

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0028b.2
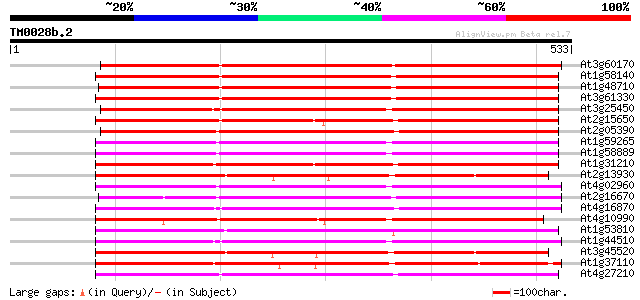
(533 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g60170 putative protein 468 e-132
At1g58140 hypothetical protein 461 e-130
At1g48710 hypothetical protein 459 e-129
At3g61330 copia-type polyprotein 457 e-129
At3g25450 hypothetical protein 414 e-116
At2g15650 putative retroelement pol polyprotein 402 e-112
At2g05390 putative retroelement pol polyprotein 398 e-111
At1g59265 polyprotein, putative 343 2e-94
At1g58889 polyprotein, putative 343 2e-94
At1g31210 putative reverse transcriptase 332 2e-91
At2g13930 putative retroelement pol polyprotein 331 7e-91
At4g02960 putative polyprotein of LTR transposon 329 2e-90
At2g16670 putative retroelement pol polyprotein 327 1e-89
At4g16870 retrotransposon like protein 323 2e-88
At4g10990 putative retrotransposon polyprotein 322 3e-88
At1g53810 321 6e-88
At1g44510 polyprotein, putative 321 6e-88
At3g45520 copia-like polyprotein 318 4e-87
At1g37110 317 1e-86
At4g27210 putative protein 317 1e-86
>At3g60170 putative protein
Length = 1339
Score = 468 bits (1205), Expect = e-132
Identities = 232/439 (52%), Positives = 323/439 (72%), Gaps = 5/439 (1%)
Query: 87 GLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPPGFEIQ 146
G+DY+EVFAPVAR++T+R +++I+S NW IFQLDVKSAFL+G L+EEVYV QP GF +
Sbjct: 901 GIDYTEVFAPVARLDTVRTILAISSQFNWEIFQLDVKSAFLHGELKEEVYVRQPEGFIRE 960
Query: 147 GKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTGLLLIC 206
G+E V+KL KALYGLKQAPRAW RI+ + L+ +F RC E+ ++ + G+ +L++
Sbjct: 961 GEEEKVYKLRKALYGLKQAPRAWYSRIEAYFLKEEFERCPSEHTLFTKTRVGN--ILIVS 1018
Query: 207 LYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQKYIKD 266
LYVDDL+ TGSD DE K+ + EFEMSDLGK+ +FLG++ + + GIF+ Q++Y ++
Sbjct: 1019 LYVDDLIFTGSDKAMCDEFKKSMMLEFEMSDLGKMKHFLGIEVKQSDGGIFICQRRYARE 1078
Query: 267 ILKRFNMGNCHPVTTPSEVNIKLRRDDEGD-VDASLYKQLVGSLRFLCNSRLDISYGVGL 325
+L RF M + V P KL +D+ G+ VD +++KQLVGSL +L +R D+ YGV L
Sbjct: 1079 VLARFGMDESNAVKNPIVPGTKLTKDENGEKVDETMFKQLVGSLMYLTVTRPDLMYGVCL 1138
Query: 326 ISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSGDLDDR 385
IS+FM P+ SH AAKRILRYL+GT + GI + R+K + L+L+ +TDSDY+GDL+DR
Sbjct: 1139 ISRFMSNPRMSHWLAAKRILRYLKGTVELGIFYRRRKN--RSLKLMAFTDSDYAGDLNDR 1196
Query: 386 KSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMGFVTKK 445
+STSG++F + I W SKKQPV+ALS+ EAEYIA +F ACQ VWL ++L+++G K
Sbjct: 1197 RSTSGFVFLMASGAICWASKKQPVVALSTTEAEYIAAAFCACQCVWLRKVLEKLGAEEKS 1256
Query: 446 PMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKADIFTK 505
+ DN S I L+K+P+ HG+SKHIEVRFH+LR+ VNG ++L YCPT++Q ADIFTK
Sbjct: 1257 ATVINCDNSSTIQLSKHPVLHGKSKHIEVRFHYLRDLVNGDVVKLEYCPTEDQVADIFTK 1316
Query: 506 PLKTERFVILRKEIGVVSL 524
PLK E+F LR +G+V++
Sbjct: 1317 PLKLEQFEKLRALLGMVNM 1335
>At1g58140 hypothetical protein
Length = 1320
Score = 461 bits (1187), Expect = e-130
Identities = 219/441 (49%), Positives = 316/441 (70%), Gaps = 7/441 (1%)
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
+S + G+DY EVFAPVAR+ET+RL++S+A+ W I Q+DVKSAFLNG LEEEVY+ QP
Sbjct: 875 YSQRAGIDYDEVFAPVARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLEEEVYIEQPQ 934
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTG 201
G+ ++G+E+ V +L KALYGLKQAPRAWN RID + + F +C E+ +Y++ D
Sbjct: 935 GYIVKGEEDKVLRLKKALYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALYIKIQKED-- 992
Query: 202 LLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQ 261
+L+ CLYVDDL+ TG++P +E K+ + EFEM+D+G + Y+LG++ + GIF+ Q+
Sbjct: 993 ILIACLYVDDLIFTGNNPSMFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQE 1052
Query: 262 KYIKDILKRFNMGNCHPVTTPSEVNIKLRRDDEGD-VDASLYKQLVGSLRFLCNSRLDIS 320
Y K++LK+F M + +PV TP E IKL + +EG+ VD + +K LVGSLR+L +R DI
Sbjct: 1053 GYAKEVLKKFKMDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYLTCTRPDIL 1112
Query: 321 YGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSG 380
Y VG++S++M P +H AAKRILRY++GT ++G+ + + +LVGY+DSD+ G
Sbjct: 1113 YAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHYSTTS----DYKLVGYSDSDWGG 1168
Query: 381 DLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMG 440
D+DDRKSTSG++F + +W SKKQP++ LS+CEAEY+A + C A+WL LLKE+
Sbjct: 1169 DVDDRKSTSGFVFYIGDTAFTWMSKKQPIVTLSTCEAEYVAATSCVCHAIWLRNLLKELS 1228
Query: 441 FVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKA 500
++P ++ VDN SAI+LAKNP+ H RSKHI+ R+H++RE V+ ++L Y T +Q A
Sbjct: 1229 LPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYIRECVSKKDVQLEYVKTHDQVA 1288
Query: 501 DIFTKPLKTERFVILRKEIGV 521
DIFTKPLK E F+ +R +GV
Sbjct: 1289 DIFTKPLKREDFIKMRSLLGV 1309
>At1g48710 hypothetical protein
Length = 1352
Score = 459 bits (1181), Expect = e-129
Identities = 218/438 (49%), Positives = 314/438 (70%), Gaps = 7/438 (1%)
Query: 85 KPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPPGFE 144
+ G+DY EVFAPVAR+ET+RL++S+A+ W I Q+DVKSAFLNG LEEEVY+ QP G+
Sbjct: 910 RAGIDYDEVFAPVARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLEEEVYIEQPQGYI 969
Query: 145 IQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTGLLL 204
++G+E+ V +L KALYGLKQAPRAWN RID + + F +C E+ +Y++ D +L+
Sbjct: 970 VKGEEDKVLRLKKALYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALYIKIQKED--ILI 1027
Query: 205 ICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQKYI 264
CLYVDDL+ TG++P +E K+ + EFEM+D+G + Y+LG++ + GIF+ Q+ Y
Sbjct: 1028 ACLYVDDLIFTGNNPSMFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQEGYA 1087
Query: 265 KDILKRFNMGNCHPVTTPSEVNIKLRRDDEGD-VDASLYKQLVGSLRFLCNSRLDISYGV 323
K++LK+F M + +PV TP E IKL + +EG+ VD + +K LVGSLR+L +R DI Y V
Sbjct: 1088 KEVLKKFKMDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYLTCTRPDILYAV 1147
Query: 324 GLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSGDLD 383
G++S++M P +H AAKRILRY++GT ++G+ + + +LVGY+DSD+ GD+D
Sbjct: 1148 GVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHYSTTS----DYKLVGYSDSDWGGDVD 1203
Query: 384 DRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMGFVT 443
DRKSTSG++F + +W SKKQP++ LS+CEAEY+A + C A+WL LLKE+
Sbjct: 1204 DRKSTSGFVFYIGDTAFTWMSKKQPIVVLSTCEAEYVAATSCVCHAIWLRNLLKELSLPQ 1263
Query: 444 KKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKADIF 503
++P ++ VDN SAI+LAKNP+ H RSKHI+ R+H++RE V+ ++L Y T +Q ADIF
Sbjct: 1264 EEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYIRECVSKKDVQLEYVKTHDQVADIF 1323
Query: 504 TKPLKTERFVILRKEIGV 521
TKPLK E F+ +R +GV
Sbjct: 1324 TKPLKREDFIKMRSLLGV 1341
>At3g61330 copia-type polyprotein
Length = 1352
Score = 457 bits (1175), Expect = e-129
Identities = 216/441 (48%), Positives = 314/441 (70%), Gaps = 7/441 (1%)
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
+S + G+DY EVFAPVAR+ET+RL++S+A+ W I Q+DVKSAFLNG LEEEVY+ QP
Sbjct: 907 YSQRVGIDYDEVFAPVARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLEEEVYIEQPQ 966
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTG 201
G+ ++G+E+ V +L K LYGLKQAPRAWN RID + + F +C E+ +Y++ D
Sbjct: 967 GYIVKGEEDKVLRLKKVLYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALYIKIQKED-- 1024
Query: 202 LLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQ 261
+L+ CLYVDDL+ TG++P +E K+ + EFEM+D+G + Y+LG++ + GIF+ Q+
Sbjct: 1025 ILIACLYVDDLIFTGNNPSIFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQE 1084
Query: 262 KYIKDILKRFNMGNCHPVTTPSEVNIKLRRDDEGD-VDASLYKQLVGSLRFLCNSRLDIS 320
Y K++LK+F + + +PV TP E IKL + +EG+ VD + +K LVGSLR+L +R DI
Sbjct: 1085 GYAKEVLKKFKIDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYLTCTRPDIL 1144
Query: 321 YGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSG 380
Y VG++S++M P +H AAKRILRY++GT ++G+ + + +LVGY+DSD+ G
Sbjct: 1145 YAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHYSTTS----DYKLVGYSDSDWGG 1200
Query: 381 DLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMG 440
D+DDRKSTSG++F + +W SKKQP++ LS+CEAEY+A + C A+WL LLKE+
Sbjct: 1201 DVDDRKSTSGFVFYIGDTAFTWMSKKQPIVTLSTCEAEYVAATSCVCHAIWLRNLLKELS 1260
Query: 441 FVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKA 500
++P ++ VDN SAI+LAKNP+ H RSKHI+ R+H++RE V+ ++L Y T +Q A
Sbjct: 1261 LPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYIRECVSKKDVQLEYVKTHDQVA 1320
Query: 501 DIFTKPLKTERFVILRKEIGV 521
D FTKPLK E F+ +R +GV
Sbjct: 1321 DFFTKPLKRENFIKMRSLLGV 1341
>At3g25450 hypothetical protein
Length = 1343
Score = 414 bits (1063), Expect = e-116
Identities = 211/436 (48%), Positives = 308/436 (70%), Gaps = 7/436 (1%)
Query: 87 GLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPPGFEIQ 146
G+D+ EVFAPVARIET+RL++++A++ W I LDVK+AFL+G L E+VYVSQP GF +
Sbjct: 894 GVDFEEVFAPVARIETVRLIIALAASNGWEIHHLDVKTAFLHGELREDVYVSQPEGFTNK 953
Query: 147 GKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTGLLLIC 206
+ V+KL KALYGL+QAPRAWN +++ L +LKF +C E +Y RK G+ +L++
Sbjct: 954 ESKEKVYKLHKALYGLRQAPRAWNTKLNEILKELKFEKCHKEPSLY-RKQEGEN-ILVVA 1011
Query: 207 LYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQKYIKD 266
+YVDDLL+TGS+ + K+ + +FEMSDLGKL Y+LG++ + GI + Q++Y K
Sbjct: 1012 VYVDDLLVTGSNLDIILNFKKGMVGKFEMSDLGKLTYYLGIEVLQSKDGITLKQERYAKK 1071
Query: 267 ILKRFNMGNCHPVTTPSEVNIKLRR-DDEGDVDASLYKQLVGSLRFLCNSRLDISYGVGL 325
IL+ M C+ V TP +++L + DE +D + Y++ +G LR+L ++R D+SY VG+
Sbjct: 1072 ILEEAGMSKCNTVNTPMIASLELSKAQDEKRIDETDYRRNIGCLRYLLHTRPDLSYNVGI 1131
Query: 326 ISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSGDLDDR 385
+S+++ P++SH AA K+ILRYLQGTT +G+ F K G+ L+GY+DS ++ DLDD
Sbjct: 1132 LSRYLQEPRESHGAALKQILRYLQGTTSHGLYF----KKGENAGLIGYSDSSHNVDLDDG 1187
Query: 386 KSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMGFVTKK 445
KST G++F L+ PI+W S+KQ V+ LSSCEAE++A + AA QA+WL+ELL E+ +
Sbjct: 1188 KSTGGHIFYLNDCPITWCSQKQQVVTLSSCEAEFMAATEAAKQAIWLQELLAEVIGTECE 1247
Query: 446 PMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKADIFTK 505
+ + VDN SAI+L KNP+ HGRSKHI R+HF+RE V G++E+ + P QKADI TK
Sbjct: 1248 KVTIRVDNKSAIALTKNPVFHGRSKHIHRRYHFIRECVENGQIEVEHVPGVRQKADILTK 1307
Query: 506 PLKTERFVILRKEIGV 521
L +F+ +R+ IGV
Sbjct: 1308 ALGKIKFLEMRELIGV 1323
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 402 bits (1032), Expect = e-112
Identities = 204/444 (45%), Positives = 297/444 (65%), Gaps = 11/444 (2%)
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
FS + G+DY E FAPV+R +TIR +++ A+ W ++Q+DVKSAFLNG LEEEVYV+QPP
Sbjct: 906 FSQEYGIDYLETFAPVSRYDTIRALLAYAAQMKWRLYQMDVKSAFLNGELEEEVYVTQPP 965
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTG 201
GF I+GKE V +L KALYGLKQAPRAW +RID + +Q F R + +Y +K D
Sbjct: 966 GFVIEGKEEKVLRLYKALYGLKQAPRAWYERIDSYFIQNGFARSMNDAALYSKKKGED-- 1023
Query: 202 LLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQ 261
+L++ LYVDDL+ITG++ ++ K+ +K EFEM+DLG L YFLGM+ +GIF+ Q+
Sbjct: 1024 VLIVSLYVDDLIITGNNTHLINTFKKNMKDEFEMTDLGLLNYFLGMEVNQDDSGIFLSQE 1083
Query: 262 KYIKDILKRFNMGNCHPVTTPSEVNIKLRRDDEGD----VDASLYKQLVGSLRFLCNSRL 317
KY ++ +F M V+TP K R+ EGD D + Y+++VG L +LC SR
Sbjct: 1084 KYANKLIDKFGMKESKSVSTPLTPQGK-RKGVEGDDKEFADPTKYRRIVGGLLYLCASRP 1142
Query: 318 DISYGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSD 377
D+ Y +S++M +P H AKR+LRY++GT+++G+LF ++ LVGY+DSD
Sbjct: 1143 DVMYASSYLSRYMSSPSIQHYQEAKRVLRYVKGTSNFGVLFTSKETP----RLVGYSDSD 1198
Query: 378 YSGDLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLK 437
+ G L+D+KST+GY+F L A W S KQ +A S+ EAEYIA A QA+WL+ L +
Sbjct: 1199 WGGSLEDKKSTTGYVFTLGLAMFCWQSCKQQTVAQSTAEAEYIAVCAATNQAIWLQRLFE 1258
Query: 438 EMGFVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDE 497
+ G K+ + ++ DN SAI++ +NP+ H R+KHIE+++HF+RE + G ++L YC ++
Sbjct: 1259 DFGLKFKEGIPILCDNKSAIAIGRNPVQHRRTKHIEIKYHFVREAEHKGLIQLEYCKGED 1318
Query: 498 QKADIFTKPLKTERFVILRKEIGV 521
Q AD+ TK L RF LR+++GV
Sbjct: 1319 QLADVLTKALSVSRFEGLRRKLGV 1342
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 398 bits (1023), Expect = e-111
Identities = 203/436 (46%), Positives = 300/436 (68%), Gaps = 7/436 (1%)
Query: 87 GLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPPGFEIQ 146
G+DY EVFA VARIETIR+++++A++ W + LDVK+AFL+G L E+VYV+QP GF +
Sbjct: 855 GIDYDEVFAHVARIETIRVIIALAASNGWEVHHLDVKTAFLHGELREDVYVTQPEGFTNK 914
Query: 147 GKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTGLLLIC 206
E V+KL KALYGLKQAPRAWN +++ L +L F +CS E VY R+ + LL++
Sbjct: 915 DNEGKVYKLHKALYGLKQAPRAWNTKLNKILQELNFVKCSKEPSVYRRQE--EKKLLIVA 972
Query: 207 LYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQKYIKD 266
+YVDDLL+TGS + K+ + +FEMSDLG+L Y+LG++ + GI + Q++Y
Sbjct: 973 IYVDDLLVTGSSLDLILCFKKDMAGKFEMSDLGQLTYYLGIEVLHRKNGIILRQERYAMK 1032
Query: 267 ILKRFNMGNCHPVTTPSEVNIKL-RRDDEGDVDASLYKQLVGSLRFLCNSRLDISYGVGL 325
I++ M NC+PV P ++L + +E + Y++++G LR++ ++R D+SY VG+
Sbjct: 1033 IIEEAGMSNCNPVLIPMAAGLELCKAQEEKCITERDYRRMIGCLRYIVHTRPDLSYCVGV 1092
Query: 326 ISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSGDLDDR 385
+S+++ P++SH A K++LRYL+GT +G+ R K G LVGY+DS +S DLDD
Sbjct: 1093 LSRYLQQPRESHGNALKQVLRYLKGTMSHGLYLKRGFKSG----LVGYSDSSHSADLDDG 1148
Query: 386 KSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMGFVTKK 445
KST+G++F LH PI+W S+KQ V+ALSSCEAE++A + AA QA+WL++L E+ T +
Sbjct: 1149 KSTAGHIFYLHQCPITWCSQKQQVVALSSCEAEFMAATEAAKQAIWLQDLFAEVCGTTSE 1208
Query: 446 PMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKADIFTK 505
+ + VDN SAI+L KN + HGRSKHI R+HF+RE V +E+ + P EQ+ADI TK
Sbjct: 1209 KVMIRVDNKSAIALTKNLVFHGRSKHIHRRYHFIRECVENNLVEVDHVPGVEQRADILTK 1268
Query: 506 PLKTERFVILRKEIGV 521
PL +F +R+ +GV
Sbjct: 1269 PLGRIKFREMRELVGV 1284
>At1g59265 polyprotein, putative
Length = 1466
Score = 343 bits (879), Expect = 2e-94
Identities = 188/441 (42%), Positives = 266/441 (59%), Gaps = 7/441 (1%)
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
++ +PGLDY+E F+PV + +IR+V+ +A R+WPI QLDV +AFL G L ++VY+SQPP
Sbjct: 1026 YNQRPGLDYAETFSPVIKSTSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDDVYMSQPP 1085
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTG 201
GF + + N V KL KALYGLKQAPRAW + +LL + F + ++V +
Sbjct: 1086 GFIDKDRPNYVCKLRKALYGLKQAPRAWYVELRNYLLTIGFVNSVSDTSLFVLQR--GKS 1143
Query: 202 LLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQ 261
++ + +YVDD+LITG+DP L L F + D +L YFLG++ + G+ + Q+
Sbjct: 1144 IVYMLVYVDDILITGNDPTLLHNTLDNLSQRFSVKDHEELHYFLGIEAKRVPTGLHLSQR 1203
Query: 262 KYIKDILKRFNMGNCHPVTTPSEVNIKLRRDDEGDV-DASLYKQLVGSLRFLCNSRLDIS 320
+YI D+L R NM PVTTP + KL + D + Y+ +VGSL++L +R DIS
Sbjct: 1204 RYILDLLARTNMITAKPVTTPMAPSPKLSLYSGTKLTDPTEYRGIVGSLQYLAFTRPDIS 1263
Query: 321 YGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSG 380
Y V +S+FM P + HL A KRILRYL GT ++GI K G L L Y+D+D++G
Sbjct: 1264 YAVNRLSQFMHMPTEEHLQALKRILRYLAGTPNHGIFL----KKGNTLSLHAYSDADWAG 1319
Query: 381 DLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMG 440
D DD ST+GY+ L PISW+SKKQ + SS EAEY + + + + W+ LL E+G
Sbjct: 1320 DKDDYVSTNGYIVYLGHHPISWSSKKQKGVVRSSTEAEYRSVANTSSEMQWICSLLTELG 1379
Query: 441 FVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKA 500
+P + DN+ A L NP+ H R KHI + +HF+R QV G L +V+ T +Q A
Sbjct: 1380 IRLTRPPVIYCDNVGATYLCANPVFHSRMKHIAIDYHFIRNQVQSGALRVVHVSTHDQLA 1439
Query: 501 DIFTKPLKTERFVILRKEIGV 521
D TKPL F +IGV
Sbjct: 1440 DTLTKPLSRTAFQNFASKIGV 1460
>At1g58889 polyprotein, putative
Length = 1466
Score = 343 bits (879), Expect = 2e-94
Identities = 188/441 (42%), Positives = 266/441 (59%), Gaps = 7/441 (1%)
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
++ +PGLDY+E F+PV + +IR+V+ +A R+WPI QLDV +AFL G L ++VY+SQPP
Sbjct: 1026 YNQRPGLDYAETFSPVIKSTSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDDVYMSQPP 1085
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTG 201
GF + + N V KL KALYGLKQAPRAW + +LL + F + ++V +
Sbjct: 1086 GFIDKDRPNYVCKLRKALYGLKQAPRAWYVELRNYLLTIGFVNSVSDTSLFVLQR--GKS 1143
Query: 202 LLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQ 261
++ + +YVDD+LITG+DP L L F + D +L YFLG++ + G+ + Q+
Sbjct: 1144 IVYMLVYVDDILITGNDPTLLHNTLDNLSQRFSVKDHEELHYFLGIEAKRVPTGLHLSQR 1203
Query: 262 KYIKDILKRFNMGNCHPVTTPSEVNIKLRRDDEGDV-DASLYKQLVGSLRFLCNSRLDIS 320
+YI D+L R NM PVTTP + KL + D + Y+ +VGSL++L +R DIS
Sbjct: 1204 RYILDLLARTNMITAKPVTTPMAPSPKLSLYSGTKLTDPTEYRGIVGSLQYLAFTRPDIS 1263
Query: 321 YGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSG 380
Y V +S+FM P + HL A KRILRYL GT ++GI K G L L Y+D+D++G
Sbjct: 1264 YAVNRLSQFMHMPTEEHLQALKRILRYLAGTPNHGIFL----KKGNTLSLHAYSDADWAG 1319
Query: 381 DLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMG 440
D DD ST+GY+ L PISW+SKKQ + SS EAEY + + + + W+ LL E+G
Sbjct: 1320 DKDDYVSTNGYIVYLGHHPISWSSKKQKGVVRSSTEAEYRSVANTSSEMQWICSLLTELG 1379
Query: 441 FVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKA 500
+P + DN+ A L NP+ H R KHI + +HF+R QV G L +V+ T +Q A
Sbjct: 1380 IRLTRPPVIYCDNVGATYLCANPVFHSRMKHIAIDYHFIRNQVQSGALRVVHVSTHDQLA 1439
Query: 501 DIFTKPLKTERFVILRKEIGV 521
D TKPL F +IGV
Sbjct: 1440 DTLTKPLSRTAFQNFASKIGV 1460
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 332 bits (852), Expect = 2e-91
Identities = 181/440 (41%), Positives = 265/440 (60%), Gaps = 7/440 (1%)
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
F + G+DY E F+PV R TIRLV+ +++++ WPI QLDV +AFL+G L+E V++ QP
Sbjct: 929 FDQEEGVDYLETFSPVVRTATIRLVLDVSTSKGWPIKQLDVSNAFLHGELQEPVFMYQPS 988
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTG 201
GF K V +L+KA+YGLKQAPRAW FLL F + ++V H D
Sbjct: 989 GFIDPQKPTHVCRLTKAIYGLKQAPRAWFDTFSNFLLDYGFVCSKSDPSLFV--CHQDGK 1046
Query: 202 LLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQ 261
+L + LYVDD+L+TGSD L++L Q LK+ F M DLG YFLG+Q E A G+F++Q
Sbjct: 1047 ILYLLLYVDDILLTGSDQSLLEDLLQALKNRFSMKDLGPPRYFLGIQIEDYANGLFLHQT 1106
Query: 262 KYIKDILKRFNMGNCHPVTTPSEVNIKLRRDDEGDVDASLYKQLVGSLRFLCNSRLDISY 321
Y DIL++ M +C+P+ TP + + E + + ++ L G L++L +R DI +
Sbjct: 1107 AYATDILQQAGMSDCNPMPTPLPQQLD-NLNSELFAEPTYFRSLAGKLQYLTITRPDIQF 1165
Query: 322 GVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSGD 381
V I + M +P S KRILRY++GT G+ R L L Y+DSD++G
Sbjct: 1166 AVNFICQRMHSPTTSDFGLLKRILRYIKGTIGMGLPIKRNSTL----TLSAYSDSDHAGC 1221
Query: 382 LDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMGF 441
+ R+ST+G+ L ISW++K+QP ++ SS EAEY A ++AA + W+ LL+++G
Sbjct: 1222 KNTRRSTTGFCILLGSNLISWSAKRQPTVSNSSTEAEYRALTYAAREITWISFLLRDLGI 1281
Query: 442 VTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKAD 501
P ++ DN+SA+ L+ NP H RSKH + +H++REQV G +E + Q AD
Sbjct: 1282 PQYLPTQVYCDNLSAVYLSANPALHNRSKHFDTDYHYIREQVALGLIETQHISATFQLAD 1341
Query: 502 IFTKPLKTERFVILRKEIGV 521
+FTK L FV LR ++GV
Sbjct: 1342 VFTKSLPRRAFVDLRSKLGV 1361
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 331 bits (848), Expect = 7e-91
Identities = 179/441 (40%), Positives = 280/441 (62%), Gaps = 16/441 (3%)
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
F+ K G+DY+E+F+PV + +IR ++S+ N + Q+DVK+AFL+G LEEE+Y++QP
Sbjct: 889 FTQKEGVDYNEIFSPVVKHVSIRYLLSMVVHYNMELQQMDVKTAFLHGFLEEEIYMAQPE 948
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTG 201
GFEI+ N V L ++LYGLKQ+PR WN R D F+ +K+ R + + VY +K +GDT
Sbjct: 949 GFEIKRGSNKVCLLKRSLYGLKQSPRQWNLRFDEFMRGIKYTRSAYDSCVYFKKCNGDTY 1008
Query: 202 LLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQF--EYTAAGIFMY 259
+ L+ LYVDD+LI ++ E++ELKQ+L EFEM DLG LGM+ + A + +
Sbjct: 1009 IYLL-LYVDDMLIASANKSEVNELKQLLSREFEMKDLGDAKKILGMEISRDRDAGLLTLS 1067
Query: 260 QQKYIKDILKRFNMGNCHPVTTPSEVNIKLRRDDEGDVDASL-------YKQLVGSLRF- 311
Q+ Y+K +L+ F M N PV+TP ++ KL+ + + + Y +GS+ +
Sbjct: 1068 QEGYVKKVLRSFQMDNAKPVSTPLGIHFKLKAATDKEYEEQFERMKIVPYANTIGSIMYS 1127
Query: 312 LCNSRLDISYGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELV 371
+ +R D++Y +G+IS+FM P K H A K +LRY++GT + F +Q ++ L
Sbjct: 1128 MIGTRPDLAYSLGVISRFMSKPLKDHWQAVKWVLRYMRGTEKKKLCFRKQ----EDFLLR 1183
Query: 372 GYTDSDYSGDLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVW 431
GY DSDY + D R+S +GY+F + G ISW SK Q V+A+SS EAEY+A + A +A+W
Sbjct: 1184 GYCDSDYGSNFDTRRSITGYVFTVGGNTISWKSKLQKVVAISSTEAEYMALTEAVKEALW 1243
Query: 432 LEELLKEMGFVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELV 491
L+ E+G ++ +E+ D+ SAI+LAKN + H R+KHI++R HF+R+ + G +++V
Sbjct: 1244 LKGFAAELGH-SQDYVEVHSDSQSAITLAKNSVHHERTKHIDIRLHFIRDIICAGLIKVV 1302
Query: 492 YCPTDEQKADIFTKPLKTERF 512
T+ A+IFTK + +F
Sbjct: 1303 KIATECNPANIFTKTVPLAKF 1323
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 329 bits (844), Expect = 2e-90
Identities = 179/444 (40%), Positives = 262/444 (58%), Gaps = 7/444 (1%)
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
++ +PGLDY+E F+PV + +IR+V+ +A R+WPI QLDV +AFL G L +EVY+SQPP
Sbjct: 1009 YNQRPGLDYAETFSPVIKSTSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDEVYMSQPP 1068
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTG 201
GF + + + V +L KA+YGLKQAPRAW + +LL + F + ++V +
Sbjct: 1069 GFVDKDRPDYVCRLRKAIYGLKQAPRAWYVELRTYLLTVGFVNSISDTSLFVLQR--GRS 1126
Query: 202 LLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQ 261
++ + +YVDD+LITG+D L L F + + L YFLG++ + G+ + Q+
Sbjct: 1127 IIYMLVYVDDILITGNDTVLLKHTLDALSQRFSVKEHEDLHYFLGIEAKRVPQGLHLSQR 1186
Query: 262 KYIKDILKRFNMGNCHPVTTPSEVNIKLRRDDEGDV-DASLYKQLVGSLRFLCNSRLDIS 320
+Y D+L R NM PV TP + KL + D + Y+ +VGSL++L +R D+S
Sbjct: 1187 RYTLDLLARTNMLTAKPVATPMATSPKLTLHSGTKLPDPTEYRGIVGSLQYLAFTRPDLS 1246
Query: 321 YGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSG 380
Y V +S++M P H A KR+LRYL GT D+GI K G L L Y+D+D++G
Sbjct: 1247 YAVNRLSQYMHMPTDDHWNALKRVLRYLAGTPDHGIFL----KKGNTLSLHAYSDADWAG 1302
Query: 381 DLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMG 440
D DD ST+GY+ L PISW+SKKQ + SS EAEY + + + + W+ LL E+G
Sbjct: 1303 DTDDYVSTNGYIVYLGHHPISWSSKKQKGVVRSSTEAEYRSVANTSSELQWICSLLTELG 1362
Query: 441 FVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKA 500
P + DN+ A L NP+ H R KHI + +HF+R QV G L +V+ T +Q A
Sbjct: 1363 IQLSHPPVIYCDNVGATYLCANPVFHSRMKHIALDYHFIRNQVQSGALRVVHVSTHDQLA 1422
Query: 501 DIFTKPLKTERFVILRKEIGVVSL 524
D TKPL F ++IGV+ +
Sbjct: 1423 DTLTKPLSRVAFQNFSRKIGVIKV 1446
>At2g16670 putative retroelement pol polyprotein
Length = 1333
Score = 327 bits (837), Expect = 1e-89
Identities = 178/441 (40%), Positives = 265/441 (59%), Gaps = 8/441 (1%)
Query: 85 KPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPPGFE 144
K G DY E FAPVA++ T+RL + +A RNW I Q+DV +AFL+G L EEVY+ PPGFE
Sbjct: 896 KEGEDYGETFAPVAKMATVRLFLDVAVKRNWEIHQMDVHNAFLHGDLREEVYMKLPPGFE 955
Query: 145 IQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTGLLL 204
N V +L KALYGLKQAPR W +++ L + F + +Y ++ + +
Sbjct: 956 AS-HPNKVCRLRKALYGLKQAPRCWFEKLTTALKRYGFQQSLADYSLFTLVK--GSVRIK 1012
Query: 205 ICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQKYI 264
I +YVDDL+ITG+ + + K+ L S F M DLG L YFLG++ + GI++ Q+KY
Sbjct: 1013 ILIYVDDLIITGNSQRATQQFKEYLASCFHMKDLGPLKYFLGIEVARSTTGIYICQRKYA 1072
Query: 265 KDILKRFNMGNCHPVTTPSEVNIKLRRDDEGDV-DASLYKQLVGSLRFLCNSRLDISYGV 323
DI+ + P P E N KL + D Y++LVG L +L +RLD+++ V
Sbjct: 1073 LDIISETGLLGVKPANFPLEQNHKLGLSTSPLLTDPQRYRRLVGRLIYLAVTRLDLAFSV 1132
Query: 324 GLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSGDLD 383
++++FM P++ H AAA R++RYL+ G+ R + ++ G+ DSD++GD
Sbjct: 1133 HILARFMQEPREDHWAAALRVVRYLKADPGQGVFLRRSG----DFQITGWCDSDWAGDPM 1188
Query: 384 DRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMGFVT 443
R+S +GY Q +PISW +KKQ ++ SS EAEY A SF A + +WL++LL +G
Sbjct: 1189 SRRSVTGYFVQFGDSPISWKTKKQDTVSKSSAEAEYRAMSFLASELLWLKQLLFSLGVSH 1248
Query: 444 KKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKADIF 503
+PM + D+ SAI +A NP+ H R+KHIE+ +HF+R++ G + + T Q ADIF
Sbjct: 1249 VQPMIMCCDSKSAIYIATNPVFHERTKHIEIDYHFVRDEFVKGVITPRHVGTTSQLADIF 1308
Query: 504 TKPLKTERFVILRKEIGVVSL 524
TKPL + F R ++G+ +L
Sbjct: 1309 TKPLGRDCFSAFRIKLGIRNL 1329
>At4g16870 retrotransposon like protein
Length = 1474
Score = 323 bits (827), Expect = 2e-88
Identities = 181/444 (40%), Positives = 259/444 (57%), Gaps = 7/444 (1%)
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
F+ + G+DY+E F+PV + TIRLV+ +A ++W I QLDV +AFL G L EEVY++QPP
Sbjct: 1034 FNQQYGVDYAETFSPVIKSTTIRLVLDVAVKKDWEIKQLDVNNAFLQGTLTEEVYMAQPP 1093
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTG 201
GF + + V +L KA+YGLKQAPRAW + L + F + +++ HG T
Sbjct: 1094 GFIDKDRPTHVCRLRKAIYGLKQAPRAWYMELKQHLFNIGFVNSLSDASLFIY-CHGTT- 1151
Query: 202 LLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQ 261
+ + +YVDD+++TGSD +D + L F + D L YFLG++ T G+ + Q+
Sbjct: 1152 FVYVLVYVDDIIVTGSDKSSIDAVLTSLAERFSIKDPTDLHYFLGIEATRTKQGLHLMQR 1211
Query: 262 KYIKDILKRFNMGNCHPVTTPSEVNIKLRRDDEGDV-DASLYKQLVGSLRFLCNSRLDIS 320
KYIKD+L + NM + PV TP + KL + DAS Y+ +VGSL++L +R DI+
Sbjct: 1212 KYIKDLLAKHNMADAKPVLTPLPTSPKLTLHGGTKLNDASEYRSVVGSLQYLAFTRPDIA 1271
Query: 321 YGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSG 380
Y V +S+ M P + H AAKR+LRYL GT+ +GI L L ++D+D++G
Sbjct: 1272 YAVNRLSQLMPQPTEDHWQAAKRVLRYLAGTSTHGIFLDTTSPLN----LHAFSDADWAG 1327
Query: 381 DLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMG 440
D DD ST+ Y+ L PISW+SKKQ +A SS E+EY A + AA + WL LL ++
Sbjct: 1328 DSDDYVSTNAYVIYLGKNPISWSSKKQRGVARSSTESEYRAVANAASEVKWLCSLLSKLH 1387
Query: 441 FVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKA 500
+ DNI A L NP+ H R KHI + +HF+R + G L + + T +Q A
Sbjct: 1388 IRLPIRPSIFCDNIGATYLCANPVFHSRMKHIAIDYHFVRNMIQSGALRVSHVSTRDQLA 1447
Query: 501 DIFTKPLKTERFVILRKEIGVVSL 524
D TKPL F R +IGV L
Sbjct: 1448 DALTKPLSRAHFQSARFKIGVRQL 1471
>At4g10990 putative retrotransposon polyprotein
Length = 1203
Score = 322 bits (825), Expect = 3e-88
Identities = 165/432 (38%), Positives = 274/432 (63%), Gaps = 13/432 (3%)
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
F+ + G+DY E F+PVA+ +++L++ +A+A W + Q+DV +AFL+G L+EE+Y+S P
Sbjct: 594 FTQQEGIDYMETFSPVAKFGSVKLLLGLAAATGWSLTQMDVSNAFLHGELDEEIYMSLPQ 653
Query: 142 GFE----IQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSH 197
G+ I V +L K+LYGLKQA R W KR+ L F + + ++V+ S
Sbjct: 654 GYTPPTGISLPSKPVCRLLKSLYGLKQASRQWYKRLSSVFLGANFIQSPADNTMFVKVSC 713
Query: 198 GDTGLLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIF 257
T ++++ +YVDDL+I +D ++ LK++L+SEF++ DLG +FLG++ ++ GI
Sbjct: 714 --TSIIVVLVYVDDLMIASNDSSAVENLKELLRSEFKIKDLGPARFFLGLEIARSSEGIS 771
Query: 258 MYQQKYIKDILKRFNMGNCHPVTTPSEVNIKLRRDDEGDV--DASLYKQLVGSLRFLCNS 315
+ Q+KY +++L+ + C P + P + N+ L ++ G + +A+ Y++LVG L +LC +
Sbjct: 772 VCQRKYAQNLLEDVGLSGCKPSSIPMDPNLHLTKE-MGTLLPNATSYRELVGRLLYLCIT 830
Query: 316 RLDISYGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTD 375
R DI++ V +S+F+ AP H+ AA ++LRYL+G G+++ EL L G++D
Sbjct: 831 RPDITFAVHTLSQFLSAPTDIHMQAAHKVLRYLKGNPGQGLMY----SASSELCLNGFSD 886
Query: 376 SDYSGDLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEEL 435
+D+ D R+S +G+ L + I+W SKKQ V++ SS E+EY + + A C+ +WL++L
Sbjct: 887 ADWGTCKDSRRSVTGFCIYLGTSLITWKSKKQSVVSRSSTESEYRSLAQATCEIIWLQQL 946
Query: 436 LKEMGFVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPT 495
LK++ P +L DN SA+ LA NP+ H R+KHIE+ H +R+Q+ GKL+ ++ PT
Sbjct: 947 LKDLHVTMTCPAKLFCDNKSALHLATNPVFHERTKHIEIDCHTVRDQIKAGKLKTLHVPT 1006
Query: 496 DEQKADIFTKPL 507
Q ADI TKPL
Sbjct: 1007 GNQLADILTKPL 1018
>At1g53810
Length = 1522
Score = 321 bits (823), Expect = 6e-88
Identities = 178/446 (39%), Positives = 259/446 (57%), Gaps = 8/446 (1%)
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
F + G+DY E ++PV R T+RL++ +A+ W + Q+DVK+AFL+G L E VY+ QP
Sbjct: 985 FKQEEGIDYLETYSPVVRTPTVRLILHVATVLKWELKQMDVKNAFLHGDLTETVYMRQPA 1044
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTG 201
GF + K + V L K+LYGLKQ+PRAW R FLL+ F + ++V S+ D
Sbjct: 1045 GFVDKSKPDHVCLLHKSLYGLKQSPRAWFDRFSNFLLEFGFICSLFDPSLFVYSSNNDVI 1104
Query: 202 LLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQ 261
LLL LYVDD++ITG++ + L L L EF M D+G++ YFLG+Q + G+FM QQ
Sbjct: 1105 LLL--LYVDDMVITGNNSQSLTHLLAALNKEFRMKDMGQVHYFLGIQIQTYDGGLFMSQQ 1162
Query: 262 KYIKDILKRFNMGNCHPVTTPSEVNI-KLRRDDEGDVDASLYKQLVGSLRFLCNSRLDIS 320
KY +D+L +M NC P+ TP + + ++ DE D + ++ L G L++L +R DI
Sbjct: 1163 KYAEDLLITASMANCSPMPTPLPLQLDRVSNQDEVFSDPTYFRSLAGKLQYLTLTRPDIQ 1222
Query: 321 YGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKK-----LGKELELVGYTD 375
+ V + + M P S KRILRY++GT GI + + +L Y+D
Sbjct: 1223 FAVNFVCQKMHQPSVSDFNLLKRILRYIKGTVSMGIQYNSNSSSVVSAYESDYDLSAYSD 1282
Query: 376 SDYSGDLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEEL 435
SDY+ + R+S GY + ISW+SKKQP ++ SS EAEY + S A + W+ +
Sbjct: 1283 SDYANCKETRRSVGGYCTFMGQNIISWSSKKQPTVSRSSTEAEYRSLSETASEIKWMSSI 1342
Query: 436 LKEMGFVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPT 495
L+E+G EL DN+SA+ L NP H R+KH +V H++RE+V L + + P
Sbjct: 1343 LREIGVSLPDTPELFCDNLSAVYLTANPAFHARTKHFDVDHHYIRERVALKTLVVKHIPG 1402
Query: 496 DEQKADIFTKPLKTERFVILRKEIGV 521
Q ADIFTK L E F LR ++GV
Sbjct: 1403 HLQLADIFTKSLPFEAFTRLRFKLGV 1428
>At1g44510 polyprotein, putative
Length = 1459
Score = 321 bits (823), Expect = 6e-88
Identities = 179/444 (40%), Positives = 262/444 (58%), Gaps = 7/444 (1%)
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
F+ + G+DY+E F+PV + TIR+V+ +A +NWP+ QLDV +AFL G L EEVY++QPP
Sbjct: 1019 FNQQYGVDYAETFSPVIKATTIRVVLDVAVKKNWPLKQLDVNNAFLQGTLTEEVYMAQPP 1078
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTG 201
GF + + + V +L KA+YGLKQAPRAW + LL + F + +++ SHG T
Sbjct: 1079 GFVDKDRPSHVCRLRKAIYGLKQAPRAWYMELKQHLLNIGFVNSLADTSLFI-YSHGTT- 1136
Query: 202 LLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQ 261
LL + +YVDD+++TGSD K + + L F + D L YFLG++ T G+ + Q+
Sbjct: 1137 LLYLLVYVDDIIVTGSDHKSVSAVLSSLAERFSIKDPTDLHYFLGIEATRTNTGLHLMQR 1196
Query: 262 KYIKDILKRFNMGNCHPVTTPSEVNIKLRRDDEGDV-DASLYKQLVGSLRFLCNSRLDIS 320
KY+ D+L + NM + PV TP + KL + DAS Y+ +VGSL++L +R DI+
Sbjct: 1197 KYMTDLLAKHNMLDAKPVATPLPTSPKLTLHGGTKLNDASEYRSVVGSLQYLAFTRPDIA 1256
Query: 321 YGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSG 380
+ V +S+FM P H AAKR+LRYL GTT +GI + L ++D+D++G
Sbjct: 1257 FAVNRLSQFMHQPTSDHWQAAKRVLRYLAGTTTHGIFL----NSSSPIHLHAFSDADWAG 1312
Query: 381 DLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMG 440
D D ST+ Y+ L PISW+SKKQ ++ SS E+EY A + AA + WL LL E+
Sbjct: 1313 DSADYVSTNAYVIYLGRNPISWSSKKQRGVSRSSTESEYRAVANAASEIRWLCSLLTELH 1372
Query: 441 FVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKA 500
+ DNI A + NP+ H R KHI + +HF+R + L + + T++Q A
Sbjct: 1373 IRLPHGPTIFCDNIGATYICANPVFHSRMKHIALDYHFVRGMIQSRALRVSHVSTNDQLA 1432
Query: 501 DIFTKPLKTERFVILRKEIGVVSL 524
D TK L F+ R +IGV L
Sbjct: 1433 DALTKSLSRPHFLSARSKIGVRQL 1456
>At3g45520 copia-like polyprotein
Length = 1363
Score = 318 bits (816), Expect = 4e-87
Identities = 170/441 (38%), Positives = 274/441 (61%), Gaps = 16/441 (3%)
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
++ + G+DY E+FAPV + +IR+++SI + N + QLDVK+AFL+G L+E++Y+ P
Sbjct: 917 YAQREGVDYHEIFAPVVKHVSIRILLSIVAQENLELEQLDVKTAFLHGELKEKIYMMPPE 976
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTG 201
G E KEN V L+K+LYGLKQAPR WN++ + ++ ++ F R + Y +K D+
Sbjct: 977 GCESLFKENEVCLLNKSLYGLKQAPRQWNEKFNHYMTEIGFKRSDYDSCAYTKKLSDDST 1036
Query: 202 LLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQ--FEYTAAGIFMY 259
+ L+ YVDD+L+ ++ + +D LK+ L +FEM DLG LG++ + A +++
Sbjct: 1037 MYLL-FYVDDMLVAANNMQAIDALKKELSIKFEMKDLGAAKKILGIEIIIDREAGVLWLS 1095
Query: 260 QQKYIKDILKRFNMGNCHPVTTPSEVNIKLR-------RDDEGDVDASLYKQLVGSLRF- 311
Q+ Y+ +LK FNM P TP ++K++ +E +++ Y VGS+ +
Sbjct: 1096 QESYLNKVLKTFNMLESKPALTPLGAHLKMKSATEEKLSTEEEYMNSVPYSSAVGSIMYA 1155
Query: 312 LCNSRLDISYGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELV 371
+ +R D++Y VG++S+FM P K H K +LRY++GT D + + R + +
Sbjct: 1156 MIGTRPDLAYPVGVVSRFMSQPAKEHWLGVKWVLRYIKGTVDTRLCYKR----NSDFSIC 1211
Query: 372 GYTDSDYSGDLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVW 431
GY D+DY+ DLD R+S +G +F L G ISW S Q V+A SS E EY++ + A +A+W
Sbjct: 1212 GYCDADYAADLDKRRSITGLVFTLGGNTISWKSGLQRVVAQSSTECEYMSLTEAVKEAIW 1271
Query: 432 LEELLKEMGFVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELV 491
L+ LLK+ G+ +K +E+ D+ SAI+L+KN + H R+KHI+V+FHF+RE + GK+E+
Sbjct: 1272 LKGLLKDFGY-EQKNVEIFCDSQSAIALSKNNVHHERTKHIDVKFHFIREIIADGKVEVS 1330
Query: 492 YCPTDEQKADIFTKPLKTERF 512
T++ ADIFTK L +F
Sbjct: 1331 KISTEKNPADIFTKVLPVNKF 1351
>At1g37110
Length = 1356
Score = 317 bits (812), Expect = 1e-86
Identities = 173/450 (38%), Positives = 273/450 (60%), Gaps = 16/450 (3%)
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
++ + G+DY E+FAPV + +IR+++S+ ++ + Q+DVK+ FL+G LEEE+Y+ QP
Sbjct: 912 YTQREGVDYQEIFAPVVKHVSIRILMSLVVDKDLELEQMDVKTTFLHGDLEEELYMEQPE 971
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTG 201
GF EN V +L K+LYGLKQ+PR WNKR D F+ +F R + VYV K +
Sbjct: 972 GFVSDSSENKVCRLKKSLYGLKQSPRQWNKRFDRFMSSQQFIRSEHDACVYV-KHVSEHD 1030
Query: 202 LLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAG--IFMY 259
+ + LYVDD+LI G+ E++ +K+ L +EFEM D+G LG+ G + +
Sbjct: 1031 FIYLLLYVDDMLIAGASKAEINRVKEQLSTEFEMKDMGGASRILGIDIYRDRKGGVLKLS 1090
Query: 260 QQKYIKDILKRFNMGNCHPVTTPSEVNIKL---RRDDE-GDVDASLYKQLVGSLRF-LCN 314
Q+ YI+ +L RFNM P + KL R +DE D D Y VGS+ + +
Sbjct: 1091 QEIYIRKVLDRFNMSGAKMTNAPVGAHFKLAAVREEDECVDTDVVPYSSAVGSIMYAMLG 1150
Query: 315 SRLDISYGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYT 374
+R D++Y + LIS++M P H A K ++RYL+G D ++F ++ K+ + GY
Sbjct: 1151 TRPDLAYAICLISRYMSKPGSMHWEAVKWVMRYLKGAQDLNLVFTKE----KDFTVTGYC 1206
Query: 375 DSDYSGDLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEE 434
DS+Y+ DLD R+S SGY+F + G +SW + QPV+A+S+ EAEYIA + AA +A+W++
Sbjct: 1207 DSNYAADLDRRRSISGYVFTIGGNTVSWKASLQPVVAMSTTEAEYIALAEAAKEAMWIKG 1266
Query: 435 LLKEMGFVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCP 494
LL++MG K +++ D+ SAI L+KN + H R+KHI+VRF+++R+ V G ++++
Sbjct: 1267 LLQDMGMQQDK-VKIWCDSQSAICLSKNSVYHERTKHIDVRFNYIRDVVESGDVDVLKIH 1325
Query: 495 TDEQKADIFTKPLKTERFVILRKEIGVVSL 524
T D TK + +F + +GV+ L
Sbjct: 1326 TSRNPVDALTKCIPVNKF---KSALGVLKL 1352
>At4g27210 putative protein
Length = 1318
Score = 317 bits (811), Expect = 1e-86
Identities = 176/441 (39%), Positives = 261/441 (58%), Gaps = 7/441 (1%)
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
F + G+DY E ++PV R T+RLV+ +A+A NW I Q+DVK+AFL+G L+E VY++QP
Sbjct: 783 FLQEEGIDYLETYSPVVRTPTVRLVLHLATALNWDIKQMDVKNAFLHGDLKETVYMTQPA 842
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTG 201
GF K + V L K++YGLKQ+PRAW + FLL+ F + +++ + +
Sbjct: 843 GFVDPSKPDHVCLLHKSIYGLKQSPRAWFDKFSTFLLEFGFFCSKSDPSLFIYAHNNN-- 900
Query: 202 LLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQ 261
L+L+ LYVDD++ITG+ + L L L EF M+D+G+L YFLG+Q + G+FM QQ
Sbjct: 901 LILLLLYVDDMVITGNSSQTLTSLLAALNKEFRMTDMGQLHYFLGIQVQRQQNGLFMSQQ 960
Query: 262 KYIKDILKRFNMGNCHPVTTPSEVNI-KLRRDDEGDVDASLYKQLVGSLRFLCNSRLDIS 320
KY +D+L +M +C P+ TP V + ++ +E D + ++ + G L++L +R DI
Sbjct: 961 KYAEDLLIAASMEHCTPLPTPLPVQLDRVPHQEELFSDPTYFRSIAGKLQYLTLTRPDIQ 1020
Query: 321 YGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSG 380
+ V + + M P S KRILRY++GT GI + R L Y+DSD+
Sbjct: 1021 FAVNFVCQKMHQPTISDFHLLKRILRYIKGTITMGISYSRDSPT----LLQAYSDSDWGN 1076
Query: 381 DLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMG 440
R+S G + +SW+SKK P ++ SS EAEY + S AA + +WL LL+E+
Sbjct: 1077 CKQTRRSVGGLCTFMGTNLVSWSSKKHPTVSRSSTEAEYKSLSDAASEILWLSTLLRELR 1136
Query: 441 FVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKA 500
EL DN+SA+ L NP H R+KH ++ FHF+RE+V L + + P EQ A
Sbjct: 1137 IPLPDTPELFCDNLSAVYLTANPAFHARTKHFDIDFHFVRERVALKALVVKHIPGSEQIA 1196
Query: 501 DIFTKPLKTERFVILRKEIGV 521
DIFTK L E F+ LR ++GV
Sbjct: 1197 DIFTKSLPYEAFIHLRGKLGV 1217
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.331 0.146 0.454
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,639,202
Number of Sequences: 26719
Number of extensions: 491889
Number of successful extensions: 1624
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 105
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 1209
Number of HSP's gapped (non-prelim): 154
length of query: 533
length of database: 11,318,596
effective HSP length: 104
effective length of query: 429
effective length of database: 8,539,820
effective search space: 3663582780
effective search space used: 3663582780
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0028b.2


