

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0026.5
(1710 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
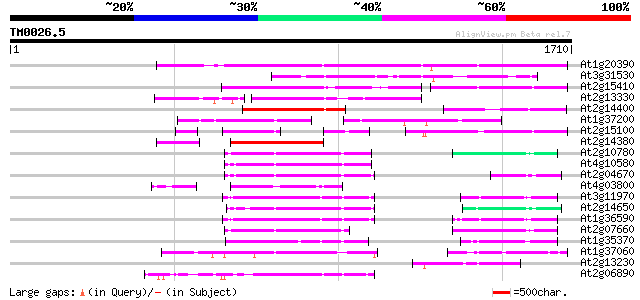
Score E
Sequences producing significant alignments: (bits) Value
At1g20390 hypothetical protein 748 0.0
At3g31530 hypothetical protein 437 e-122
At2g15410 putative retroelement pol polyprotein 370 e-102
At2g13330 F14O4.9 321 3e-87
At2g14400 putative retroelement pol polyprotein 279 9e-75
At1g37200 hypothetical protein 268 2e-71
At2g15100 putative retroelement pol polyprotein 253 5e-67
At2g14380 putative retroelement pol polyprotein 250 6e-66
At2g10780 pseudogene 234 3e-61
At4g10580 putative reverse-transcriptase -like protein 231 2e-60
At2g04670 putative retroelement pol polyprotein 230 6e-60
At4g03800 229 1e-59
At3g11970 hypothetical protein 219 1e-56
At2g14650 putative retroelement pol polyprotein 219 1e-56
At1g36590 hypothetical protein 219 1e-56
At2g07660 putative retroelement pol polyprotein 215 2e-55
At1g35370 hypothetical protein 204 4e-52
At1g37060 Athila retroelment ORF 1, putative 199 2e-50
At2g13230 pseudogene 189 2e-47
At2g06890 putative retroelement integrase 185 2e-46
>At1g20390 hypothetical protein
Length = 1791
Score = 748 bits (1931), Expect = 0.0
Identities = 453/1277 (35%), Positives = 680/1277 (52%), Gaps = 78/1277 (6%)
Query: 448 HLKPLFVWAKVDEKGVNKVLVDGGATINLMPKFMLKKLGKTEADLIPHDMVLSDYEGKTG 507
H PL V + + V +VL+D G++++L+ K +L + T+ + P L+ ++G
Sbjct: 567 HNDPLVVELIISDSRVTRVLIDTGSSVDLIFKDVLTAMNITDRQIKPVSKPLAGFDGDFV 626
Query: 508 SSLGAIMLNITVGTVARSTLFIVVPSKANYNLLLGREWIHGVGAVPSTLHQRIS------ 561
++G I L I VG + F+V+ A YN++LG WIH + A+PST HQ +
Sbjct: 627 MTIGTIKLPIFVGGLIAWVKFVVIGKPAVYNVILGTPWIHQMQAIPSTYHQCVKFPTHNG 686
Query: 562 IWKLDGVVENVQADQSYYLAETGYVGKKNFEKSLATIAPLDTVANQYFNPYSEYSVMLDP 621
I+ L E +SY +E N ++S DP
Sbjct: 687 IFTLRAPKEAKTPSRSYEESELCRTEMVNIDES-------------------------DP 721
Query: 622 IRGLNLNEAYKPDAMSGWHNDEEKDATTEWQNEMVKLLKEFKDCFAWDYDEMPGLSRDLV 681
R + + P + E++ LLK FAW ++M G+ +
Sbjct: 722 TRCVGVGAEISPSI----------------RLELIALLKRNSKTFAWSIEDMKGIDPAIT 765
Query: 682 ELKLPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLKCKFIRTARYVDWLANVVPVIKKN 741
+L + KPVKQ R+ P+ + EE+E+LLK I +Y +WLAN V V KKN
Sbjct: 766 AHELNVDPTFKPVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKN 825
Query: 742 GKMRVCIDFRDLNAATPKDEYHMPVAEMMVDSTAGHEYLSLLDGYSGYNQIFIAEEDVSK 801
GK RVC+D+ DLN A PKD Y +P + +V++T+G+ LS +D +SGYNQI + ++D K
Sbjct: 826 GKWRVCVDYTDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEK 885
Query: 802 TAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDD 861
T+F GTY + VM FGLKNAGATYQR +N + D I ++VYIDD++VKS +D
Sbjct: 886 TSFVTDR--GTYCYKVMSFGLKNAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPED 943
Query: 862 HLAHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDTSPPT 921
H+ HL K F+ + YG+K+NP KC FGV +G+FLG+VV K+GIE N + +AIL+ P
Sbjct: 944 HVEHLSKCFDVLNTYGMKLNPTKCTFGVTSGEFLGYVVTKRGIEANPKQIRAILELPSPR 1003
Query: 922 SKKQLQSLLGKINFLRRFIANLSEKTKSFSPLLRLKKEDAFRWEAEHQKAFDELKVYLSS 981
+ +++Q L G+I L RFI+ ++K F L LK+ F W+ + ++AF++LK YLS+
Sbjct: 1004 NAREVQRLTGRIAALNRFISRSTDKCLPFYNL--LKRRAQFDWDKDSEEAFEKLKDYLST 1061
Query: 982 PHVMAPPIRGKPMKLYISATDGTIGSMLAQEDEDSKERAIFYLSRVLNDAETRYTMIEKL 1041
P ++ P G+ + LYI+ +D + S+L +ED ++R IFY S+ L +AETRY +IEK
Sbjct: 1062 PPILVKPEVGETLYLYIAVSDHAVSSVLVREDR-GEQRPIFYTSKSLVEAETRYPVIEKA 1120
Query: 1042 CLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHSRIGKWALALTEYSLTYAPLK 1101
L + S KL+ Y + + V + ++ L P R+ KWA+ L+EY + + P
Sbjct: 1121 ALAVVTSARKLRPYFQSHTIAVLTDQP-LRVALHSPSQSGRMTKWAVELSEYDIDFRPRP 1179
Query: 1102 AVKGQAIADFLVDHTL--PKEIVTYVGIQPWKLFFDGSSHKNGTGIGMFIVSPGGIPTKF 1159
A+K Q +ADFL++ L + V+ + W L+ DGSS G+GIG+ +VSP +
Sbjct: 1180 AMKSQVLADFLIELPLQSAERAVSGNRGEEWSLYVDGSSSARGSGIGIRLVSPTAEVLEQ 1239
Query: 1160 KFRIKKNCSNNEAEYEALISGLEILIAFGAKNVVIKGDSELVIKQLTKEYKCVSENLARY 1219
FR++ +NN AEYE LI+GL + + DS+L+ QL+ EY+ +E + Y
Sbjct: 1240 SFRLRFVATNNVAEYEVLIAGLRLAAGMQITTIHAFTDSQLIAGQLSGEYEAKNEKMDAY 1299
Query: 1220 YTKANNLLAKFDEARLSHVSRVDNQEANELAQIASGYMVDKCRLKELIEVK--EKLNLSD 1277
+ F+ +LS + R DN A+ LA +A + L+ +I V+ +K ++
Sbjct: 1300 LKIVQLMTKDFENFKLSKIPRGDNAPADALAALA---LTSDSDLRRIIPVESIDKPSIDS 1356
Query: 1278 LNILVI----------DNMAPNDWRKPIVDYLQNPVGTTD----RKTKYRAMSYVIMGNE 1323
+ + I D P DWR I DYL + +D R+ + +A Y +M
Sbjct: 1357 TDAVEIVNTIRSSNAPDPADPTDWRVEIRDYLSDGTLPSDKWTARRLRIKAAKYTLMKEH 1416
Query: 1324 LFKKNVDGTLLKCLSEDDAFIAISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCME 1383
L K + G +L CL + + H+G G H G + L + G YWPT++ DC
Sbjct: 1417 LLKVSAFGAMLNCLHGTEINEIMKETHEGAAGNHSGGRALALKLKKLGFYWPTMISDCKT 1476
Query: 1384 YDKGCQDFQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYF 1443
+ C+ QRHA H P L + + P+PF WA+D++G + +SRQ ++I+V DYF
Sbjct: 1477 FTAKCEQCQRHAPTIHQPTELLRAGVAPYPFMRWAMDIVGPMP--ASRQKRFILVMTDYF 1534
Query: 1444 TKWVEAIPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKVASFAESWGIKLL 1503
TKWVEA + + V +F+ I+ R GLP + TD G+ F+ +F SW I+L
Sbjct: 1535 TKWVEAESYATIRANDVQNFVWKFIICRHGLPYEIITDNGSQFISLSFENFCASWKIRLN 1594
Query: 1504 NSTPYYAQANGQVEAANKTLISLIKKHVGRKPKRWHQTLGQVLWAYRNSPKEATGATPFR 1563
STP Y Q NGQ EA NKT++S +KK + K W L VLW+YR +P+ AT TPF
Sbjct: 1595 KSTPRYPQGNGQAEATNKTILSGLKKRLDEKKGAWADELDGVLWSYRTTPRSATDQTPFA 1654
Query: 1564 LAYGQEAVLPAEVYLQSCRIQRQEEIPSEDYWNMMLDELVNLDEERLSALDILTRQKDRV 1623
AYG EA+ PAEV S R + P E MMLD L +L+E R +AL + +
Sbjct: 1655 HAYGMEAMAPAEVGYSSLRRSMMVKNP-ELNDRMMLDRLDDLEEIRNAALCRIQNYQLAA 1713
Query: 1624 AKAYNKKVRAKSFMPGDYVWKVVLPVDKRDKRYGKWAPNWEGPFTVEKILLNNAYSIKEL 1683
AK YN+KV + F GD V + V + + GK NWEG + V KI+ Y + +
Sbjct: 1714 AKHYNQKVHNRHFDVGDLVLRKVFE-NTAEINAGKLGANWEGSYQVSKIVRPGDYELLTM 1772
Query: 1684 GGRNRQMTVNGKYLKTY 1700
G T N +LK Y
Sbjct: 1773 SGTAVPRTWNSMHLKRY 1789
Score = 42.4 bits (98), Expect = 0.002
Identities = 54/235 (22%), Positives = 88/235 (36%), Gaps = 44/235 (18%)
Query: 4 FPSSLTKNAFTWFTTLAPRSVHTWAQLERIFHEQFF-RGECKVSLKDLASVKRKPAESID 62
F LT A WF+ L P S+ ++ QL F + + E + S DL S+ + ES+
Sbjct: 212 FIEHLTGPAHNWFSRLKPNSIDSFHQLTSSFLKHYAPLIENQTSNADLWSISQGAKESLR 271
Query: 63 DYLNRFRMLKSRCFTHVPEHELVVLAAGGLEYSIRKKIDTQYIRDMSLLADRPRENKLGE 122
+++RF+++ + VP+ +V + Y R + D +L R ++ E
Sbjct: 272 SFVDRFKLVVTN--ITVPDEAAIVALRNAVWYDSRFRDDITLHAPSTLEDALHRASRFIE 329
Query: 123 ENPKSTIPAVKTYTFDVTKCDAIYDILVSDGLLVVPKDQKLP------------------ 164
+ I A K + TK A D +V + V P D P
Sbjct: 330 LEEEKLILARK---HNSTKTPACKDAVV---IKVGPDDSNEPRQHLDRNPSAGRKPTSFL 383
Query: 165 ----SPEQR-------------KGKKYCKYHNFFGHWTSQCVRFRDLVQGALDSG 202
+P+ + G YC+YH H T C + L+ SG
Sbjct: 384 VSTETPDAKPWNKYIRDADSPAAGPMYCEYHKSRAHSTENCRFLQGLLMAKYKSG 438
>At3g31530 hypothetical protein
Length = 831
Score = 437 bits (1124), Expect = e-122
Identities = 282/831 (33%), Positives = 421/831 (49%), Gaps = 118/831 (14%)
Query: 797 EDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKS 856
ED KTAF + Y VMPFGLKNAGATY+R +N IF I M+VYIDD++VKS
Sbjct: 99 EDQEKTAFYTEQGIFCYR--VMPFGLKNAGATYERFVNKIFALQIGKTMEVYIDDMLVKS 156
Query: 857 PSRDDHLAHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILD 916
+ DH++HLR+ F+++ Y +K+NP KC FGV + GIE N + +A+L
Sbjct: 157 MTEKDHISHLRECFKQLNLYNVKLNPAKCRFGV-----------RSGIEANPKQIEALLG 205
Query: 917 TSPPTSKKQLQSLLGKINFLRRFIANLSEKTKSFSPLLRLKKEDAFRWEAEHQKAFDELK 976
+ P +K+++Q L G++ L RFI+ +EK +F +LR K+ F W ++AF ELK
Sbjct: 206 MASPQNKREVQCLTGRVAALNRFISRSTEKCLAFYDVLRRNKK--FEWTTRCEEAFQELK 263
Query: 977 VYLSSPHVMAPPIRGKPMKLYISATDGTIGSMLAQEDEDSKERAIFYLSRVLNDAETRYT 1036
YL++P ++A P+ G+P LY++ +D + L +ED +++ IFY+S+ AE+RY
Sbjct: 264 KYLATPPILAKPVIGEPQYLYVAVSDTAVSGELVREDR-GEQKLIFYVSQTFTSAESRYP 322
Query: 1037 MIEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHSRIGKWALALTEYSLT 1096
+EKL L + S KL+ Y + ++V ++ +L P R+ KW + L+EY +
Sbjct: 323 QMEKLALAVVMSAQKLRPYFQSHSIIVMGSMP-LRVILHSPSQSGRLAKWTIELSEYDIE 381
Query: 1097 YAPLKAVKGQAIADFLVDHTLP-KEIVTYVGIQPWKLFFDGSSHKNGTGIGMFIVSPGGI 1155
Y K Q +ADF+V+ LP KE W L DGSS K G+G+G+ + SP G
Sbjct: 382 YQNKTCAKSQVLADFIVE--LPTKEARENPLDTTWLLHVDGSSSKQGSGVGIRLTSPTG- 438
Query: 1156 PTKFKFRIKKNCSNNEAEYEALISGLEILIAFGAKNVVIKG--DSELVIKQLTKEYKCVS 1213
+NN AEYEAL++GL +A+G K + DS+L+ Q EY
Sbjct: 439 ----------EATNNVAEYEALVAGLN--LAWGLKIGKTRAFCDSQLIANQFNGEYTTQD 486
Query: 1214 ENLARYYTKANNLLAKFDEARLSHVSRVDNQEANELAQIASGYMVDKCRLKELIEVK--E 1271
+ + Y NL FDE L+ + R +N A+ LA +AS LK +I V+ E
Sbjct: 487 KKMEAYLIHVQNLAKNFDEFELTRIPRGENTSADALAALAS---TSDTSLKRVIPVEFIE 543
Query: 1272 KLNL---SDLNILVIDNMAPND--------WRKPIVDYLQNPVGTTD----RKTKYRAMS 1316
K ++ + ++L I A D W +PI+ Y+ +D RK K +A
Sbjct: 544 KPSIELGKEEHVLPIQISADQDDPDDCNSEWMEPIISYISEGKLPSDKWKARKLKAQAAR 603
Query: 1317 YVIMGNELFKKNVDGTLLKCLSEDDAFIAISAVHDGLCGAHQAGIKMKWILFRQGMYWPT 1376
+V++ +L+K + G L+ C+ + + +H G
Sbjct: 604 FVLVDTKLYKWRLSGPLMTCVEAEAICKIMKEIHSG------------------------ 639
Query: 1377 IMKDCMEYDKGCQDFQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSRQHKYI 1436
HA H PA L SI P+PF W++D+IG ++P S+Q K +
Sbjct: 640 ----------------SHAPTIHQPAELLSSIASPYPFMRWSMDIIGPMHP--SKQKKLV 681
Query: 1437 IVAIDYFTKWVEAIPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKVASFAE 1496
+V DYF+KW+EA ++ V +F+ HI+ R G+P + TD G+ F+ + F +
Sbjct: 682 LVLTDYFSKWIEAESYASIKDAQVENFVLKHILCRHGIPYEIVTDNGSQFISTRFQGFCD 741
Query: 1497 SWGIKLLNSTPYYAQANGQVEAANKTLISLIKKHVGRKPKRWHQTLGQVLWAYRNSPKEA 1556
WGI+L STP Y Q NGQ EAAN+ L VLW +R +P+ A
Sbjct: 742 KWGIRLSKSTPRYPQGNGQAEAANE--------------------LEGVLWLHRTTPRRA 781
Query: 1557 TGATPFRLAYGQEAVLPAEVYLQSCRIQRQEEIPSEDYWNMMLDELVNLDE 1607
T TPF YG E V+PAE+ + S R E D LDEL +DE
Sbjct: 782 TRETPFASIYGTECVIPAEMIVPSLRRSLSPE-NDPDNTQTPLDELDLIDE 831
Score = 32.3 bits (72), Expect = 2.4
Identities = 22/83 (26%), Positives = 33/83 (39%), Gaps = 6/83 (7%)
Query: 509 SLGAIMLNITVGTVARSTLFIVVPSKANYNLLLGREWIHGVGAVPSTLHQRI------SI 562
+LG I L + V + F + YN ++G W++ AV ST H + S+
Sbjct: 2 TLGTIKLPVRAKGVTKIVDFSITDQPTVYNAIIGTPWLNQFRAVASTYHLCLKFPTSDSV 61
Query: 563 WKLDGVVENVQADQSYYLAETGY 585
W+ G L ET Y
Sbjct: 62 WEKLGHERAEAVKAENQLLETSY 84
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 370 bits (949), Expect = e-102
Identities = 218/609 (35%), Positives = 326/609 (52%), Gaps = 64/609 (10%)
Query: 646 DATTEWQNEMVKLLKEFKDCFAWDYDEMPGLSRDLVELKLPIKEDKKPVKQLPRRFHPDV 705
D +E QNE+V L++ FAW ++MPG+ + +L + KP+KQ R+ PD
Sbjct: 735 DLPSELQNELVNFLRQNAATFAWSVEDMPGIDSAVTCHELNVDPTYKPLKQKRRKLGPDR 794
Query: 706 LVKIKEEIERLLKCKFIRTARYVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMP 765
+ EE+++LL I RY DWL N V V KKNGK RVCIDF DLN A PKD + +P
Sbjct: 795 TKDVNEEVKKLLDAGSIVEVRYPDWLRNPVVVKKKNGKWRVCIDFTDLNKACPKDSFPLP 854
Query: 766 VAEMMVDSTAGHEYLSLLDGYSGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNA 825
+ +V++TAG+E LS +D +SGYNQI + + D KT F GTY + VMPFGLKNA
Sbjct: 855 HIDRLVEATAGNELLSFMDAFSGYNQILMHQNDREKTVFITDQ--GTYCYKVMPFGLKNA 912
Query: 826 GATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDHLAHLRKSFERMRKYGLKMNPLKC 885
GATY R++N +F D ++ M+VYIDD++VKS ++H+ HLR+ F+ + +Y +K+NP KC
Sbjct: 913 GATYPRLVNQMFTDQLDHSMEVYIDDMLVKSLRAEEHITHLRQCFQVLNRYNMKLNPSKC 972
Query: 886 AFGVIAGDFLGFVVHKKGIEINKNKAKAILDTSPPTSKKQLQSLLGKINFLRRFIANLSE 945
FGV +G+FLG++V ++GIE N + AI+D P + +++Q L+G+I L RFI+ ++
Sbjct: 973 TFGVTSGEFLGYLVTRRGIEANPKQISAIIDLPSPRNTREVQRLIGRIAALNRFISRSTD 1032
Query: 946 KTKSFSPLLRLKKEDAFRWEAEHQKAFDELKVYLSSPHVMAPPIRGKPMKLYISATDGTI 1005
K F LLR K F W+ + ++ G+ + LYI+ + +
Sbjct: 1033 KCLPFYQLLRANKR--FEWDEKCEE--------------------GETLYLYIAVSTSAV 1070
Query: 1006 GSMLAQEDEDSKERAIFYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFS 1065
+L +ED ++ IFY+S+ L+ AE RY +EKL + S KL+ Y K V V +
Sbjct: 1071 SGVLVREDR-GEQHPIFYVSKTLDGAELRYPTLEKLAFAVVISARKLRPYFKSYTVEVLT 1129
Query: 1066 HYDIIKHMLSKPILHSRIGKWALALTEYSLTYAPLKAVKGQAIADFLVDHTLPKEIVTYV 1125
+ ++ +L P R+ KWA+ L+EY + Y K H P
Sbjct: 1130 NQP-LRTILHSPSQSGRLAKWAVELSEYDIAYKNRTCAK---------SHRAP------- 1172
Query: 1126 GIQPWKLFFDGSSHKNGTGIGMFIVSPGGIPTKFKFRIKKNCSNNEAEYEALISGLEILI 1185
+P++ IP + + SNNEAEYEALI+GL +
Sbjct: 1173 -TRPYR---------------------RSIPCRKVDLARFPASNNEAEYEALIAGLRLAH 1210
Query: 1186 AFGAKNVVIKGDSELVIKQLTKEYKCVSENLARYYTKANNLLAKFDEARLSHVSRVDNQE 1245
K + DS+LV Q + Y+ +E + Y L F+ L+ + R DN
Sbjct: 1211 GIEVKKIQAYCDSQLVASQFSGNYEAKNERMDAYLKVVRELSYNFEVFELTKIPRSDNAP 1270
Query: 1246 ANELAQIAS 1254
A+ LA +AS
Sbjct: 1271 ADALAVLAS 1279
Score = 262 bits (669), Expect = 1e-69
Identities = 144/421 (34%), Positives = 217/421 (51%), Gaps = 8/421 (1%)
Query: 1284 DNMAPNDWRKPIVDYLQNPVGTTD----RKTKYRAMSYVIMGNELFKKNVDGTLLKCLSE 1339
D+ A +W + I Y+ + + D R+ + R+ Y + L + + G LL CL +
Sbjct: 1369 DSPASTNWIEEIRSYIADRIVPKDKWAARRLRARSAQYTLPHEHLLRWSATGVLLSCLDD 1428
Query: 1340 DDAFIAISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYDKGCQDFQRHAGIQH 1399
++A + +H+G G H G + + + G YWPT+ DC ++ C+ Q HA H
Sbjct: 1429 EEAQQVMREIHEGAGGNHSGGRALALKIRKHGQYWPTMNADCEKFVARCEKCQGHAPFIH 1488
Query: 1400 VPASELHSIIKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVEAIPLQNVTQDT 1459
+P+ L + + P+PF WA+D+I +SRQ KYI+V DYFTKW EA +
Sbjct: 1489 IPSEVLQTAMPPYPFMRWAMDIIRPFP--ASRQKKYILVMTDYFTKWFEAESYARIQSKE 1546
Query: 1460 VIDFIQNHIVYRFGLPESLTTDQGTVFVGQKVASFAESWGIKLLNSTPYYAQANGQVEAA 1519
V +F+ +I+ GLP + TD GT F + F W I+L STP Y Q NGQ EA
Sbjct: 1547 VQNFVWKNIICHHGLPYEIVTDNGTQFTSLQFEGFCAKWKIRLSKSTPRYPQCNGQAEAT 1606
Query: 1520 NKTLISLIKKHVGRKPKRWHQTLGQVLWAYRNSPKEATGATPFRLAYGQEAVLPAEVYLQ 1579
NKT++ +KK + K W L VLW+YR +P+ +T TPF L YG EA+ P E L
Sbjct: 1607 NKTILDGLKKRLDEKKGAWADELDGVLWSYRTTPRRSTDRTPFSLTYGMEALAPCEAGLP 1666
Query: 1580 SCRIQRQEEIPSEDYWNMMLDELVNLDEERLSALDILTRQKDRVAKAYNKKVRAKSFMPG 1639
+ R P+ + M+LD L L+E+R AL + + AK YNKKV+ + F G
Sbjct: 1667 TVRRSMLINDPTLND-QMLLDNLDTLEEQRDQALLRIQNYQQAAAKFYNKKVKNRLFAEG 1725
Query: 1640 DYVWKVVLPVDKRDKRYGKWAPNWEGPFTVEKILLNNAYSIKELGGRNRQMTVNGKYLKT 1699
D V + V + ++ GK +WEGP+ + K++ Y + + G + N LK
Sbjct: 1726 DLVLRKVFE-NTVEQDAGKLGAHWEGPYLISKVVKPGVYELLTMDGTPIPRSWNSANLKR 1784
Query: 1700 Y 1700
Y
Sbjct: 1785 Y 1785
>At2g13330 F14O4.9
Length = 889
Score = 321 bits (822), Expect = 3e-87
Identities = 188/519 (36%), Positives = 286/519 (54%), Gaps = 41/519 (7%)
Query: 738 IKKNGKMRVCIDFRDLNAATPKDEYHMPVAEMMVDSTAGHEYLSLLDGYSGYNQIFIAEE 797
++KNGK RVCIDFRDLN A PKD + +P + +V++T HE LS +D + GYNQI + +
Sbjct: 288 LEKNGKWRVCIDFRDLNKACPKDSFPLPHIDRLVEATVEHEKLSFMDAFYGYNQILMRRD 347
Query: 798 DVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSP 857
KTAF GTY + VMPFGLKNAG TYQR++N +F D + ++VYIDD++VKS
Sbjct: 348 GQEKTAFITDR--GTYYYKVMPFGLKNAGTTYQRLVNRMFVDQLGKTIEVYIDDMLVKSA 405
Query: 858 SRDDHLAHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDT 917
DH+ LR+ F+ + K+ +K+NP KC+F V + +FL ++V ++GIE N + A ++
Sbjct: 406 HEKDHVPQLRECFKILIKFEMKLNPEKCSFEVQSREFLEYLVTERGIEANPKQIAAFIEM 465
Query: 918 SPPTSKKQLQSLLGKINFLRRFIANLSEKTKSFSPLLRLKKEDAFRWEAEHQKAFDELKV 977
P +++Q L G+I L FI+ + K F LR KE F W + ++AF +LK
Sbjct: 466 PSPKMAREVQRLTGRIPALNGFISRSANKCVPFYQPLRKGKE--FDWNKDCEQAFKQLKA 523
Query: 978 YLSSPHVMAPPIRGKPMKLYISATDGTIGSMLAQEDEDSKERAIFYLSRVLNDAETRYTM 1037
YL+ P ++A P +G+P+ LY + + R
Sbjct: 524 YLTEPPILAKPEKGEPLYLY-------------------------------TNRDKRCPA 552
Query: 1038 IEKLCLCLYFSCVKLKYYIK--PIDVMVFSHYDIIKHMLSKPILHSRIGKWALALTEYSL 1095
+EKL L + S KL+ Y + PI VM I H L++ R+ KWA+ L EY +
Sbjct: 553 MEKLALTVVMSVRKLRLYFQSHPIVVMTSQPIRTILHSLTQS---GRLAKWAIELREYDI 609
Query: 1096 TYAPLKAVKGQAIADFLVDHTLPKEIVTYVGIQPWKLFFDGSSHKNGTGIGMFIVSPGGI 1155
Y ++K Q +ADF++ L T + W L DGSS++ G+G+G+ + SP G
Sbjct: 610 EYRTRTSLKAQVLADFVIKLPLADLDGTNSN-KKWLLHVDGSSNRQGSGVGIQLTSPTGE 668
Query: 1156 PTKFKFRIKKNCSNNEAEYEALISGLEILIAFGAKNVVIKGDSELVIKQLTKEYKCVSEN 1215
+ ++ N SNNE+EYEALI+G+++ G + + DS+LV Q EY+ E
Sbjct: 669 VIEQSLQLGFNASNNESEYEALIAGIKLAQEKGIREIHAYSDSQLVTSQFHGEYEAKDER 728
Query: 1216 LARYYTKANNLLAKFDEARLSHVSRVDNQEANELAQIAS 1254
+ Y L +F+ +L+ + R +N A+ LA +AS
Sbjct: 729 MEAYLELVKTLAQQFESFKLTRIPRGENTSADTLAALAS 767
Score = 75.1 bits (183), Expect = 3e-13
Identities = 75/296 (25%), Positives = 123/296 (41%), Gaps = 41/296 (13%)
Query: 442 TEQMKSHLKPLFVWAKVDEKGVNKVLVDGGATINLMPKFMLKKLGKTEADLIPHDMVLSD 501
T+ K H L + V ++ ++VD G++++++ K+ G ++ L L+
Sbjct: 55 TDLDKPHDDALVIRIDVGNYELSCIMVDTGSSVDVLFYDAFKRTGHLDSKLQGRKTPLTG 114
Query: 502 YEGKTGSSLGAIMLNITVGTVARSTLFIVVPSKANYNLLLGREWIHGVGAVPSTLHQRIS 561
+ G T S+G I L V + T F+VV KA +N +LGR W+H + AVPST HQ I
Sbjct: 115 FAGDTTFSIGTIQLPTIARGVRQLTNFLVVDKKAPFNAILGRPWLHVMKAVPSTYHQCIK 174
Query: 562 IWKLDGVVENVQADQSYYLAETGYVGKKNFEKSLATIAPLDTVANQYFNPYSEYSVM--L 619
G+ + +S +K + S I D V + +E + L
Sbjct: 175 FPSYKGIAVVYGSQRS---------SRKCYMGSYEDIKKADPVVLMIEDELAEMKTVRSL 225
Query: 620 DP---------IRGLNLNEAYKPDAMSGWHNDEEKDATTEWQNEMVKLLKEFKDCFAWDY 670
DP I + ++E+ + H D + +++ LKE KD FAW
Sbjct: 226 DPSQRGTRKSLITQVCIDESDPKRCVGIGH-----DLDLTVREDLITFLKENKDSFAWSS 280
Query: 671 DEMPGLSR----------DLVELKLPIKEDKKPVKQLPRRFHPDVLVKIKEEIERL 716
+ G+S D +L +D P+ H D LV+ E E+L
Sbjct: 281 ANLQGISLEKNGKWRVCIDFRDLNKACPKDSFPLP------HIDRLVEATVEHEKL 330
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 279 bits (714), Expect = 9e-75
Identities = 140/316 (44%), Positives = 211/316 (66%), Gaps = 5/316 (1%)
Query: 709 IKEEIERLLKCKFIRTARYVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPVAE 768
+ +E+++LLK IR +Y DWLAN V V KKNGK RVCIDF DLN A PKD + +P +
Sbjct: 480 VNDEVDKLLKIGSIREVQYPDWLANTVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHID 539
Query: 769 MMVDSTAGHEYLSLLDGYSGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGAT 828
+V+STAG+E LS +D +SGYNQI + ED KT F G Y + VMPFGL+NAGAT
Sbjct: 540 RLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDR--GIYCYKVMPFGLRNAGAT 597
Query: 829 YQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDHLAHLRKSFERMRKYGLKMNPLKCAFG 888
Y R++N +F + + M+VYIDD+++KS ++DH+ HL + F + +Y +K+NP KC FG
Sbjct: 598 YPRLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFAILNQYQMKLNPAKCTFG 657
Query: 889 VIAGDFLGFVVHKKGIEINKNKAKAILDTSPPTSKKQLQSLLGKINFLRRFIANLSEKTK 948
V +G+FLG++V K+GIE N N+ A L+ P + K++Q L G+I L RFI+ ++K+
Sbjct: 658 VPSGEFLGYIVTKRGIEANPNQINAFLNMPSPKNFKEVQRLTGRIAALNRFISRSTDKSL 717
Query: 949 SFSPLLRLKKEDAFRWEAEHQKAFDELKVYLSSPHVMAPPIRGKPMKLYISATDGTIGSM 1008
F +L+ KE F W+ + ++AF +LK YL+SP V+ P + + LY+S ++ + +
Sbjct: 718 PFYQILKGNKE--FLWDEKCEEAFGQLKAYLTSPPVLFKPELDEKLYLYVSVSNHAVSGV 775
Query: 1009 LAQEDEDSKERAIFYL 1024
L +ED +++ I+Y+
Sbjct: 776 LIREDR-GEQKPIYYI 790
Score = 225 bits (573), Expect = 2e-58
Identities = 131/379 (34%), Positives = 199/379 (51%), Gaps = 51/379 (13%)
Query: 1322 NELFKKNVDGTLLKCLSEDDAFIAISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDC 1381
N L+++ V L + I +S VH+GLCG+H +G M + + + YWPT++ DC
Sbjct: 951 NSLYRRGVSDPYLLSIFGPVVEIVMSEVHEGLCGSHSSGRAMAFKIKKMCYYWPTMITDC 1010
Query: 1382 MEYDKGCQDFQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSRQHKYIIVAID 1441
++Y + C+ Q HA + H P+ SI P+PF W++D+IG ++ S+R +Y++V D
Sbjct: 1011 VKYAQRCKRCQLHAPLIHQPSELFSSISAPYPFMRWSMDIIGPLHR-STRGVQYLLVLTD 1069
Query: 1442 YFTKWVEAIPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKVASFAESWGIK 1501
YF+KW+EA + +WGIK
Sbjct: 1070 YFSKWIEA------------------------------------------EAILLNWGIK 1087
Query: 1502 LLNSTPYYAQANGQVEAANKTLISLIKKHVGRKPKRWHQTLGQVLWAYRNSPKEATGATP 1561
+ ST Y Q NGQ EAANKT++S +KK + W+ L VLWAYR +P+ +TG TP
Sbjct: 1088 VSYSTLRYPQGNGQAEAANKTILSNLKKRLNHLKGGWYDELQPVLWAYRTTPRRSTGETP 1147
Query: 1562 FRLAYGQEAVLPAEVYLQSCRIQRQEEIPSEDYWN--MMLDELVNLDEERLSALDILTRQ 1619
F L YG +AV+PAE+ + R+ E P + N M+ D L ++E R AL +
Sbjct: 1148 FSLVYGMKAVVPAELNVPGL---RRTEAPLNEKENSAMLDDSLDTINERRDQALIQIQNY 1204
Query: 1620 KDRVAKAYNKKVRAKSFMPGDYVWKVVLPVDKRDKRYGKWAPNWEGPFTVEKILLNNAYS 1679
+ A+ YN KV+++ F GDYV K V +K+++ GK NWEGP+ V +++ N Y
Sbjct: 1205 QHAAARYYNSKVKSRPFFVGDYVLKRVFD-NKKEEGAGKLGINWEGPYIVTEVVRNGVYR 1263
Query: 1680 IKELGGR--NRQMTVNGKY 1696
+K+L R R VNG +
Sbjct: 1264 LKDLEDRPVQRPWNVNGHH 1282
>At1g37200 hypothetical protein
Length = 1564
Score = 268 bits (685), Expect = 2e-71
Identities = 158/414 (38%), Positives = 233/414 (56%), Gaps = 16/414 (3%)
Query: 512 AIMLNITVGTVARSTLFIVVPS---KANYNLLLGREWIHGVGAVPSTLHQRISIWKLDGV 568
AI++ I VG S + I S KA +N +LGR W+H + AVPST HQ I G+
Sbjct: 504 AIVIRIDVGNYKLSRIMIDTGSSVDKAPFNAILGRPWLHAMKAVPSTYHQCIKFPSEKGI 563
Query: 569 VENVQADQSYYLAETGYVGKKNFEKSLATIAPL--DTVANQYFNPYSEYSVMLDPIRGLN 626
+ +S + Y+G K + + D +A S+ S P + +
Sbjct: 564 AVIYGSQRS---SRRCYMGSYELIKKAELVVLMIEDKLAEMKTVRSSDPS-QCGPQKS-S 618
Query: 627 LNEAYKPDAMSGWHNDEEKDATTEWQNEMVKLLKEFKDCFAWDYDEMPGLSRDLVELKLP 686
+ + Y ++ +D + + + LKE KD FAW + G+S ++ +L
Sbjct: 619 ITQVYIDESDPKQCVGVGQDLDPAIREDFITFLKENKDSFAWSSANLQGISLEVTSHELN 678
Query: 687 IKEDKKPVKQLPRRFHPDVLVKIKEEIERLLKCKFIRTARYVDWLANVVPVIKKNGKMRV 746
+ +P+KQ R+ P+ +++E++RLLK IR +Y DWLAN V V KNGK RV
Sbjct: 679 VDPTYRPIKQKRRKLGPERAKAVQDEVDRLLKIGSIREVKYPDWLANPVVVKNKNGKWRV 738
Query: 747 CIDFRDLNAATPKDEYHMPVAEMMVDSTAGHEYLSLLDGYSGYNQIFIAEEDVSKTAFRC 806
CIDF DLN A PKD + +P + +V +TAGHE LS +D +SGYNQI + +D KTAF
Sbjct: 739 CIDFMDLNKACPKDSFPLPHIDRLVKATAGHELLSFMDAFSGYNQILMRPDDQEKTAFI- 797
Query: 807 PGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDHLAHL 866
T + VMPFGLKN GATYQR++N +F D + M++YIDD++VKS DHL L
Sbjct: 798 -----TDCYKVMPFGLKNTGATYQRLVNRMFADQLGKTMELYIDDMLVKSAHEKDHLPQL 852
Query: 867 RKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDTSPP 920
R+ F+ + K+ +K+NP KC+FGV +G+FLG++V ++GIE N + A +D P
Sbjct: 853 RECFKILNKFEMKLNPEKCSFGVPSGEFLGYLVTQRGIEANPKQIAAFIDMPSP 906
Score = 218 bits (554), Expect = 3e-56
Identities = 152/531 (28%), Positives = 244/531 (45%), Gaps = 63/531 (11%)
Query: 1018 ERAIFYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKP 1077
++ I+Y+S+ L DAET Y +EKL L + S KL+ Y++ ++V + I + +L
Sbjct: 931 QKPIYYVSKTLIDAETHYRAMEKLALAVVMSARKLRQYLQSHPIVVMTSQPI-RTILHST 989
Query: 1078 ILHSRIGKWALALTEYSLTYAPLKAVKGQAIADFLVDHTLPKEIVTYVGIQPWKLFFDGS 1137
SR+ KWA+ L+EY + Y + K Q +ADF+++ L T + W L DGS
Sbjct: 990 TQSSRLAKWAIELSEYDIEYRTRTSFKAQVLADFVIELPLADLDGTNSN-KKWLLHVDGS 1048
Query: 1138 SHKNGTGIGMFIVSPGGIPTKFKFRIKKNCSNNEAEYEALISGLEILIAFGAKNVVIKGD 1197
S++ G+G G+ + SP G + FR+ N SNNE+EYEALI G+++ +++ D
Sbjct: 1049 SNRQGSGEGIQLTSPTGEVIEQSFRLGFNASNNESEYEALIDGIKLAQGMRIRDIHAHSD 1108
Query: 1198 SEL---------------------VIKQLTKEYKCVS-ENLARYYTKANNLLAKFDEARL 1235
S+L ++K LT++++ + R + + LA
Sbjct: 1109 SQLVTSQFHGEYEAKDERMEAYLELVKTLTQQFESFELTRIPRGENTSTDALAALASTLD 1168
Query: 1236 SHVSRVDNQEANELAQI------ASGYMVDKCRLKELIE-----------------VKEK 1272
V R+ E E I A C + +E+
Sbjct: 1169 PFVKRIIPVEGIEHPSIDLTVKHAGMETSATCNFTRVTRSTTAAARALAAAAEAGAAEEE 1228
Query: 1273 LNLSDLNILVIDNMAP-NDWRKPIVDYLQNPVGTTD----RKTKYRAMSYVIMGNELFKK 1327
L L P NDWR PI+DY++ + D RK K ++ Y IM L K+
Sbjct: 1229 AKLRTPEQLASYEPEPYNDWRIPIIDYIERGITPPDKWETRKLKAQSARYCIMEGRLMKR 1288
Query: 1328 NVDGTLLKCLSEDDAFIAISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYDKG 1387
+V G + C + ++H+G CG+H +G ++ I+ + DC+ +
Sbjct: 1289 SVAGPYMVCTYGQQTKDLMKSMHEGQCGSHYSGRTLRCIM----------LADCIAHSLR 1338
Query: 1388 CQDFQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSRQH-KYIIVAIDYFTKW 1446
C QRHA H P E+ SI P+PF W++D++ + ++ K ++V DY TKW
Sbjct: 1339 CDKCQRHAPTLHQPPEEMSSISSPYPFMKWSMDVVRPMEASGGKKRLKNLLVLTDYSTKW 1398
Query: 1447 VEAIPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKVASFAES 1497
+E Q VT+ V F+ +IVYR G+P + TD G +K FA S
Sbjct: 1399 IEGKAFQQVTEKQVEVFLLENIVYRHGIPYEIVTDNGLTSPREKSKHFATS 1449
Score = 43.5 bits (101), Expect = 0.001
Identities = 25/97 (25%), Positives = 45/97 (45%), Gaps = 2/97 (2%)
Query: 1605 LDEERLSALDILTRQKDRVAKAYNKKVRAKSFMPGDYVWKVVLPVDKRDKRYGKWAPNWE 1664
+DE R A+ + + + YN ++ + F G+ V + V + R+ GK NWE
Sbjct: 1467 IDERRDHAMIQVQNYQQPAVRYYNSNIKIRRFEVGELVLRKVFS-NTRELNAGKLGTNWE 1525
Query: 1665 GPFTVEKILLNNAYS-IKELGGRNRQMTVNGKYLKTY 1700
GP+ + +++ + Y +K G N +LK Y
Sbjct: 1526 GPYRITEVVRDGIYKLVKVFNGVPELRPWNAMHLKKY 1562
Score = 38.9 bits (89), Expect = 0.026
Identities = 48/208 (23%), Positives = 84/208 (40%), Gaps = 26/208 (12%)
Query: 42 ECKVSLKDLASVKRKPAESIDDYLNRFRMLKSRCFTHVPEHELVVLAAGGL--------E 93
E + S DL S+ ES Y+ +F+ +KS+ ++ E + GL E
Sbjct: 185 EQRTSEADLWSLTLLQNESHRSYVEKFKAIKSK-IANLNEEVAIAALRNGLWFSSRFREE 243
Query: 94 YSIRKKIDTQYIRDMSLLADRPREN------KLGEENPKSTIPAVKTYTFDVTKCDAIYD 147
++R+ I +L + E K E ++ A+KT + +
Sbjct: 244 LTVRQPISLDDALHKALHFAKGEEELAVLALKFKESKTQNAPLAIKTPFKKENQTQGQHT 303
Query: 148 ILVSDGLLVVPKDQKLPSPEQRKGKKYCKYHNFFGHWTSQCVRFRDLVQGALDSGRLKRL 207
L + + + SPE GK YCKYHN GH T +C + L+ G+ K+
Sbjct: 304 ------LFAIEEAAEDESPELDLGK-YCKYHNKRGHSTEECRAVKKLIAA---GGKTKKG 353
Query: 208 KYPHEEWVERDQELDGHKGQKLRSKDKT 235
P E D++ + + K + +++T
Sbjct: 354 SNPKVETPPPDEQ-EEEQTPKQKKRERT 380
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 253 bits (647), Expect = 5e-67
Identities = 162/550 (29%), Positives = 253/550 (45%), Gaps = 74/550 (13%)
Query: 1208 EYKCVSENLARYYTKANNLLAKFDEARLSHVSRVDNQEANELAQIASGYMVD-------- 1259
EY+ + Y + +F++ L+ + R +N AN LA +AS + V
Sbjct: 795 EYEAKDACMEAYLNLVREVSGRFEQFELTRIPRAENSAANALAALASTFEVTLPRVIPVE 854
Query: 1260 -----KCRLKEL------------------------------------IEVKEKLNLSDL 1278
RL E+ E+ E +L D
Sbjct: 855 TISQPSIRLDEISFVTTRAMRRRLDAQSAENGLHQLGDDEEISDAVHPTEIVENQSLPDN 914
Query: 1279 NILVIDNMAPNDW----RKPIVDYLQNPVGTTD----RKTKYRAMSYVIMGNELFKKNVD 1330
+ + + P+DW R+PI DY+ N + RK K + I + L+++
Sbjct: 915 HNAPLPDQPPHDWGADWREPIRDYILNGTLPAEKWAARKLKATCARFCIANDILYRRIFS 974
Query: 1331 GTLLKCLSEDDAFIAISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYDKGCQD 1390
C+ + + VHDG CG H G + + + + G YWPT++ DC Y + C+
Sbjct: 975 APDAVCIFGEQTRTVMKEVHDGTCGNHTGGRSLAFKVRKYGYYWPTLVADCEAYARKCEQ 1034
Query: 1391 FQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVEAI 1450
Q+HA + PA L ++ P+PF W +D++G ++ T+ VEA
Sbjct: 1035 CQKHAPLILQPAELLTTVSAPYPFMKWLMDIVGPLHVS---------------TRGVEAA 1079
Query: 1451 PLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKVASFAESWGIKLLNSTPYYA 1510
N+T V +FI I+ R GLP + TD G+ F+ ++ F E W I+L +STP Y
Sbjct: 1080 AYSNITHVQVWNFIWKDIICRHGLPYEIVTDNGSQFISEQFEVFCEEWQIRLSHSTPRYP 1139
Query: 1511 QANGQVEAANKTLISLIKKHVGRKPKRWHQTLGQVLWAYRNSPKEATGATPFRLAYGQEA 1570
Q NGQ EA NKT+IS +KK + W L VLWA R +P+ AT TPF L YG EA
Sbjct: 1140 QGNGQAEAMNKTIISNLKKKLNAYKGAWFGELQNVLWAVRTTPRRATDETPFSLIYGMEA 1199
Query: 1571 VLPAEVYLQSCRIQRQEEIPSEDYWNMMLDELVNLDEERLSALDILTRQKDRVAKAYNKK 1630
V+PAE+ + S R R + +E+ M++D + +DE R AL + + A+ YN
Sbjct: 1200 VIPAEIKVPSARRIRNPQNETENN-EMIIDVIDTIDERRNRALARMQNYHNAGARYYNSN 1258
Query: 1631 VRAKSFMPGDYVWKVVLPVDKRDKRYGKWAPNWEGPFTVEKILLNNAYSIKELGGRNRQM 1690
VR +SF G V + V +K +K GK +WEGP+ + ++ N Y + + G+ +
Sbjct: 1259 VRNRSFEVGTLVLRRV-QQNKAEKGAGKLGISWEGPYKITHVVRNGVYRLINMEGKTVRR 1317
Query: 1691 TVNGKYLKTY 1700
N +LK +
Sbjct: 1318 AWNSMHLKRF 1327
Score = 145 bits (365), Expect = 3e-34
Identities = 80/179 (44%), Positives = 106/179 (58%), Gaps = 4/179 (2%)
Query: 649 TEWQNEMVKLLKEFKDC--FAWDYDEMPGLSRDLVELKLPIKEDKKPVKQLPRRFHPDVL 706
T W EM + + C F +M G+S +++ +L + KPVKQ R+ PD
Sbjct: 386 TPWLYEMKVVPSTYHQCVKFPTPVGKMTGISTEVISHELNVDPTFKPVKQKRRKLGPDRA 445
Query: 707 VKIKEEIERLLKCKFIRTARYVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPV 766
+ E+ RLL+ IR +Y +WLAN V V KKNGK RVC+DF DLN A KD + +P
Sbjct: 446 QAVNIEVVRLLEVGRIREVKYPEWLANPVVVKKKNGKWRVCVDFTDLNKACSKDFFPLPH 505
Query: 767 AEMMVDSTAGHEYLSLLDGYSGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNA 825
+ +V+ST GHE LS +D +SGYNQI + ED KT+F GTY + VMPFGLKNA
Sbjct: 506 IDRLVESTTGHEMLSFMDAFSGYNQILMNPEDQEKTSFIT--ECGTYYYKVMPFGLKNA 562
Score = 73.6 bits (179), Expect = 1e-12
Identities = 45/141 (31%), Positives = 76/141 (52%), Gaps = 9/141 (6%)
Query: 957 KKEDAFRWEAEHQKAFDELKVYLSSPHVMAPPIRGKPMKLYISATDGTIGSMLAQEDEDS 1016
K F W ++AF +LK YLS P V+A G+ + LYI+ ++ + + + E S
Sbjct: 624 KISKGFIWNESCEEAFKQLKRYLSEPSVLAKREFGEQLFLYIAVSESAVTGVQVRV-ERS 682
Query: 1017 KERAIFYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSK 1076
+R IFY+ +TRY M+EKL L + + KL+ Y + ++V + ++ +L
Sbjct: 683 DKRPIFYV-------KTRYPMMEKLALAVVTAARKLRPYFQSHPIVVLTSLP-LRTILHS 734
Query: 1077 PILHSRIGKWALALTEYSLTY 1097
P R+GKWA+ L+E+ L +
Sbjct: 735 PTQSGRLGKWAIELSEFDLEF 755
Score = 46.2 bits (108), Expect = 2e-04
Identities = 24/72 (33%), Positives = 38/72 (52%), Gaps = 4/72 (5%)
Query: 505 KTGSSLGAIMLNITVGTVARSTLFIVVPSKANYNLLLGREWIHGVGAVPSTLHQRIS--- 561
+T SLG ++L +T V + F V A YN++LG W++ + VPST HQ +
Sbjct: 348 ETSMSLGTVVLPVTAQGVVKMVEFTVFDRPAAYNVILGTPWLYEMKVVPSTYHQCVKFPT 407
Query: 562 -IWKLDGVVENV 572
+ K+ G+ V
Sbjct: 408 PVGKMTGISTEV 419
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 250 bits (638), Expect = 6e-66
Identities = 130/284 (45%), Positives = 184/284 (64%), Gaps = 2/284 (0%)
Query: 672 EMPGLSRDLVELKLPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLKCKFIRTARYVDWL 731
+M G+ ++ +L + K VKQ R+ P+ + +E+++LL I +Y +WL
Sbjct: 476 DMVGIDPEVACHELNVDPTFKLVKQKRRKLGPERSKAVNDEVDKLLDAGSIVEVKYPEWL 535
Query: 732 ANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPVAEMMVDSTAGHEYLSLLDGYSGYNQ 791
AN V V KKN K RVCIDF DLN A PKD + +P + MV++T G+E LS +D +SGYNQ
Sbjct: 536 ANPVVVKKKNDKWRVCIDFTDLNKACPKDSFPLPHIDRMVEATTGNELLSFMDAFSGYNQ 595
Query: 792 IFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDD 851
I + ++D KT+F GTY + VMPFGLKN GA YQR++N +F + M+VYIDD
Sbjct: 596 IPMHKDDQEKTSFIIDR--GTYCYKVMPFGLKNVGARYQRLVNQMFAPQLGKTMEVYIDD 653
Query: 852 IVVKSPSRDDHLAHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKA 911
++VKS DH+ HL+ FE + KY +K+NP KC FGV +G+FLG++V K+GIE N +
Sbjct: 654 MLVKSTRSADHIDHLKACFETLNKYNMKLNPAKCLFGVTSGEFLGYIVTKRGIEANPKQI 713
Query: 912 KAILDTSPPTSKKQLQSLLGKINFLRRFIANLSEKTKSFSPLLR 955
+AILD P +KK++Q L G+I L RFIA ++K+ F LLR
Sbjct: 714 RAILDLQSPRNKKEVQRLTGRIAGLNRFIARSTDKSLPFYQLLR 757
Score = 75.9 bits (185), Expect = 2e-13
Identities = 42/130 (32%), Positives = 73/130 (55%), Gaps = 1/130 (0%)
Query: 448 HLKPLFVWAKVDEKGVNKVLVDGGATINLMPKFMLKKLGKTEADLIPHDMVLSDYEGKTG 507
H PL + + E V ++L+D G+++N++ K +L+K+ + + P L+ ++G T
Sbjct: 344 HNDPLVIELHIGESEVTRILIDTGSSVNVVFKDVLQKMKVHDRHIKPSVRPLTGFDGNTM 403
Query: 508 SSLGAIMLNITVGTVARSTLFIVVPSKANYNLLLGREWIHGVGAVPSTLHQRISIWKLDG 567
+ G I L I +G A F+VV YN++LG WIH + A+PS+ HQ I I G
Sbjct: 404 MTNGTIKLPIYLGGAATWHKFVVVDKPTIYNIILGTPWIHDMQAIPSSYHQCIKIPTSIG 463
Query: 568 VVENVQADQS 577
+E ++ +Q+
Sbjct: 464 -IETIRGNQN 472
>At2g10780 pseudogene
Length = 1611
Score = 234 bits (597), Expect = 3e-61
Identities = 148/451 (32%), Positives = 244/451 (53%), Gaps = 19/451 (4%)
Query: 656 VKLLKEFKDCFAWDYDEMPGLSRDLVE-LKLPIKEDKKPVKQLPRRFHPDVLVKIKEEIE 714
+ ++ EF D FA + G+ D + + ++ P+ + P R P + K+K+++E
Sbjct: 625 IPIVNEFSDVFA----AVSGVPPDRSDPFTIELEPGTTPISKAPYRMAPAEMAKLKKQLE 680
Query: 715 RLLKCKFIRTARYVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPVAEMMVDST 774
LL FIR + W A V+ V KK+G R+CID+R LN T K++Y +P + ++D
Sbjct: 681 ELLDKGFIRPSSS-PWGAPVLFVKKKDGSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQL 739
Query: 775 AGHEYLSLLDGYSGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMN 834
G ++ S +D SGY+QI I DV KTAFR +E+VVMPFGL NA A + ++MN
Sbjct: 740 GGAQWFSKIDLASGYHQIPIEPTDVRKTAFRT--RYDHFEFVVMPFGLTNAPAAFMKMMN 797
Query: 835 TIFHDFIETFMQVYIDDIVVKSPSRDDHLAHLRKSFERMRKYGLKMNPLKCAFGVIAGDF 894
+F DF++ F+ ++I+DI+V S S + H HLR ER+R++ L KC+F + F
Sbjct: 798 GVFRDFLDEFVIIFINDILVYSKSWEAHQEHLRAVLERLREHELFAKLSKCSFWQRSVGF 857
Query: 895 LGFVVHKKGIEINKNKAKAILDTSPPTSKKQLQSLLGKINFLRRFIANLSEKTKSFSPLL 954
LG V+ +G+ ++ K ++I + P + +++S LG + RRF+ + + + PL
Sbjct: 858 LGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMSFASMAQ---PLT 914
Query: 955 RLK-KEDAFRWEAEHQKAFDELKVYLSSPHVMAPPIRGKPMKLYISATDGTIGSMLAQED 1013
RL K+ AF W E +K+F ELK L++ V+ P G+P +Y A+ +G +L Q
Sbjct: 915 RLTGKDTAFNWSDECEKSFLELKAMLTNAPVLVLPEEGEPYTVYTDASIVGLGCVLMQ-- 972
Query: 1014 EDSKERAIFYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHM 1073
K I Y SR L E Y + + F + Y+ V +++ + +K++
Sbjct: 973 ---KGSVIAYASRQLRKHEKNYPTHDLEMAAVVFFLKIWRSYLYGAKVQIYTDHKSLKYI 1029
Query: 1074 LSKPILHSRIGKWALALTEYSL--TYAPLKA 1102
++P L+ R +W + +Y+L Y P KA
Sbjct: 1030 FTQPELNLRQRRWMELVADYNLDIAYHPGKA 1060
Score = 70.5 bits (171), Expect = 8e-12
Identities = 76/325 (23%), Positives = 130/325 (39%), Gaps = 19/325 (5%)
Query: 1350 HDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYDKGCQDFQRHAGIQHVPASELHSI- 1408
H H KM L R +W + KD + C Q VP+ L ++
Sbjct: 1168 HQSKFSIHPGSNKMYRDLKRY-YHWVGMKKDVARWVAKCPTCQLVKAEHQVPSGLLQNLP 1226
Query: 1409 IKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVEAIPLQNVTQDTVI-DFIQNH 1467
I W + +D + + +H + V +D TK + + + +I + +
Sbjct: 1227 IPEWKWDHITMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDGAEIIAEKYIDE 1286
Query: 1468 IVYRFGLPESLTTDQGTVFVGQKVASFAESWGIKLLNSTPYYAQANGQVEAANKTLISLI 1527
IV G+P S+ +D+ T F + +F ++ G ++ ST Y+ Q + Q E +TL ++
Sbjct: 1287 IVRLHGIPVSIVSDRDTRFTSKFWKAFQKALGTRVNLSTAYHPQTDEQSERTIQTLEDML 1346
Query: 1528 KKHVGRKPKRWHQTLGQVLWAYRNSPKEATGATPFRLAYGQEAVLPAEVYLQSCRIQRQE 1587
+ V W + L V +AY NS + + G +P+ YG+ P C E
Sbjct: 1347 RACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYGRACRTPL------CWTPVGE 1400
Query: 1588 EIPSEDYWNMMLDELVNLDEERLSALDI-LTRQKDRVAKAYNKKVRAKSFMPGDYVWKVV 1646
+ +V+ ER+ L I L +DR NK+ + F GD V+
Sbjct: 1401 R-------RLFGPTIVDETTERMKFLKIKLKEAQDRQKSYANKRRKELEFQVGDLVYLKA 1453
Query: 1647 LPVD--KRDKRYGKWAPNWEGPFTV 1669
+ R K +P + GP+ V
Sbjct: 1454 MTYKGAGRFTSRKKLSPRYVGPYKV 1478
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 231 bits (590), Expect = 2e-60
Identities = 149/450 (33%), Positives = 244/450 (54%), Gaps = 17/450 (3%)
Query: 656 VKLLKEFKDCFAWDYDEMPGLSRDLVELKLPIKEDKKPVKQLPRRFHPDVLVKIKEEIER 715
+++++EF+D F +P D ++L + P+ + P R P + ++K++++
Sbjct: 457 IRVVQEFQDVFQ-SLQGLPPSQSDPFTIEL--EPGTAPLSKAPYRMAPAEMAELKKQLKD 513
Query: 716 LLKCKFIRTARYVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPVAEMMVDSTA 775
LL FIR + W A V+ V KK+G R+CID+R+LN T K+ Y +P + ++D
Sbjct: 514 LLGKGFIRPSTS-PWGAPVLFVKKKDGSFRLCIDYRELNRVTVKNRYPLPRIDELLDQLR 572
Query: 776 GHEYLSLLDGYSGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNT 835
G S +D SGY+QI IAE DV KTAFR G +E+VVMPFGL NA A + R+MN+
Sbjct: 573 GATCFSKIDLTSGYHQIPIAEADVRKTAFRT--RYGHFEFVVMPFGLTNAPAVFMRLMNS 630
Query: 836 IFHDFIETFMQVYIDDIVVKSPSRDDHLAHLRKSFERMRKYGLKMNPLKCAFGVIAGDFL 895
+F +F++ F+ ++IDDI+V S S ++ HLR+ E++R+ L KC+F FL
Sbjct: 631 VFQEFLDEFVIIFIDDILVYSKSPEEQEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFL 690
Query: 896 GFVVHKKGIEINKNKAKAILDTSPPTSKKQLQSLLGKINFLRRFIANLSEKTKSFSPLLR 955
G +V +G+ ++ K +AI D PT+ +++S LG + RRF+ + + P+ +
Sbjct: 691 GHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFLGWAGYYRRFVKGFASMAQ---PMTK 747
Query: 956 LKKEDA-FRWEAEHQKAFDELKVYLSSPHVMAPPIRGKPMKLYISATDGTIGSMLAQEDE 1014
L +D F W E ++ F LK L+S V+A P G+P +Y A+ +G +L Q
Sbjct: 748 LTGKDVPFVWSQECEEGFVSLKEMLTSTPVLALPEHGQPYMVYTDASRVGLGCVLMQHG- 806
Query: 1015 DSKERAIFYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHML 1074
+ I Y SR L E Y + + F+ + Y+ V VF+ + +K++
Sbjct: 807 ----KVIAYASRQLMKHEGNYPTHDLEMAAVIFALKIWRSYLYGGKVQVFTDHKSLKYIF 862
Query: 1075 SKPILHSRIGKWALALTEYSL--TYAPLKA 1102
++P L+ R +W + +Y L Y P KA
Sbjct: 863 TQPELNLRQRRWMELVADYDLEIAYHPGKA 892
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 230 bits (586), Expect = 6e-60
Identities = 152/460 (33%), Positives = 248/460 (53%), Gaps = 20/460 (4%)
Query: 656 VKLLKEFKDCFAWDYDEMPGLSRDLVELKLPIKEDKKPVKQLPRRFHPDVLVKIKEEIER 715
+++++EF+D F +P D ++L + P+ + P R P + ++K+++E
Sbjct: 483 IRVIQEFEDVFQ-SLQGLPPSRSDPFTIEL--EPGTAPLSKAPYRMAPAEMTELKKQLED 539
Query: 716 LLKCKFIRTARYVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPVAEMMVDSTA 775
LL FIR + W A V+ V KK+G R+CID+R LN T K++Y +P + ++D
Sbjct: 540 LLGKGFIRPSTS-PWGAPVLFVKKKDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLR 598
Query: 776 GHEYLSLLDGYSGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNT 835
G S +D SGY+QI IAE DV KTAFR G +E+VVMPF L NA A + R+MN+
Sbjct: 599 GATCFSKIDLTSGYHQIPIAEADVRKTAFRT--RYGHFEFVVMPFALTNAPAAFMRLMNS 656
Query: 836 IFHDFIETFMQVYIDDIVVKSPSRDDHLAHLRKSFERMRKYGLKMNPLKCAFGVIAGDFL 895
+F +F++ F+ ++IDDI+V S S ++H HLR+ E++R+ L KC+F FL
Sbjct: 657 VFQEFLDEFVIIFIDDILVYSKSPEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREIGFL 716
Query: 896 GFVVHKKGIEINKNKAKAILDTSPPTSKKQLQSLLGKINFLRRFIANLSEKTKSFSPLLR 955
G +V +G+ ++ K +AI D PT+ +++S L + RRF+ + + P+ +
Sbjct: 717 GHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFLRLTGYYRRFVKGFASMAQ---PMTK 773
Query: 956 LKKEDA-FRWEAEHQKAFDELKVYLSSPHVMAPPIRGKPMKLYISATDGTIGSMLAQEDE 1014
L +D F W E ++ F LK L+S V+A P G+P +Y A+ +G +L Q
Sbjct: 774 LTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALPEHGQPYMVYTDASRVGLGCVLMQ--- 830
Query: 1015 DSKERAIFYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHML 1074
+ + I Y SR L E Y + + F+ + Y+ V VF+ + +K++
Sbjct: 831 --RGKVIAYASRQLRKHEGNYPTHDLEMAVVIFALKIWRSYLYGGKVQVFTDHKSLKYIF 888
Query: 1075 SKPILHSRIGKWALALTEYSL--TYAPLKAVKGQAIADFL 1112
++P L+ R +W + +Y L Y P KA +AD L
Sbjct: 889 NQPELNLRQMRWMELVADYDLEIAYHPGKA---NVVADAL 925
Score = 62.4 bits (150), Expect = 2e-09
Identities = 57/219 (26%), Positives = 97/219 (44%), Gaps = 16/219 (7%)
Query: 1466 NHIVYRFGLPESLTTDQGTVFVGQKVASFAESWGIKLLNSTPYYAQANGQVEAANKTLIS 1525
+ IV G+P S+ +D+ + F +F G K+ ST Y+ Q +GQ E +TL
Sbjct: 1108 SEIVKLHGVPVSIVSDRDSKFTFAFWRAFQAKMGTKVQMSTAYHPQTDGQSERTIQTLED 1167
Query: 1526 LIKKHVGRKPKRWHQTLGQVLWAYRNSPKEATGATPFRLAYGQEAVLPAEVYLQSCRIQR 1585
+++ V W L V +AY NS + + G PF YG+ P L+ +++
Sbjct: 1168 MLRMCVLDWGGHWADHLSLVEFAYNNSYQASIGMAPFEALYGRPCWTP----LRWTQVE- 1222
Query: 1586 QEEIPSEDYWNMMLDELVNLDEERLSALDILTRQKDRVAKAY-NKKVRAKSFMPGD--YV 1642
+ I DY V ER+ L + ++ ++Y +K+ R F GD Y+
Sbjct: 1223 ERSIYGADY--------VQETTERIRVLKLNMKEAQARQRSYADKRRRELEFEVGDRVYL 1274
Query: 1643 WKVVLPVDKRDKRYGKWAPNWEGPFTVEKILLNNAYSIK 1681
+L R K +P + GPF + + + AY ++
Sbjct: 1275 KMAMLRGPNRSILETKLSPRYMGPFRIVERVGPVAYRLE 1313
>At4g03800
Length = 637
Score = 229 bits (583), Expect = 1e-59
Identities = 125/341 (36%), Positives = 201/341 (58%), Gaps = 25/341 (7%)
Query: 673 MPGLSRDLVELKLPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLKCKFIRTARYVDWLA 732
MP + ++ +L + KP+KQ R+ + + +I++LLK IR +Y DW+A
Sbjct: 321 MPDIDPSIICHELNVDPRFKPLKQKRRKLGVERAKAVNGKIDKLLKIGSIREVQYPDWVA 380
Query: 733 NVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPVAEMMVDSTAGHEYLSLLDGYSGYNQI 792
V V KKNGK RVCIDF DLN A PKD + +P + +V+STAG+E L+ +D + GYNQI
Sbjct: 381 ITVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHIDRLVESTAGNELLTFMDAFLGYNQI 440
Query: 793 FIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDI 852
+ ED KT+F + + GATYQ ++N +F++ + M+V IDD
Sbjct: 441 MMNPEDQEKTSF-----------------ITDRGATYQWLVNKMFNEHLRKTMEVSIDDT 483
Query: 853 VVKSPSRDDHLAHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAK 912
+VKS ++DH+ HL + FE + +Y +K+N KC FGV +G+FLG++V K+GIE N N+
Sbjct: 484 LVKSLKKEDHVKHLGECFEILNQYQMKLNLAKCTFGVPSGEFLGYIVTKRGIEANPNQIN 543
Query: 913 AILDTSPPTSKKQLQSLLGKINFLRRFIANLSEKTKSFSPLLRLKKEDAFRWEAEHQKAF 972
A L T + K++Q L G+I + ++K+ F + LK + F W+ + ++AF
Sbjct: 544 AFLKTPSLRNFKEVQRLTGRI------ASRSTDKSLPFYQI--LKGNNGFLWDEKCEEAF 595
Query: 973 DELKVYLSSPHVMAPPIRGKPMKLYISATDGTIGSMLAQED 1013
+LK YL++P V++ P + + LY+ ++ + +L +ED
Sbjct: 596 RQLKAYLTTPPVLSKPEADEKLYLYVFVSNHAVSGVLVRED 636
Score = 54.3 bits (129), Expect = 6e-07
Identities = 37/137 (27%), Positives = 63/137 (45%), Gaps = 20/137 (14%)
Query: 432 EDERAIFQRPTEQMKSHLKPLFVWAKVDEKGVNKVLVDGGATINLMPKFMLKKLGKTEAD 491
E+E +RP H L + V ++++LVD G++++L+
Sbjct: 189 EEETRHIERP------HDDALIITLDVANFKISRILVDTGSSVDLI-------------- 228
Query: 492 LIPHDMVLSDYEGKTGSSLGAIMLNITVGTVARSTLFIVVPSKANYNLLLGREWIHGVGA 551
+ L + ++ SLG I L + G +++ F+V A YN++LG WI + A
Sbjct: 229 FLGPPSPLVAFTSESAMSLGTIKLPVLAGKMSKIVDFVVFDKPATYNIILGTPWIFQMKA 288
Query: 552 VPSTLHQRISIWKLDGV 568
VPST HQ + +GV
Sbjct: 289 VPSTYHQWLKFPTSNGV 305
>At3g11970 hypothetical protein
Length = 1499
Score = 219 bits (558), Expect = 1e-56
Identities = 153/469 (32%), Positives = 244/469 (51%), Gaps = 20/469 (4%)
Query: 648 TTEWQNEMV--KLLKEFKDCFAWDYDEMPGLSRDLVELKLPIKEDKKPVKQLPRRFHPDV 705
T+E E V ++L E+ D F + +P R+ K+ + E PV Q P R+
Sbjct: 556 TSELGEESVVEEVLNEYPDIFI-EPTALPPF-REKHNHKIKLLEGSNPVNQRPYRYSIHQ 613
Query: 706 LVKIKEEIERLLKCKFIRTARYVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMP 765
+I + +E LL ++ A + + VV V KK+G R+C+D+R+LN T KD + +P
Sbjct: 614 KNEIDKLVEDLLTNGTVQ-ASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIP 672
Query: 766 VAEMMVDSTAGHEYLSLLDGYSGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNA 825
+ E ++D G S +D +GY+Q+ + +D+ KTAF+ G +E++VMPFGL NA
Sbjct: 673 LIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAFKTHS--GHFEYLVMPFGLTNA 730
Query: 826 GATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDHLAHLRKSFERMRKYGLKMNPLKC 885
AT+Q +MN IF F+ F+ V+ DDI+V S S ++H HL++ FE MR L KC
Sbjct: 731 PATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKC 790
Query: 886 AFGVIAGDFLGFVVHKKGIEINKNKAKAILDTSPPTSKKQLQSLLGKINFLRRFIANLSE 945
AF V ++LG + +GIE + K KA+ + PT+ KQL+ LG + RRF+ +
Sbjct: 791 AFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGV 850
Query: 946 KTKSFSPLLRLKKEDAFRWEAEHQKAFDELKVYLSSPHVMAPPIRGKPMKLYISATDGTI 1005
PL L K DAF W A Q+AF++LK L V++ P+ K + A I
Sbjct: 851 IA---GPLHALTKTDAFEWTAVAQQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGI 907
Query: 1006 GSMLAQEDEDSKERAIFYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFS 1065
G++L QE + Y+SR L + ++ EK L + F+ K ++Y+ ++ +
Sbjct: 908 GAVLMQEG-----HPLAYISRQLKGKQLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKT 962
Query: 1066 HYDIIKHMLSKPILHSRIGKWALALTE--YSLTYAPLKAVKGQAIADFL 1112
+K++L + + +W L E Y + Y + K +AD L
Sbjct: 963 DQRSLKYLLEQRLNTPIQQQWLPKLLEFDYEIQY---RQGKENVVADAL 1008
Score = 67.8 bits (164), Expect = 5e-11
Identities = 69/306 (22%), Positives = 133/306 (42%), Gaps = 27/306 (8%)
Query: 1373 YWPTIMKDCMEYDKGCQDFQRHAGIQHVPASELHSIIKPWPFRG--WA---LDLIGEINP 1427
YW ++KD Y + C Q+ + PA+ +++P P W+ +D I + P
Sbjct: 1111 YWKGMIKDIQAYIRSCGTCQQ---CKSDPAAS-PGLLQPLPIPDTIWSEVSMDFIEGL-P 1165
Query: 1428 CSSRQHKYIIVAIDYFTKWVEAIPLQN-VTQDTVIDFIQNHIVYRFGLPESLTTDQGTVF 1486
S + I+V +D +K I L + + TV +++ G P S+ +D+ VF
Sbjct: 1166 VSGGK-TVIMVVVDRLSKAAHFIALSHPYSALTVAHAYLDNVFKLHGCPTSIVSDRDVVF 1224
Query: 1487 VGQKVASFAESWGIKLLNSTPYYAQANGQVEAANKTLISLIKKHVGRKPKRWHQTLGQVL 1546
+ F G+ L ++ Y+ Q++GQ E N+ L + ++ +P+ W + L
Sbjct: 1225 TSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCMCHDRPQLWSKWLALAE 1284
Query: 1547 WAYRNSPKEATGATPFRLAYGQEAVLPAEVYLQSCRIQRQEEIPSEDYWNMMLDELVNLD 1606
+ Y + ++ TPF + YGQ V P + +P E ++ L +
Sbjct: 1285 YWYNTNYHSSSRMTPFEIVYGQ--VPPVHL----------PYLPGESKVAVVARSLQERE 1332
Query: 1607 EERLSALDILTRQKDRVAKAYNKKVRAKSFMPGDYVWKVVLPVDKRD---KRYGKWAPNW 1663
+ L L R + R+ + ++ + F GDYV+ + P ++ + K +P +
Sbjct: 1333 DMLLFLKFHLMRAQHRMKQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQKLSPKY 1392
Query: 1664 EGPFTV 1669
GP+ +
Sbjct: 1393 FGPYKI 1398
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 219 bits (558), Expect = 1e-56
Identities = 148/455 (32%), Positives = 239/455 (52%), Gaps = 32/455 (7%)
Query: 661 EFKDCFAWDYDEMPGLSRDLVELKLPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLKCK 720
EF+D F +P D ++L + P+ + P R P + ++K+++E LL
Sbjct: 449 EFEDVFQ-SLQGLPPSRSDPFTIEL--EPGTAPLSKAPYRMAPAEMAELKKQLEDLLG-- 503
Query: 721 FIRTARYVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPVAEMMVDSTAGHEYL 780
A V+ V KK+G R+CID+R LN T K++Y +P + ++D G
Sbjct: 504 -----------APVLFVKKKDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCF 552
Query: 781 SLLDGYSGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDF 840
S +D SGY+ I IAE DV KTAFR G +E+VVMPFGL NA A + R+MN++F +
Sbjct: 553 SKIDLTSGYHLIPIAEADVRKTAFRT--RYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEV 610
Query: 841 IETFMQVYIDDIVVKSPSRDDHLAHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVH 900
++ F+ ++IDDI+V S S ++H HLR+ E++R+ L KC+F FLG +V
Sbjct: 611 LDEFVIIFIDDILVYSKSLEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVS 670
Query: 901 KKGIEINKNKAKAILDTSPPTSKKQLQSLLGKINFLRRFIANLSEKTKSFSPLLRLKKED 960
+G+ ++ K +AI D PT+ +++S LG + RRF+ + + P+ +L +D
Sbjct: 671 AEGVSVDPEKIEAIRDWHTPTNATEIRSFLGLAGYYRRFVKGFASMAQ---PMTKLTGKD 727
Query: 961 A-FRWEAEHQKAFDELKVYLSSPHVMAPPIRGKPMKLYISATDGTIGSMLAQEDEDSKER 1019
F W E ++ F LK L+S V+A P G+P +Y A+ +G +L Q + +
Sbjct: 728 VPFVWSPECEEGFVSLKEMLTSTPVLALPEHGEPYMVYTDASGVGLGCVLMQ-----RGK 782
Query: 1020 AIFYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPIL 1079
I Y SR L E Y + + F+ + Y+ +V VF+ + +K++ ++P L
Sbjct: 783 VIAYASRQLRKHEGNYPTHDLEMAAVIFALKIWRSYLYGGNVQVFTDHKSLKYIFTQPEL 842
Query: 1080 HSRIGKWALALTEYSL--TYAPLKAVKGQAIADFL 1112
+ R +W + +Y L Y P KA +AD L
Sbjct: 843 NLRQRQWMELVADYDLEIAYHPGKA---NVVADAL 874
Score = 63.5 bits (153), Expect = 1e-09
Identities = 76/309 (24%), Positives = 120/309 (38%), Gaps = 22/309 (7%)
Query: 1379 KDCMEYDKGCQDFQRHAGIQHVPASELHSI-IKPWPFRGWALDLIGEINPCSSRQHKYII 1437
+D + C Q VP L S+ I W + +D + + SR I
Sbjct: 941 EDVANWVAECDVCQLVKAEHQVPGGMLQSLPIPEWKWDFITIDFV--VGLPVSRTKDAIW 998
Query: 1438 VAIDYFTKWVEAIPLQNVTQDTVI-DFIQNHIVYRFGLPESLTTDQGTVFVGQKVASFAE 1496
V +D TK + ++ V+ + IV G+P S+ +D+ + F +F
Sbjct: 999 VIVDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQA 1058
Query: 1497 SWGIKLLNSTPYYAQANGQVEAANKTLISLIKKHVGRKPKRWHQTLGQVLWAYRNSPKEA 1556
G K+ ST Y+ Q GQ E +TL +++ V W L V +AY NS +
Sbjct: 1059 EMGTKVQMSTAYHPQTYGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYPAS 1118
Query: 1557 TGATPFRLAYGQEAVLPAEVYLQSCRIQ-RQEEIPSEDYWNMMLDELVNLDEERLSALDI 1615
G PF Y + P C Q + I DY V ER+ L +
Sbjct: 1119 IGMAPFEALYERPCRTPL------CLTQVGERSIYGADY--------VQETTERIRVLKL 1164
Query: 1616 -LTRQKDRVAKAYNKKVRAKSFMPGD--YVWKVVLPVDKRDKRYGKWAPNWEGPFTVEKI 1672
+ +DR +K+ R F GD Y+ +L R K +P + GPF + +
Sbjct: 1165 NMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLSPRYMGPFRIVER 1224
Query: 1673 LLNNAYSIK 1681
+ AY ++
Sbjct: 1225 VGPVAYRLE 1233
>At1g36590 hypothetical protein
Length = 1499
Score = 219 bits (558), Expect = 1e-56
Identities = 153/469 (32%), Positives = 244/469 (51%), Gaps = 20/469 (4%)
Query: 648 TTEWQNEMV--KLLKEFKDCFAWDYDEMPGLSRDLVELKLPIKEDKKPVKQLPRRFHPDV 705
T+E E V ++L E+ D F + +P R+ K+ + E PV Q P R+
Sbjct: 556 TSELGEESVVEEVLNEYPDIFI-EPTALPPF-REKHNHKIKLLEGSNPVNQRPYRYSIHQ 613
Query: 706 LVKIKEEIERLLKCKFIRTARYVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMP 765
+I + +E LL ++ A + + VV V KK+G R+C+D+R+LN T KD + +P
Sbjct: 614 KNEIDKLVEDLLTNGTVQ-ASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIP 672
Query: 766 VAEMMVDSTAGHEYLSLLDGYSGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNA 825
+ E ++D G S +D +GY+Q+ + +D+ KTAF+ G +E++VMPFGL NA
Sbjct: 673 LIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAFKTHS--GHFEYLVMPFGLTNA 730
Query: 826 GATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDHLAHLRKSFERMRKYGLKMNPLKC 885
AT+Q +MN IF F+ F+ V+ DDI+V S S ++H HL++ FE MR L KC
Sbjct: 731 PATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKC 790
Query: 886 AFGVIAGDFLGFVVHKKGIEINKNKAKAILDTSPPTSKKQLQSLLGKINFLRRFIANLSE 945
AF V ++LG + +GIE + K KA+ + PT+ KQL+ LG + RRF+ +
Sbjct: 791 AFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGV 850
Query: 946 KTKSFSPLLRLKKEDAFRWEAEHQKAFDELKVYLSSPHVMAPPIRGKPMKLYISATDGTI 1005
PL L K DAF W A Q+AF++LK L V++ P+ K + A I
Sbjct: 851 IA---GPLHALTKTDAFEWTAVAQQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGI 907
Query: 1006 GSMLAQEDEDSKERAIFYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFS 1065
G++L QE + Y+SR L + ++ EK L + F+ K ++Y+ ++ +
Sbjct: 908 GAVLMQEG-----HPLAYISRQLKGKQLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKT 962
Query: 1066 HYDIIKHMLSKPILHSRIGKWALALTE--YSLTYAPLKAVKGQAIADFL 1112
+K++L + + +W L E Y + Y + K +AD L
Sbjct: 963 DQRSLKYLLEQRLNTPIQQQWLPKLLEFDYEIQY---RQGKENVVADAL 1008
Score = 66.2 bits (160), Expect = 2e-10
Identities = 74/329 (22%), Positives = 142/329 (42%), Gaps = 35/329 (10%)
Query: 1350 HDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYDKGCQDFQRHAGIQHVPASELHSII 1409
H G HQ ++K + + +GM +KD Y + C Q+ + PA+ ++
Sbjct: 1096 HSGRDVTHQ---RVKGLFYSKGM-----IKDIQAYIRSCGTCQQ---CKSDPAAS-PGLL 1143
Query: 1410 KPWPFRG--WA---LDLIGEINPCSSRQHKYIIVAIDYFTKWVEAIPLQN-VTQDTVIDF 1463
+P P W+ +D I + P S + I+V +D +K I L + + TV
Sbjct: 1144 QPLPIPDTIWSEVSMDFIEGL-PVSGGK-TVIMVVVDRLSKAAHFIALSHPYSALTVAQA 1201
Query: 1464 IQNHIVYRFGLPESLTTDQGTVFVGQKVASFAESWGIKLLNSTPYYAQANGQVEAANKTL 1523
+++ G P S+ +D+ VF + F G+ L ++ Y+ Q++GQ E N+ L
Sbjct: 1202 YLDNVFKLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCL 1261
Query: 1524 ISLIKKHVGRKPKRWHQTLGQVLWAYRNSPKEATGATPFRLAYGQEAVLPAEVYLQSCRI 1583
+ ++ +P+ W + L + Y + ++ TPF + YGQ V P +
Sbjct: 1262 ETYLRCMCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQ--VPPVHL------- 1312
Query: 1584 QRQEEIPSEDYWNMMLDELVNLDEERLSALDILTRQKDRVAKAYNKKVRAKSFMPGDYVW 1643
+P E ++ L ++ L L R + R+ + ++ + F GDYV+
Sbjct: 1313 ---PYLPGESKVAVVARSLQEREDMLLFLKFHLMRAQHRMKQFADQHRTEREFEIGDYVY 1369
Query: 1644 KVVLPVDKRD---KRYGKWAPNWEGPFTV 1669
+ P ++ + K +P + GP+ +
Sbjct: 1370 VKLQPYRQQSVVMRANQKLSPKYFGPYKI 1398
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 215 bits (547), Expect = 2e-55
Identities = 134/382 (35%), Positives = 211/382 (55%), Gaps = 17/382 (4%)
Query: 656 VKLLKEFKDCFAWDYDEMPGLSRDLVE-LKLPIKEDKKPVKQLPRRFHPDVLVKIKEEIE 714
+ ++ EF D FA + G+ D + + ++ P+ + P R P + ++K+++E
Sbjct: 120 ILIVNEFSDVFA----AVSGVPPDRSDPFTIELEPGTTPISKAPYRMAPAEMAELKKQLE 175
Query: 715 RLLKCKFIRTARYVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPVAEMMVDST 774
LL FIR + W A V+ V KK+G R+CID+R LN T K++Y +P + ++D
Sbjct: 176 ELLAKGFIRPSSS-PWGAPVLFVKKKDGSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQL 234
Query: 775 AGHEYLSLLDGYSGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMN 834
G ++ S +D SGY+QI I DV KTAFR G +E+VVMPFGL NA A + ++MN
Sbjct: 235 GGAQWFSKIDLASGYHQIPIEPTDVRKTAFRT--RYGHFEFVVMPFGLTNAPAAFMKMMN 292
Query: 835 TIFHDFIETFMQVYIDDIVVKSPSRDDHLAHLRKSFERMRKYGLKMNPLKCAFGVIAGDF 894
+F DF++ F+ ++IDDI+V S S + H HLR ER+R++ L K +F + F
Sbjct: 293 GVFRDFLDEFVIIFIDDILVHSKSWEAHQEHLRAVLERLREHELFAKLSKFSFWQRSVGF 352
Query: 895 LGFVVHKKGIEINKNKAKAILDTSPPTSKKQLQSLLGKINFLRRFIANLSEKTKSFSPLL 954
LG V+ +G+ ++ K ++I + P + +++S LG + RRF+ + + + PL
Sbjct: 353 LGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMSFASMAQ---PLT 409
Query: 955 RLK-KEDAFRWEAEHQKAFDELKVYLSSPHVMAPPIRGKPMKLYISATDGTIGSMLAQED 1013
RL K+ AF W E +K+F ELK L + V+ P G+P +Y A+ +G +L Q
Sbjct: 410 RLTGKDTAFNWSDECEKSFLELKAMLINAPVLVLPEEGEPYTVYTDASIVGLGCVLMQ-- 467
Query: 1014 EDSKERAIFYLSRVLNDAETRY 1035
K I Y SR L E Y
Sbjct: 468 ---KGSVIAYASRQLRKHEKNY 486
Score = 72.8 bits (177), Expect = 2e-12
Identities = 75/325 (23%), Positives = 131/325 (40%), Gaps = 19/325 (5%)
Query: 1350 HDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYDKGCQDFQRHAGIQHVPASELHSI- 1408
H H KM L R +W + KD + C Q VP+ L ++
Sbjct: 560 HQSKFSIHPGSNKMYRDLKRY-YHWVGMRKDVARWVAKCPTCQLVKAEHQVPSGLLQNLP 618
Query: 1409 IKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVEAIPLQNVTQDTVI-DFIQNH 1467
I W + +D + + +H + V +D TK + + + +I + +
Sbjct: 619 ISEWKWDHITMDFVTRLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDGAEIIAEKYIDE 678
Query: 1468 IVYRFGLPESLTTDQGTVFVGQKVASFAESWGIKLLNSTPYYAQANGQVEAANKTLISLI 1527
I+ G+P S+ +D+ T F + +F ++ G ++ ST Y+ Q +GQ E +TL ++
Sbjct: 679 IMRLHGIPVSIVSDRDTRFTSKFWNAFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDML 738
Query: 1528 KKHVGRKPKRWHQTLGQVLWAYRNSPKEATGATPFRLAYGQEAVLPAEVYLQSCRIQRQE 1587
+ V W + L + +AY NS + + G +P+ YG+ P C E
Sbjct: 739 RACVLDWGGNWEKYLRLIEFAYNNSFQASIGMSPYEALYGRACRTPL------CWTPVGE 792
Query: 1588 EIPSEDYWNMMLDELVNLDEERLSALDI-LTRQKDRVAKAYNKKVRAKSFMPGDYVWKVV 1646
+ +V+ ER+ L I L +DR NK+ + F GD V+
Sbjct: 793 R-------RLFGPTIVDETTERMKFLKIKLKEAQDRQKSYANKRRKELEFQVGDLVYLKA 845
Query: 1647 LPVD--KRDKRYGKWAPNWEGPFTV 1669
+ R K +P + GP+ V
Sbjct: 846 MTYKGAGRFTSRKKLSPRYVGPYKV 870
>At1g35370 hypothetical protein
Length = 1447
Score = 204 bits (519), Expect = 4e-52
Identities = 136/436 (31%), Positives = 225/436 (51%), Gaps = 21/436 (4%)
Query: 658 LLKEFKDCFAWDYDEMPGLSRDLVELKLPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLL 717
+++EF D FA D P R+ + K+ + E PV Q P R+ +I + ++ ++
Sbjct: 547 IVEEFPDVFAEPTDLPP--FREKHDHKIKLLEGANPVNQRPYRYVVHQKDEIDKIVQDMI 604
Query: 718 KCKFIRTARYVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPVAEMMVDSTAGH 777
K I+ + + + VV V KK+G R+C+D+ +LN T KD + +P+ E ++D G
Sbjct: 605 KSGTIQVSSS-PFASPVVLVKKKDGTWRLCVDYTELNGMTVKDRFLIPLIEDLMDELGGS 663
Query: 778 EYLSLLDGYSGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIF 837
S +D +GY+Q+ + +D+ KTAF+ G +E++VM FGL NA AT+Q +MN++F
Sbjct: 664 VVFSKIDLRAGYHQVRMDPDDIQKTAFKTHN--GHFEYLVMLFGLTNAPATFQSLMNSVF 721
Query: 838 HDFIETFMQVYIDDIVVKSPSRDDHLAHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGF 897
DF+ F+ V+ DDI++ S S ++H HLR FE MR + L F + + LG
Sbjct: 722 RDFLRKFVLVFFDDILIYSSSIEEHKEHLRLVFEVMRLHKL--------FAKGSKEHLGH 773
Query: 898 VVHKKGIEINKNKAKAILDTSPPTSKKQLQSLLGKINFLRRFIANLSEKTKSFSPLLRLK 957
+ + IE + K +A+ + PT+ KQ++ LG + RRF+ N PL L
Sbjct: 774 FISAREIETDPAKIQAVKEWPTPTTVKQVRGFLGFAGYYRRFVRNFGVIA---GPLHALT 830
Query: 958 KEDAFRWEAEHQKAFDELKVYLSSPHVMAPPIRGKPMKLYISATDGTIGSMLAQEDEDSK 1017
K D F W E Q AFD LK L + V+A P+ K + A I ++L Q K
Sbjct: 831 KTDGFCWSLEAQSAFDTLKAVLCNAPVLALPVFDKQFMVETDACGQGIRAVLMQ-----K 885
Query: 1018 ERAIFYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKP 1077
+ Y+SR L + ++ EK L F+ K ++Y+ P ++ + +K++L +
Sbjct: 886 GHPLAYISRQLKGKQLHLSIYEKELLAFIFAVRKWRHYLLPSHFIIKTDQRSLKYLLEQR 945
Query: 1078 ILHSRIGKWALALTEY 1093
+ +W L E+
Sbjct: 946 LNTPVQQQWLPKLLEF 961
Score = 63.9 bits (154), Expect = 8e-10
Identities = 62/304 (20%), Positives = 127/304 (41%), Gaps = 23/304 (7%)
Query: 1373 YWPTIMKDCMEYDKGCQDFQRHAGIQHVPASELHSIIKPWPFRGW---ALDLIGEINPCS 1429
YW ++KD + + C Q+ L + P P + W ++D I + +
Sbjct: 1082 YWKGMVKDIQAFIRSCGTCQQCKSDNAAYPGLLQPL--PIPDKIWCDVSMDFIEGLP--N 1137
Query: 1430 SRQHKYIIVAIDYFTKWVEAIPLQN-VTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVG 1488
S I+V +D +K + L + + TV +++ G P S+ +D+ +F
Sbjct: 1138 SGGKSVIMVVVDRLSKAAHFVALAHPYSALTVAQAFLDNVYKHHGCPTSIVSDRDVLFTS 1197
Query: 1489 QKVASFAESWGIKLLNSTPYYAQANGQVEAANKTLISLIKKHVGRKPKRWHQTLGQVLWA 1548
F + G++L S+ Y+ Q++GQ E N+ L + ++ +P W++ L +
Sbjct: 1198 DFWKEFFKLQGVELRMSSAYHPQSDGQTEVVNRCLENYLRCMCHARPHLWNKWLPLAEYW 1257
Query: 1549 YRNSPKEATGATPFRLAYGQEAVLPAEVYLQSCRIQRQEEIPSEDYWNMMLDELVNLDEE 1608
Y + ++ TPF L YGQ + +P + ++ L +
Sbjct: 1258 YNTNYHSSSQMTPFELVYGQAPPI------------HLPYLPGKSKVAVVARSLQERENM 1305
Query: 1609 RLSALDILTRQKDRVAKAYNKKVRAKSFMPGDYVWKVVLPVDKRD---KRYGKWAPNWEG 1665
L L R + R+ + ++ ++F GD+V+ + P ++ + K +P + G
Sbjct: 1306 LLFLKFHLMRAQHRMKQFADQHRTERTFDIGDFVYVKLQPYRQQSVVLRVNQKLSPKYFG 1365
Query: 1666 PFTV 1669
P+ +
Sbjct: 1366 PYKI 1369
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 199 bits (505), Expect = 2e-50
Identities = 182/751 (24%), Positives = 311/751 (41%), Gaps = 138/751 (18%)
Query: 464 NKVLVDGGATINLMPKFMLKKLGKTEADLIPHDMVLSDYEGKTGSSLGAIMLNITVGTVA 523
+K L D GA+++LMP + KKLG + ++L+D + L L + +G V
Sbjct: 614 SKCLCDLGASVSLMPLPVAKKLGFNKYKPCNISLILADRSVRISHGL-LEDLPVMIGVVE 672
Query: 524 RSTLFIV--VPSKANYNLLLGREWIHGVGAVPSTLHQRISIWKLDGVVENVQADQSYYLA 581
T F+V + + L+LGR ++ A+ +I + N+ D
Sbjct: 673 VPTDFVVLEMDEEPKDPLILGRPFLARARAIIDVKKGKIDL--------NLGRDLKMTFD 724
Query: 582 ETGYVGKKNFEKSLATIAPLDTVANQYFNPYSEY-----------------------SVM 618
T + K E ++ I +D +A++ E +
Sbjct: 725 ITNTMKKPTIEGNIFWIEEMDMLADKMLEELGETDHLQSALTKDSKEGDLHLETLGGEYI 784
Query: 619 LDPIRGLNLNEAYKPDAMSGWHNDEEKDATTEWQN------------------------- 653
LD I + Y ++ + E + +W
Sbjct: 785 LDHITRPTAHSVYSTTSLDHNSSSEANLVSDDWSEIKAPKVDLKPLPKGLRYVFLGLNST 844
Query: 654 -----------EMVKLL----KEFKDCFAWDYDEMPGLSRDLVELKLPIKEDKKPVKQLP 698
+ V LL +++ + D++ G+S L ++ ++ + +
Sbjct: 845 YPVIVNDELTADQVNLLITELMKYRKAIGYSLDDIKGISPTLCTHRIHLENESYSSIEPQ 904
Query: 699 RRFHPDVLVKIKEEIERLLKCKFIRTARYVDWLANVVPVIKKNGKM-------------- 744
RR +P++ +K+EI +LL I W++ V V KK G
Sbjct: 905 RRLNPNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHCVPKKGGMTVVKNSKDELIPTRT 964
Query: 745 ----RVCIDFRDLNAATPKDEYHMPVAEMMVDSTAGHEYLSLLDGYSGYNQIFIAEEDVS 800
R+CI++R LN A+ K+ + +P + M++ A H Y LD YSG+ QI I D
Sbjct: 965 ITGHRMCIEYRKLNVASRKEHFPLPFIDHMLERLANHPYYCFLDSYSGFFQIPIHPNDQG 1024
Query: 801 KTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRD 860
KT F CP GT+ + MPFGL NA AT+QR M +IF D IE ++V++DD V S
Sbjct: 1025 KTTFTCP--YGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVYGSSFS 1082
Query: 861 DHLAHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDTSPP 920
L +L + +R + L +N KC F V G LG + ++GIE++K K ++ PP
Sbjct: 1083 SCLLNLCRVLKRCEETNLVLNWEKCHFMVREGIVLGRKISEEGIEVDKAKIDVMMQLQPP 1142
Query: 921 TSKKQLQSLLGKINFLRRFIANLSEKTKSFSPLLRLK-KEDAFRWEAEHQKAFDELKVYL 979
+ K ++S LG F R FI + S+ + PL RL KE F ++ E AF +K L
Sbjct: 1143 KTVKDIRSFLGHAGFYRIFIKDFSKLAR---PLTRLLCKETEFAFDDECLTAFKLIKEAL 1199
Query: 980 SSPHVMAPPIRGKPMKLYISATDGTIGSMLAQEDEDSKERAIFYLSRVLNDAETRYTMIE 1039
+ ++ P P ++ ++DA+ RY E
Sbjct: 1200 ITAPIVQAPNWDFPFEII-----------------------------TMDDAQVRYATTE 1230
Query: 1040 KLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHSRIGKWALALTEYSLTYAP 1099
K L + F+ K + Y+ V +++ + ++H+ +K R+ +W L L E+ +
Sbjct: 1231 KELLAVVFAFEKFRSYLVGSKVTIYTDHAALRHIYAKKDTKPRLLRWILLLQEFDMEIVD 1290
Query: 1100 LKAVKGQAIAD----------FLVDHTLPKE 1120
K ++ +AD L+D ++P+E
Sbjct: 1291 KKGIE-NGVADHLSRMRIEDEVLIDDSMPEE 1320
Score = 160 bits (406), Expect = 5e-39
Identities = 109/366 (29%), Positives = 168/366 (45%), Gaps = 38/366 (10%)
Query: 1336 CLSEDDAFIAISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYDKGCQDFQRHA 1395
C+SED+ + H G H A K + + G +WP++ KD E+ C F+
Sbjct: 1386 CVSEDEIEGILLHCHGFAYGGHFATFKTMSKILQAGFWWPSMFKDAQEFISKCDSFEN-- 1443
Query: 1396 GIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVEAIPLQNV 1455
F W +D +G SS +KYI+VAIDY +KWVEAI
Sbjct: 1444 ------------------FDVWGIDFMGPFP--SSYGNKYILVAIDYVSKWVEAIASHTN 1483
Query: 1456 TQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKVASFAESWGIKLLNSTPYYAQANGQ 1515
V+ + I RFG+P + +D G F+ + + + G+K + +GQ
Sbjct: 1484 DARVVLKLFKTIIFPRFGVPRIVISDGGKHFINKGFENLLKKHGVK--------HKTSGQ 1535
Query: 1516 VEAANKTLISLIKKHVGRKPKRWHQTLGQVLWAYRNSPKEATGATPFRLAYGQEAVLPAE 1575
VE +N+ + ++++K VG K W L LWAYR + K G TPF L YG+ LP E
Sbjct: 1536 VEISNREIKAILEKTVGSTRKDWSAKLNDTLWAYRTAFKTPIGTTPFNLLYGKSCHLPVE 1595
Query: 1576 VYLQSCRIQRQEEIPSEDYWNMMLDELVNLDEERLSALDILTRQKDRVAKAYNKKVRAKS 1635
+ ++ + + L +L +L++ RL A + K+R ++KK+ ++
Sbjct: 1596 LEYKAMWAVKLLNFDIKTAEEKRLIQLNDLNKIRLEAYESSKIYKERTKSFHDKKIVSRD 1655
Query: 1636 FMPGDYVWKVVLPVDKRDKRY-GKWAPNWEGPFTVEKILLNNAYSIKELGGRNRQMTVNG 1694
F GD VL + R + + GK W GPF+V + Y L G+N TVNG
Sbjct: 1656 FKVGDQ----VLLFNSRLRLFPGKLKSRWSGPFSVTAV---RPYGAITLAGKNGDFTVNG 1708
Query: 1695 KYLKTY 1700
+ LK Y
Sbjct: 1709 QRLKKY 1714
Score = 36.2 bits (82), Expect = 0.17
Identities = 21/87 (24%), Positives = 42/87 (48%), Gaps = 5/87 (5%)
Query: 1 MKYFPSSLTKNAFTWFTTLAPRSVHTWAQLERIFHEQFFRGECKVSLK-DLASVKRKPAE 59
++ FP SL A W +L S+ +W ++ F +FF L+ D++ + E
Sbjct: 88 LRLFPFSLGDKAHQWEKSLPQGSITSWNDCKKAFLAKFFSNSRTARLRNDISGFTQTNNE 147
Query: 60 SIDDYLNRFRMLKSRCFTHVPEHELVV 86
+ + RF+ +++C P HE+++
Sbjct: 148 TFYEAWERFKGYQTQC----PHHEMLL 170
>At2g13230 pseudogene
Length = 353
Score = 189 bits (479), Expect = 2e-47
Identities = 112/349 (32%), Positives = 185/349 (52%), Gaps = 29/349 (8%)
Query: 1229 KFDEARLSHVSRVDNQEANELAQIASGYMVDKCRL-------KELIEVKEKLNLSDLNIL 1281
+F E +L+ + R +N A+ LA +AS ++ R+ K IE+ + + + ++
Sbjct: 14 QFHEFKLTRIPRGENTSADALAALASTSDLNMRRVIPVEFIEKPSIELDKVEHAFPIQVV 73
Query: 1282 V-----IDNM----APNDWRKPIVDYLQNPVGTTD----RKTKYRAMSYVIMGNELFKKN 1328
DN ++W +PI +++ D RK K A +V++ +LFK
Sbjct: 74 AGYEDASDNSPSLGCDSEWIEPIRNFISKRKVRLDKWEARKLKAEAARFVLVEEKLFKWR 133
Query: 1329 VDGTLLKCLSEDDAFIAISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYDKGC 1388
+ G L+ C+ ++ A + VH G CG + G + + R G +WPT++KDC
Sbjct: 134 LSGPLMTCVEKEAARKVMKEVHGGSCGNNSGGRALAIKIKRLGYFWPTMIKDC------- 186
Query: 1389 QDFQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVE 1448
+ QR+A H+PA L SI P+P W++D+IG ++ +S+Q K ++V DYF+K E
Sbjct: 187 EKCQRNAPTIHLPAELLSSIASPYPLMRWSMDIIGPMH--ASKQKKLVLVLTDYFSKLRE 244
Query: 1449 AIPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKVASFAESWGIKLLNSTPY 1508
A ++ + V F+ HI+ R G+P + TD G+ F+ + F + WGI+L STP
Sbjct: 245 AESYASIKEAQVESFMWKHILCRHGVPYEIVTDNGSQFISTRFQGFCDKWGIRLSKSTPR 304
Query: 1509 YAQANGQVEAANKTLISLIKKHVGRKPKRWHQTLGQVLWAYRNSPKEAT 1557
Y Q NGQ +AANKT++ +KK + K W L VLW++R +P+ AT
Sbjct: 305 YLQGNGQADAANKTILDGLKKRLTAKKGSWVDELEGVLWSHRTTPRRAT 353
>At2g06890 putative retroelement integrase
Length = 1215
Score = 185 bits (469), Expect = 2e-46
Identities = 193/755 (25%), Positives = 316/755 (41%), Gaps = 146/755 (19%)
Query: 410 YYDEEEPDDNPLCYYVMQKGSVEDERAIFQRPTEQMKSHLKP-----------LFVWAKV 458
+Y E P N ++ G +E E I P+ ++ P L V K
Sbjct: 167 HYANECP--NKRVMILLDNGEIEPEEEIPDSPSSLKENEELPAQGELLVARRTLSVQTKT 224
Query: 459 DEKGVNK---------------VLVDGGATINLMPKFMLKKLGKTEADLIPHDMVLSDYE 503
DE+ K +++DGG+ N+ + M+KKLG L+D
Sbjct: 225 DEQEQRKNLFHTRCHVHGKVCSLIIDGGSCTNVASETMVKKLGLKW---------LND-S 274
Query: 504 GKTGSSLGAIMLNITVGTVARSTLFIVVPSKANYNLLLGREWIHGVGAVPSTLHQRISIW 563
GK +++ I +G L V+P +A + +LLGR W
Sbjct: 275 GKMRVK-NQVVVPIVIGKYEDEILCDVLPMEAGH-ILLGRPW------------------ 314
Query: 564 KLDGVVENVQADQSYYLAETGYVGKKNFEKS-----LATIAPLDTVANQYFNPYSEYSVM 618
Q+D+ + G+ + +FE L ++ P + +Q + V+
Sbjct: 315 ---------QSDRK--VMHDGFTNRHSFEFKGGKTILVSMTPHEVYQDQIHLKQKKEQVV 363
Query: 619 LDPIRGLNLNEAYKPDAMSGWHNDEE-------KDATTEWQN-------EMVKLLKEFKD 664
P N K + ++ ++ K+A T N EM LL+++KD
Sbjct: 364 KQP------NFFAKSGEVKSAYSSKQPMLLFVFKEALTSLTNFAPVLPSEMTSLLQDYKD 417
Query: 665 CFAWDYDE----MPGLSRDLVELKLPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLKCK 720
F D + + G+ + D P LP R +E++R
Sbjct: 418 VFPEDNPKGLPPIRGIEHQI---------DFVPGASLPNRPAYRTNPVETKELQR----- 463
Query: 721 FIRTARYVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPVAEMMVDSTAGHEYL 780
+K+G R+C D R +N T K + +P + M+D G
Sbjct: 464 ------------------QKDGSWRMCFDCRAINNVTVKYCHPIPRLDDMLDELHGSSIF 505
Query: 781 SLLDGYSGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDF 840
S +D SGY+QI + E D KTAF+ G YEW+VMPFGL +A +T+ R+MN + F
Sbjct: 506 SKIDLKSGYHQIRMNEGDEWKTAFKTKH--GLYEWLVMPFGLTHAPSTFMRLMNHVLRAF 563
Query: 841 IETFMQVYIDDIVVKSPSRDDHLAHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVH 900
I F+ VY DDI+V S S +H+ HL +RK L N KC F FLGFVV
Sbjct: 564 IGIFVIVYFDDILVYSESLREHIEHLDSVLNVLRKEELYANLKKCTFCTDNLVFLGFVVS 623
Query: 901 KKGIEINKNKAKAILDTSPPTSKKQLQSLLGKINFLRRFIANLSEKTKSFSPLLRLKKED 960
G+++++ K KAI D P + +++S G F RRF + S + + +KK+
Sbjct: 624 ADGVKVDEEKVKAIRDWPSPKTVGEVRSFHGLAGFYRRFFKDFSTIVAPLTEV--MKKDV 681
Query: 961 AFRWEAEHQKAFDELKVYLSSPHVMAPPIRGKPMKLYISATDGTIGSMLAQEDEDSKERA 1020
F+WE ++AF LK L++ V+ K ++ A+ IG++L Q+ ++
Sbjct: 682 GFKWEKAQEEAFQSLKDKLTNAPVLILSEFLKTFEIECDASGIGIGAVLMQD-----QKL 736
Query: 1021 IFYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILH 1080
I + S L A Y +K L + + ++Y+ P ++ + ++ +KH+ + L+
Sbjct: 737 IAFFSEKLGGATLNYPTYDKELYALVRALQRWQHYLWPKVFVIHTDHESLKHLKGQQKLN 796
Query: 1081 SRIGKWALALTEYSLTYA---PLKAVKGQAIADFL 1112
R +W E+ T+A K K +AD L
Sbjct: 797 KRHARW----VEFIETFAYVIKYKKGKDNVVADAL 827
Score = 35.0 bits (79), Expect = 0.38
Identities = 33/130 (25%), Positives = 54/130 (41%), Gaps = 27/130 (20%)
Query: 1560 TPFRLAYGQEAVLPAEVYLQSCRIQRQEEIPSEDYWNMMLDELVNLDEERLSALDILTRQ 1619
+PF++ YG + P ++ IP + L E V++D ++ + L +
Sbjct: 988 SPFQIVYGFNPISPFDL------------IP------LPLSERVSIDGKKKAELVQQIHE 1029
Query: 1620 KDR---------VAKAYNKKVRAKSFMPGDYVWKVVLPVDKRDKRYGKWAPNWEGPFTVE 1670
R AK NK R + F GD VW + +R K P +GPF +
Sbjct: 1030 NARRNIEEKTKLYAKQANKGRREQIFEVGDMVWIHLRKERFPAQRKSKLMPRIDGPFKII 1089
Query: 1671 KILLNNAYSI 1680
K + +NAY +
Sbjct: 1090 KRINDNAYQL 1099
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,602,477
Number of Sequences: 26719
Number of extensions: 1878970
Number of successful extensions: 5869
Number of sequences better than 10.0: 173
Number of HSP's better than 10.0 without gapping: 94
Number of HSP's successfully gapped in prelim test: 80
Number of HSP's that attempted gapping in prelim test: 5381
Number of HSP's gapped (non-prelim): 366
length of query: 1710
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1597
effective length of database: 8,299,349
effective search space: 13254060353
effective search space used: 13254060353
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0026.5


