
Y++H +F G EI E+M +D +KG G F + N+Y G
Sbjct: 219 DQYKNHTSFSSDVNGIRSEIMMKQEEIMMMMMIDHHIDQRTKGYNG--EFTQGYYNYYNG 276
Query: 174 HG 175
HG
Sbjct: 277 HG 278
>At3g60750 transketolase - like protein
Length = 741
Score = 30.0 bits (66), Expect = 1.5
Identities = 21/73 (28%), Positives = 37/73 (49%), Gaps = 11/73 (15%)
Query: 175 GIVGAQVPLGCGVAFAQK-----YSKDEA-----VTFALYGDGAANQGQLFEALNIAALW 224
G +G + G+A A+K ++K +A T+A+ GDG +G EA ++A W
Sbjct: 192 GPLGQGIANAVGLALAEKHLAARFNKPDAEVVDHYTYAILGDGCQMEGISNEACSLAGHW 251
Query: 225 DLPAILV-CENNH 236
L ++ ++NH
Sbjct: 252 GLGKLIAFYDDNH 264
>At5g11380 1-D-deoxyxylulose 5-phosphate synthase - like protein
Length = 700
Score = 29.3 bits (64), Expect = 2.6
Identities = 19/71 (26%), Positives = 32/71 (44%), Gaps = 2/71 (2%)
Query: 169 NFYGGHGIVGAQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPA 228
+F GHG LG VA K +D V A+ + GQ +EA++ A D
Sbjct: 184 SFGTGHGCNSISAGLGLAVARDMKGKRDRVV--AVIDNVTITAGQAYEAMSNAGYLDSNM 241
Query: 229 ILVCENNHYGM 239
I++ ++ + +
Sbjct: 242 IVILNDSRHSL 252
>At4g37900 putative protein
Length = 787
Score = 29.3 bits (64), Expect = 2.6
Identities = 33/120 (27%), Positives = 49/120 (40%), Gaps = 22/120 (18%)
Query: 5 RLASSSSSS---SSTVGSKFLKPLTAAF--------SLRRSISSDSNPITIETSVPFTAH 53
+L SS S S +S S+FL P+T +R + D NPI++ +V FT
Sbjct: 393 QLMSSKSKSLGFTSLSFSEFLSPVTKLSVEKWLELTPTKRGKADDPNPISLRVAVSFT-- 450
Query: 54 NCDPPSRSVETSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGME 113
PP+R S LH Q + L K +L + F + D E I ++
Sbjct: 451 ---PPTR----SPTVLH--LVQARPSLKGSCFLPMLRKVRLAKSFTRVVDETETEVINLQ 501
>At1g04060 hypothetical protein
Length = 243
Score = 29.3 bits (64), Expect = 2.6
Identities = 16/43 (37%), Positives = 27/43 (62%)
Query: 7 ASSSSSSSSTVGSKFLKPLTAAFSLRRSISSDSNPITIETSVP 49
ASSSSSSSS+ ++ LKP+ FS++++ + + T +P
Sbjct: 123 ASSSSSSSSSYNNEDLKPVFILFSVKQTQPEEEDEDDGVTELP 165
>At2g33800 30S ribosomal protein S5
Length = 303
Score = 28.9 bits (63), Expect = 3.4
Identities = 20/57 (35%), Positives = 28/57 (49%), Gaps = 2/57 (3%)
Query: 4 SRLASSSSSSSSTVGSKFLKPLTAAFSLRRSISSDSNPITIETSVPFTAHNC--DPP 58
S L SSSS+ S S FL ++ +L ++ S+D+ I E P N DPP
Sbjct: 20 SSLISSSSTKSIVSFSSFLNRRFSSLTLVKASSTDTETIFFEDETPEITANVVFDPP 76
>At2g28460 unknown protein
Length = 704
Score = 28.9 bits (63), Expect = 3.4
Identities = 33/139 (23%), Positives = 50/139 (35%), Gaps = 26/139 (18%)
Query: 19 SKFLKPLTAAFSLRRSISSDSNPITIETSVPFTAHNCDPPSRSVETSTAELHSFFRQMAT 78
S+F+ T FSL S+ SDS P + + S+ + FFRQ
Sbjct: 43 SEFMSLTTQIFSLLHSMDSDSMPKPLSNLISLL-------SQGNFDHNTDFRLFFRQTMA 95
Query: 79 MRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCTFLGRGGT 138
+ I +Y+ ++ I M TK+D +I+ C LG G
Sbjct: 96 LEPEPIFMSLIYQI-------------FSLVISMN---TKRDKLISLCPQACVVLGNNGN 139
Query: 139 LLEIFSELMGRVDGCSKGK 157
I + G+ KGK
Sbjct: 140 FEVIVKKQEGQ---AHKGK 155
>At3g13150 hypothetical protein
Length = 551
Score = 28.5 bits (62), Expect = 4.4
Identities = 21/72 (29%), Positives = 42/72 (58%), Gaps = 5/72 (6%)
Query: 4 SRLASSSSSSSSTVGSKFLKPLTAAFSLRRSISSDSNPITIETSVPFTAH--NCDPPSRS 61
S ++SS+ ++S+ V S P T++F L S+++DS+ ++S P ++ + P S S
Sbjct: 416 SPVSSSAKTTSTPVSSS---PDTSSFLLSLSLAADSSSSDSDSSSPDSSSSVSSSPDSSS 472
Query: 62 VETSTAELHSFF 73
+S+ + +S F
Sbjct: 473 SVSSSPDSYSSF 484
>At2g30340 unknown protein
Length = 268
Score = 28.5 bits (62), Expect = 4.4
Identities = 14/35 (40%), Positives = 22/35 (62%)
Query: 6 LASSSSSSSSTVGSKFLKPLTAAFSLRRSISSDSN 40
++SSSSS+SS S + P ++ S+SSD+N
Sbjct: 229 VSSSSSSNSSATNSMYNPPPSSTAGYSNSLSSDNN 263
>At5g59020 putative protein
Length = 780
Score = 27.7 bits (60), Expect = 7.5
Identities = 19/76 (25%), Positives = 31/76 (40%), Gaps = 2/76 (2%)
Query: 5 RLASSSSSSSSTVGSKFLKPLTAAFSLRRSISSDSNPITIETSVPFTAHNCDPPSRSVET 64
RL+ + +S T P T S+ S DS + + + V NC+ PS+ T
Sbjct: 336 RLSFNMGKASKTNSEGGTVPTTQLDSMTNSTKIDSQNVALLSDVD--GSNCNKPSKKDTT 393
Query: 65 STAELHSFFRQMATMR 80
+T+ L + R
Sbjct: 394 TTSHLRRLLEPLLKPR 409
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,347,779
Number of Sequences: 26719
Number of extensions: 269674
Number of successful extensions: 809
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 793
Number of HSP's gapped (non-prelim): 23
length of query: 288
length of database: 11,318,596
effective HSP length: 98
effective length of query: 190
effective length of database: 8,700,134
effective search space: 1653025460
effective search space used: 1653025460
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0028b.3


