

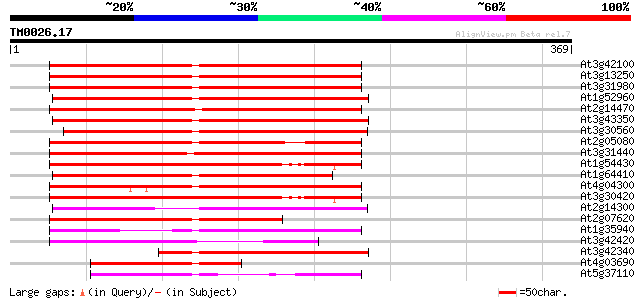
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0026.17
(369 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g42100 putative protein 218 5e-57
At3g13250 hypothetical protein 213 1e-55
At3g31980 hypothetical protein 210 9e-55
At1g52960 hypothetical protein 204 6e-53
At2g14470 pseudogene 203 1e-52
At3g43350 putative protein 202 2e-52
At3g30560 hypothetical protein 202 2e-52
At2g05080 putative helicase 196 2e-50
At3g31440 hypothetical protein 191 5e-49
At1g54430 hypothetical protein 187 6e-48
At1g64410 unknown protein 187 1e-47
At4g04300 hypothetical protein 181 7e-46
At3g30420 hypothetical protein 178 4e-45
At2g14300 pseudogene; similar to MURA transposase of maize Muta... 171 6e-43
At2g07620 putative helicase 169 3e-42
At1g35940 hypothetical protein 157 1e-38
At3g42420 putative protein 126 2e-29
At3g42340 putative protein 119 3e-27
At4g03690 hypothetical protein 107 1e-23
At5g37110 putative helicase 104 9e-23
>At3g42100 putative protein
Length = 1752
Score = 218 bits (554), Expect = 5e-57
Identities = 116/206 (56%), Positives = 150/206 (72%), Gaps = 5/206 (2%)
Query: 27 TFSAAIRSMGMIVLNVASSGIASLLLPGGRTAHSRFCIPLQTDKTATCNIKQDSLRENLL 86
T +AA+RS G IVLNVASSGIASLLL GGRTAHSRF IPL D+ + C I S NL+
Sbjct: 1311 TLAAAVRSRGQIVLNVASSGIASLLLEGGRTAHSRFAIPLNPDEFSVCKITPKSDLANLI 1370
Query: 87 ICAKFIIWDEAPMLNKNCFEALNRTLCDIMRQEDESNMDKPFGGKVVVLGGDFRQILPVI 146
A IIWDEAPM++K CFE+L+++ DI+ +D +K FGGKVVV GGDFRQ+LPVI
Sbjct: 1371 KEASLIIWDEAPMMSKFCFESLDKSFYDILNNKD----NKVFGGKVVVFGGDFRQVLPVI 1426
Query: 147 PKGGRQDIVSATVNSSDLWKYCKVLKLTKNMRLCTTG-SPELATEIKEFAD*ILKIGDGD 205
GR +IV +++N+S LW +CKVLKLTKNMRL + G S E A EI++F+D +L +GDG
Sbjct: 1427 NGAGRVEIVMSSLNASYLWDHCKVLKLTKNMRLLSGGLSSEEAKEIQQFSDWLLAVGDGR 1486
Query: 206 FESNERGESDIEIPKDLLIHDSENPL 231
GE+ I+IP++LLI ++ NP+
Sbjct: 1487 INEPNDGEALIDIPEELLIKEAGNPI 1512
>At3g13250 hypothetical protein
Length = 1419
Score = 213 bits (542), Expect = 1e-55
Identities = 113/206 (54%), Positives = 149/206 (71%), Gaps = 5/206 (2%)
Query: 27 TFSAAIRSMGMIVLNVASSGIASLLLPGGRTAHSRFCIPLQTDKTATCNIKQDSLRENLL 86
T +AA+RS G I LNVASSGIASLLL GGRTAHSRF IPL D+ + C IK S +L+
Sbjct: 979 TLAAAVRSKGQICLNVASSGIASLLLEGGRTAHSRFSIPLNPDEFSVCKIKPKSDLADLI 1038
Query: 87 ICAKFIIWDEAPMLNKNCFEALNRTLCDIMRQEDESNMDKPFGGKVVVLGGDFRQILPVI 146
A IIWDEAPM++K CFEAL+++ DI+++ D +K FGGKV+V GGDFRQ+LPVI
Sbjct: 1039 KEASLIIWDEAPMMSKFCFEALDKSFSDIIKRVD----NKVFGGKVMVFGGDFRQVLPVI 1094
Query: 147 PKGGRQDIVSATVNSSDLWKYCKVLKLTKNMRLCTTG-SPELATEIKEFAD*ILKIGDGD 205
GR +IV +++N+S LW +CKVL+LTKNMRL S + A EI+EF+D +L +GDG
Sbjct: 1095 NGAGRAEIVMSSLNASYLWDHCKVLRLTKNMRLLNNDLSVDEAKEIQEFSDWLLAVGDGR 1154
Query: 206 FESNERGESDIEIPKDLLIHDSENPL 231
GE I+IP++LLI +++NP+
Sbjct: 1155 VNEPNDGEVIIDIPEELLIQEADNPI 1180
>At3g31980 hypothetical protein
Length = 1099
Score = 210 bits (535), Expect = 9e-55
Identities = 114/206 (55%), Positives = 146/206 (70%), Gaps = 5/206 (2%)
Query: 27 TFSAAIRSMGMIVLNVASSGIASLLLPGGRTAHSRFCIPLQTDKTATCNIKQDSLRENLL 86
T SAA+R G+I +NVASSGIASLLL GGRTAHSRF IP+ D TC+I +S N+L
Sbjct: 657 TLSAAVRMRGLISVNVASSGIASLLLQGGRTAHSRFGIPINPDDFTTCHIVPNSDLANML 716
Query: 87 ICAKFIIWDEAPMLNKNCFEALNRTLCDIMRQEDESNMDKPFGGKVVVLGGDFRQILPVI 146
A IIWDEAPM+++ CFE+L+R+L D++ D KPFGGKVVV GGDFRQ+LPVI
Sbjct: 717 KEASLIIWDEAPMMSRYCFESLDRSLNDVIGNID----GKPFGGKVVVFGGDFRQVLPVI 772
Query: 147 PKGGRQDIVSATVNSSDLWKYCKVLKLTKNMRLCTTG-SPELATEIKEFAD*ILKIGDGD 205
GR +IV A +NSS LW++CKVL LTKNMRL + + A EIKEF++ +L +GDG
Sbjct: 773 HGAGRAEIVLAALNSSYLWEHCKVLTLTKNMRLMSNDLDKDEAEEIKEFSNWLLAVGDGR 832
Query: 206 FESNERGESDIEIPKDLLIHDSENPL 231
GE I+IP++LLI D+ +P+
Sbjct: 833 VSEPNDGEVLIDIPEELLIKDANDPI 858
>At1g52960 hypothetical protein
Length = 924
Score = 204 bits (519), Expect = 6e-53
Identities = 109/208 (52%), Positives = 144/208 (68%), Gaps = 4/208 (1%)
Query: 29 SAAIRSMGMIVLNVASSGIASLLLPGGRTAHSRFCIPLQTDKTATCNIKQDSLRENLLIC 88
SAAIRS G I LNVASSGIA+LLL GGRT HSRF IP+ ++++TCNI + S L+
Sbjct: 513 SAAIRSKGDISLNVASSGIAALLLDGGRTTHSRFGIPINPNESSTCNISRGSDLGELVKE 572
Query: 89 AKFIIWDEAPMLNKNCFEALNRTLCDIMRQEDESNMDKPFGGKVVVLGGDFRQILPVIPK 148
A IIWDE PM++K+CFE+L+RTL DIM DKPFGGK +V GGDFRQ+LPVI
Sbjct: 573 ANLIIWDETPMMSKHCFESLDRTLRDIMNNPG----DKPFGGKGIVFGGDFRQVLPVING 628
Query: 149 GGRQDIVSATVNSSDLWKYCKVLKLTKNMRLCTTGSPELATEIKEFAD*ILKIGDGDFES 208
GR++IV A +NSS +W++CKVL+LTKNMRL S +I+ F+ IL +GDG
Sbjct: 629 AGREEIVFAALNSSYIWEHCKVLELTKNMRLLANISEHEKRDIEYFSKWILDVGDGKISQ 688
Query: 209 NERGESDIEIPKDLLIHDSENPLLDLID 236
G + I+IP++ LI+ +P+ +I+
Sbjct: 689 PNDGIALIDIPEEFLINGDNDPVESIIE 716
>At2g14470 pseudogene
Length = 1265
Score = 203 bits (516), Expect = 1e-52
Identities = 110/206 (53%), Positives = 141/206 (68%), Gaps = 5/206 (2%)
Query: 27 TFSAAIRSMGMIVLNVASSGIASLLLPGGRTAHSRFCIPLQTDKTATCNIKQDSLRENLL 86
T SA IR IVLNVASSGIASLLL GGRTAHSRF IPL D+ + C IK S NL+
Sbjct: 841 TLSATIRYRDQIVLNVASSGIASLLLEGGRTAHSRFGIPLNPDEFSVCKIKPKSDLANLV 900
Query: 87 ICAKFIIWDEAPMLNKNCFEALNRTLCDIMRQEDESNMDKPFGGKVVVLGGDFRQILPVI 146
A +IWDEAPM+++ CFEAL+++ DI++ D + FGGKVVV GGDFRQ+ PVI
Sbjct: 901 KKASLVIWDEAPMMSRFCFEALDKSFSDIIKNTDNT----VFGGKVVVFGGDFRQVFPVI 956
Query: 147 PKGGRQDIVSATVNSSDLWKYCKVLKLTKNMRLCTTGSPEL-ATEIKEFAD*ILKIGDGD 205
GR +IV +++N+S LW CKVLKLTKN RL E A EI+EF+D +L +GDG
Sbjct: 957 NGAGRAEIVMSSLNASYLWDNCKVLKLTKNTRLLANNLSETEAKEIQEFSDWLLAVGDGR 1016
Query: 206 FESNERGESDIEIPKDLLIHDSENPL 231
+ G + I+IP+DLLI +++ P+
Sbjct: 1017 INESNDGVAIIDIPEDLLITNADKPI 1042
>At3g43350 putative protein
Length = 830
Score = 202 bits (515), Expect = 2e-52
Identities = 108/208 (51%), Positives = 146/208 (69%), Gaps = 4/208 (1%)
Query: 29 SAAIRSMGMIVLNVASSGIASLLLPGGRTAHSRFCIPLQTDKTATCNIKQDSLRENLLIC 88
SAA+RS G I LNVASSGIA+L L GGRTAHSRF IP+ ++++TCNI + S L+
Sbjct: 217 SAAVRSKGDISLNVASSGIAALRLDGGRTAHSRFDIPINPNESSTCNISRGSDLGELVKE 276
Query: 89 AKFIIWDEAPMLNKNCFEALNRTLCDIMRQEDESNMDKPFGGKVVVLGGDFRQILPVIPK 148
AK IIWDEAPM++K+CFE+L+RTL DI+ DKP GGKV+V GGDFRQ+LPVI
Sbjct: 277 AKLIIWDEAPMMSKHCFESLDRTLKDIVNNPG----DKPLGGKVIVFGGDFRQVLPVING 332
Query: 149 GGRQDIVSATVNSSDLWKYCKVLKLTKNMRLCTTGSPELATEIKEFAD*ILKIGDGDFES 208
GR++IV A +NSS +W++ KVL+LTKNMRL S +I++F+ IL +GDG
Sbjct: 333 AGREEIVFAALNSSYIWEHSKVLELTKNMRLLADISEHEKRDIEDFSKWILDVGDGKISQ 392
Query: 209 NERGESDIEIPKDLLIHDSENPLLDLID 236
G + I+IP++ LI+ +P+ +I+
Sbjct: 393 PNDGIALIDIPEEFLINGDNDPVESIIE 420
>At3g30560 hypothetical protein
Length = 1473
Score = 202 bits (515), Expect = 2e-52
Identities = 106/200 (53%), Positives = 140/200 (70%), Gaps = 4/200 (2%)
Query: 36 GMIVLNVASSGIASLLLPGGRTAHSRFCIPLQTDKTATCNIKQDSLRENLLICAKFIIWD 95
G I LNVASSGIASLLL GGRTAHSRF IPL +T+TCN+++ S L+ AK IIWD
Sbjct: 1045 GDICLNVASSGIASLLLEGGRTAHSRFGIPLTPHETSTCNMERGSDLAELVTAAKLIIWD 1104
Query: 96 EAPMLNKNCFEALNRTLCDIMRQEDESNMDKPFGGKVVVLGGDFRQILPVIPKGGRQDIV 155
EAPM++K CFE+L+++L DI+ + D PFGGK+++ GGDFRQILPVI GR+ IV
Sbjct: 1105 EAPMMSKYCFESLDKSLKDILSTPE----DMPFGGKLIIFGGDFRQILPVILAAGRELIV 1160
Query: 156 SATVNSSDLWKYCKVLKLTKNMRLCTTGSPELATEIKEFAD*ILKIGDGDFESNERGESD 215
+++NSS LW+YCKV KLTKNMRL A EI++F+ IL +G+G G +
Sbjct: 1161 KSSLNSSHLWQYCKVFKLTKNMRLLQDIDINEAREIEDFSKWILAVGEGKLNQPNDGVTQ 1220
Query: 216 IEIPKDLLIHDSENPLLDLI 235
I+I D+LI + +NP+ +I
Sbjct: 1221 IQIRDDILIPEGDNPIESII 1240
>At2g05080 putative helicase
Length = 1219
Score = 196 bits (497), Expect = 2e-50
Identities = 111/205 (54%), Positives = 137/205 (66%), Gaps = 17/205 (8%)
Query: 27 TFSAAIRSMGMIVLNVASSGIASLLLPGGRTAHSRFCIPLQTDKTATCNIKQDSLRENLL 86
T SAA+RS G IVLNVASSGIASLLL GGRTAHSR IPL ++ TCN+K S R NL+
Sbjct: 786 TLSAALRSKGDIVLNVASSGIASLLLEGGRTAHSRSGIPLNPNEFTTCNMKAGSDRANLV 845
Query: 87 ICAKFIIWDEAPMLNKNCFEALNRTLCDIMRQEDESNMDKPFGGKVVVLGGDFRQILPVI 146
A IIWDEAPM++++CFE+L+R+L DI D +KPFGGKVVV GGDFRQ+LPVI
Sbjct: 846 KEASLIIWDEAPMMSRHCFESLDRSLSDICGNCD----NKPFGGKVVVFGGDFRQVLPVI 901
Query: 147 PKGGRQDIVSATVNSSDLWKYCKVLKLTKNMRLCTTGSPELATEIKEFAD*ILKIGDGDF 206
P DIV A +NSS LW +CKVL LTKNM L + + IL +GDG
Sbjct: 902 PGADTADIVMAALNSSYLWSHCKVLTLTKNMCLFS-------------EEWILAVGDGRI 948
Query: 207 ESNERGESDIEIPKDLLIHDSENPL 231
GE+ I+IP + LI +++P+
Sbjct: 949 GEPNDGEALIDIPSEFLITKAKDPI 973
>At3g31440 hypothetical protein
Length = 536
Score = 191 bits (485), Expect = 5e-49
Identities = 107/206 (51%), Positives = 139/206 (66%), Gaps = 5/206 (2%)
Query: 27 TFSAAIRSMGMIVLNVASSGIASLLLPGGRTAHSRFCIPLQTDKTATCNIKQDSLRENLL 86
T AAIR IVLN+ASSGIASLLL GGRTAHSRF I L D+ + C IK S NL+
Sbjct: 119 TLFAAIRRRDQIVLNMASSGIASLLLEGGRTAHSRFGIRLNPDEFSVCKIKPKSDLANLV 178
Query: 87 ICAKFIIWDEAPMLNKNCFEALNRTLCDIMRQEDESNMDKPFGGKVVVLGGDFRQILPVI 146
A +I D+APM+++ CFEAL+++ DI++ + +K FGGKVVV GDFRQ+LPVI
Sbjct: 179 KEASLVICDKAPMMSRFCFEALDKSFSDIIK----NTYNKVFGGKVVVFSGDFRQVLPVI 234
Query: 147 PKGGRQDIVSATVNSSDLWKYCKVLKLTKNMRLCTTGSPEL-ATEIKEFAD*ILKIGDGD 205
GR +IV +++N+S LW +CKVLKLTKNMRL E A EI EF+D +L +GDG
Sbjct: 235 NGAGRAEIVMSSLNASYLWDHCKVLKLTKNMRLLANNLSETEAKEIHEFSDWLLAVGDGR 294
Query: 206 FESNERGESDIEIPKDLLIHDSENPL 231
+ I+IPKDLLI +++ P+
Sbjct: 295 INEPNDDVAIIDIPKDLLITNADKPI 320
>At1g54430 hypothetical protein
Length = 1639
Score = 187 bits (476), Expect = 6e-48
Identities = 108/211 (51%), Positives = 142/211 (67%), Gaps = 14/211 (6%)
Query: 27 TFSAAIRSMGMIVLNVASSGIASLLLPGGRTAHSRFCIPLQTDKTATCNIKQDSLRENLL 86
T +A+RS G V+ VASS IA+LLLPGGRTAHSRF IP+ + + C+IK S+ N+L
Sbjct: 1190 TIISALRSNGKNVMPVASSAIAALLLPGGRTAHSRFKIPINVHEDSICDIKIGSMLANVL 1249
Query: 87 ICAKFIIWDEAPMLNKNCFEALNRTLCDIMRQEDESNMDKPFGGKVVVLGGDFRQILPVI 146
IIWDEAPM +++ FEA++RTL DI+ DE + K FGGK V+LGGDFRQILPVI
Sbjct: 1250 SKVDLIIWDEAPMAHRHTFEAVDRTLRDILSVGDEKALTKTFGGKTVLLGGDFRQILPVI 1309
Query: 147 PKGGRQDIVSATVNSSDLWKYCKVLKLTKNMRLCTTGSPELATEIKEFAD*ILKIGDGDF 206
P+G RQ+ VSA +N S LW+ C L++NMR+ PE EIK FA+ IL++GDG+
Sbjct: 1310 PQGTRQETVSAAINRSYLWESCHKYLLSQNMRV----QPE---EIK-FAEWILQVGDGEA 1361
Query: 207 ESNERG------ESDIEIPKDLLIHDSENPL 231
G E +I I K+LL+ ++ENPL
Sbjct: 1362 PRKTHGIDDDQEEDNIIIDKNLLLPETENPL 1392
>At1g64410 unknown protein
Length = 753
Score = 187 bits (474), Expect = 1e-47
Identities = 102/184 (55%), Positives = 128/184 (69%), Gaps = 4/184 (2%)
Query: 29 SAAIRSMGMIVLNVASSGIASLLLPGGRTAHSRFCIPLQTDKTATCNIKQDSLRENLLIC 88
SAAIRS G I LNVASSGIA+LLL GGRTAHSRF IP+ ++++TCNI + L+
Sbjct: 563 SAAIRSKGDISLNVASSGIAALLLDGGRTAHSRFGIPINPNESSTCNISRGLDFGELVKE 622
Query: 89 AKFIIWDEAPMLNKNCFEALNRTLCDIMRQEDESNMDKPFGGKVVVLGGDFRQILPVIPK 148
A IIWDEA M++K+CFE+L+RTL DIM DKPFGGKV+V GGDFRQ+L VI
Sbjct: 623 ANLIIWDEAHMMSKHCFESLDRTLRDIMNNPG----DKPFGGKVIVFGGDFRQVLSVING 678
Query: 149 GGRQDIVSATVNSSDLWKYCKVLKLTKNMRLCTTGSPELATEIKEFAD*ILKIGDGDFES 208
GR++IV A +NSS +W++CKVL+LTKNMRL S +I+ F+ IL +GDG
Sbjct: 679 AGREEIVFAALNSSYIWEHCKVLELTKNMRLLANISEHEKRDIEYFSKWILDVGDGKISQ 738
Query: 209 NERG 212
G
Sbjct: 739 PNDG 742
>At4g04300 hypothetical protein
Length = 286
Score = 181 bits (458), Expect = 7e-46
Identities = 105/221 (47%), Positives = 141/221 (63%), Gaps = 20/221 (9%)
Query: 27 TFSAAIRSMGMIVLNVASSGIASLLLPGGRTAHSRFCIPLQTDKTATCNIKQ-----DSL 81
T +A RS G LN+ASSGIASLLL GGR AH RF IPL D+ + C IK D +
Sbjct: 42 TLAAVGRSKGQTCLNIASSGIASLLLEGGRIAHYRFSIPLNPDEFSVCKIKPKSDLADLI 101
Query: 82 RENLLIC----------AKFIIWDEAPMLNKNCFEALNRTLCDIMRQEDESNMDKPFGGK 131
+E LI A IIWD+APM +K CFEAL+++ DI+++ D +K F GK
Sbjct: 102 KEASLIIWDKLVDLIKKASLIIWDKAPMKSKFCFEALDKSFSDIIKRVD----NKVFCGK 157
Query: 132 VVVLGGDFRQILPVIPKGGRQDIVSATVNSSDLWKYCKVLKLTKNMRLCTTG-SPELATE 190
V+V GGDFRQ+LPVI GR + V +++N+ +W +CKVLKLTKNMRL S + A E
Sbjct: 158 VMVFGGDFRQVLPVINGAGRAETVMSSLNAVYIWDHCKVLKLTKNMRLLNNDLSVDEAKE 217
Query: 191 IKEFAD*ILKIGDGDFESNERGESDIEIPKDLLIHDSENPL 231
I+EF D +L +GDG GE+ I+IP++LLI +++ P+
Sbjct: 218 IQEFFDWLLVVGDGRVNEPNDGEALIDIPEELLIQEADIPI 258
>At3g30420 hypothetical protein
Length = 837
Score = 178 bits (452), Expect = 4e-45
Identities = 106/211 (50%), Positives = 138/211 (65%), Gaps = 14/211 (6%)
Query: 27 TFSAAIRSMGMIVLNVASSGIASLLLPGGRTAHSRFCIPLQTDKTATCNIKQDSLRENLL 86
T +A+RS G V+ VASS IA+LLLPGGRTAHS F IP+ + C+IK S+ N+L
Sbjct: 388 TIISALRSNGKNVMPVASSAIAALLLPGGRTAHSWFKIPINVHEDFICDIKIGSMLANVL 447
Query: 87 ICAKFIIWDEAPMLNKNCFEALNRTLCDIMRQEDESNMDKPFGGKVVVLGGDFRQILPVI 146
IIWDEAPM +++ FEA++RTL DI+ DE + K GGK V+LGGDFRQILPVI
Sbjct: 448 SKVDLIIWDEAPMAHRHTFEAVDRTLRDILSVGDEKALTKTLGGKTVLLGGDFRQILPVI 507
Query: 147 PKGGRQDIVSATVNSSDLWKYCKVLKLTKNMRLCTTGSPELATEIKEFAD*ILKIGDGDF 206
P+ RQ+ VSA +N S LW+ C L++NMR+ PE EIK FA+ IL+IGDG+
Sbjct: 508 PQRTRQETVSAAINRSYLWESCHKYLLSQNMRV----QPE---EIK-FAEWILQIGDGEA 559
Query: 207 ESNERG------ESDIEIPKDLLIHDSENPL 231
G E +I I K+LL+ ++ENPL
Sbjct: 560 PRKTHGIDDDQEEDNIIIDKNLLLPETENPL 590
>At2g14300 pseudogene; similar to MURA transposase of maize Mutator
transposon
Length = 1230
Score = 171 bits (433), Expect = 6e-43
Identities = 99/207 (47%), Positives = 124/207 (59%), Gaps = 28/207 (13%)
Query: 29 SAAIRSMGMIVLNVASSGIASLLLPGGRTAHSRFCIPLQTDKTATCNIKQDSLRENLLIC 88
SAAIR G LNVASS IASLLL GGRTAHSRF IPL +T+TCN+++ S L+
Sbjct: 861 SAAIRCKGDTCLNVASSSIASLLLEGGRTAHSRFGIPLTPHETSTCNMERGSDLAELVTA 920
Query: 89 AKFIIWDEAPMLNKNCFEALNRTLCDIMRQEDESNMDKPFGGKVVVLGGDFRQILPVIPK 148
AK IIWDE D PFG KV++ GGDFRQIL VIP
Sbjct: 921 AKLIIWDE----------------------------DMPFGRKVILFGGDFRQILHVIPA 952
Query: 149 GGRQDIVSATVNSSDLWKYCKVLKLTKNMRLCTTGSPELATEIKEFAD*ILKIGDGDFES 208
GR+ IV +++NSS LW++CKVLKLTKNMRL A EI++F IL +G+G
Sbjct: 953 AGRELIVKSSLNSSYLWQHCKVLKLTKNMRLLQDIDINEAREIEDFLKWILTVGEGKLNE 1012
Query: 209 NERGESDIEIPKDLLIHDSENPLLDLI 235
G + I+IP D+LI + +NP+ +I
Sbjct: 1013 PSDGVTHIQIPDDILIPEGDNPIESII 1039
>At2g07620 putative helicase
Length = 1241
Score = 169 bits (427), Expect = 3e-42
Identities = 90/153 (58%), Positives = 110/153 (71%), Gaps = 4/153 (2%)
Query: 27 TFSAAIRSMGMIVLNVASSGIASLLLPGGRTAHSRFCIPLQTDKTATCNIKQDSLRENLL 86
T SAA+R G+I +NVASSGIA LLL GGRTAHSRF IP+ D TC+I +S N+L
Sbjct: 886 TLSAAVRMKGLISVNVASSGIAFLLLQGGRTAHSRFGIPINPDDFTTCHIVPNSDLANML 945
Query: 87 ICAKFIIWDEAPMLNKNCFEALNRTLCDIMRQEDESNMDKPFGGKVVVLGGDFRQILPVI 146
A IIWDEAPM+++ CFE+L+R+L D++ D KPFGGKVVV GGDFRQ+L VI
Sbjct: 946 KEASLIIWDEAPMMSRYCFESLDRSLNDVIGNVD----GKPFGGKVVVFGGDFRQVLHVI 1001
Query: 147 PKGGRQDIVSATVNSSDLWKYCKVLKLTKNMRL 179
GR +IV A +NSS LW++C VL LTKNM L
Sbjct: 1002 HGAGRAEIVLAALNSSYLWEHCNVLTLTKNMSL 1034
>At1g35940 hypothetical protein
Length = 1678
Score = 157 bits (396), Expect = 1e-38
Identities = 95/206 (46%), Positives = 123/206 (59%), Gaps = 38/206 (18%)
Query: 27 TFSAAIRSMGMIVLNVASSGIASLLLPGGRTAHSRFCIPLQTDKTATCNIKQDSLRENLL 86
T SAAIR G IVLNVASSGIASLLL GGRTAHSRF IPL D+ +
Sbjct: 1270 TLSAAIRCRGQIVLNVASSGIASLLLEGGRTAHSRFGIPLNHDEFSV------------- 1316
Query: 87 ICAKFIIWDEAPMLNKNCFEALNRTLCDIMRQEDESNMDKPFGGKVVVLGGDFRQILPVI 146
+L+++ DI++ + +K FGGKVVV GGDFRQ+LPVI
Sbjct: 1317 --------------------SLDKSFSDIIKNTN----NKVFGGKVVVFGGDFRQVLPVI 1352
Query: 147 PKGGRQDIVSATVNSSDLWKYCKVLKLTKNMRLCTTG-SPELATEIKEFAD*ILKIGDGD 205
GR +IV +++N+S LW +CKVLKLTKNMRL S A EI+EF+D +L + DG
Sbjct: 1353 NGAGRAEIVMSSLNASYLWDHCKVLKLTKNMRLLANNLSATEAKEIQEFSDWLLAVSDGR 1412
Query: 206 FESNERGESDIEIPKDLLIHDSENPL 231
G + I+IP+DLLI +++ P+
Sbjct: 1413 INEPNDGVATIDIPEDLLITNADKPI 1438
>At3g42420 putative protein
Length = 1018
Score = 126 bits (316), Expect = 2e-29
Identities = 79/178 (44%), Positives = 100/178 (55%), Gaps = 44/178 (24%)
Query: 27 TFSAAIRSMGMIVLNVASSGIASLLLPGGRTAHSRFCIPLQTDKTATCNIKQDSLRENLL 86
T SAAIRS G IVLNVAS+GIASLLL GGRTAHSRF IPL D+ +C IK S NL+
Sbjct: 704 TLSAAIRSRGDIVLNVASNGIASLLLEGGRTAHSRFSIPLTPDEYTSCRIKPKSDLANLI 763
Query: 87 ICAKFIIWDEAPMLNKNCFEALNRTLCDIMRQEDESNMDKPFGGKVVVLGGDFRQILPVI 146
A IIWDEAP+++K CFE+L+++ DI+ +D +
Sbjct: 764 RKASLIIWDEAPVMSKWCFESLDKSFSDIIGNKDNKD----------------------- 800
Query: 147 PKGGRQDIVSATVNSSDLWKYCKVLKLTKNMRLCTTG-SPELATEIKEFAD*ILKIGD 203
+CKVLKLTKNMRL + S E A +IKEF+D +L +G+
Sbjct: 801 --------------------HCKVLKLTKNMRLLSKDLSEEEAKDIKEFSDWLLAVGN 838
>At3g42340 putative protein
Length = 244
Score = 119 bits (297), Expect = 3e-27
Identities = 61/138 (44%), Positives = 90/138 (65%), Gaps = 4/138 (2%)
Query: 99 MLNKNCFEALNRTLCDIMRQEDESNMDKPFGGKVVVLGGDFRQILPVIPKGGRQDIVSAT 158
M++K+CFE+L+RTL D M + DKPFGGKV+V GGDFRQ+L VI GR++IV A
Sbjct: 1 MMSKHCFESLDRTLRDFMNNLE----DKPFGGKVIVFGGDFRQVLLVINGAGREEIVFAA 56
Query: 159 VNSSDLWKYCKVLKLTKNMRLCTTGSPELATEIKEFAD*ILKIGDGDFESNERGESDIEI 218
+NSS +W++CKV +LTK M+L S +I++F+ IL +GDG G S I+I
Sbjct: 57 LNSSYIWEHCKVFELTKKMKLLANISEHEKRDIEDFSKWILDVGDGKISQCNEGISLIDI 116
Query: 219 PKDLLIHDSENPLLDLID 236
++ I+ +P+ +I+
Sbjct: 117 REEFFINGDNDPVESIIE 134
>At4g03690 hypothetical protein
Length = 570
Score = 107 bits (266), Expect = 1e-23
Identities = 54/99 (54%), Positives = 70/99 (70%), Gaps = 4/99 (4%)
Query: 54 GGRTAHSRFCIPLQTDKTATCNIKQDSLRENLLICAKFIIWDEAPMLNKNCFEALNRTLC 113
GGRTAHSRF IPL +T+TCNI++ L+ AK IIWDEAPM++K CF++L++ L
Sbjct: 258 GGRTAHSRFGIPLTPHETSTCNIERGIDLAELVTAAKLIIWDEAPMMSKYCFKSLDKRLR 317
Query: 114 DIMRQEDESNMDKPFGGKVVVLGGDFRQILPVIPKGGRQ 152
DI+ + DKP GGKV++ GGDFRQIL VI GR+
Sbjct: 318 DIISTPE----DKPLGGKVILFGGDFRQILHVIVAAGRE 352
>At5g37110 putative helicase
Length = 1307
Score = 104 bits (259), Expect = 9e-23
Identities = 68/178 (38%), Positives = 92/178 (51%), Gaps = 49/178 (27%)
Query: 54 GGRTAHSRFCIPLQTDKTATCNIKQDSLRENLLICAKFIIWDEAPMLNKNCFEALNRTLC 113
GGRTAHSRF IPL ++ TCN+K S R NL+ A IIWDEAPM+++ CFE+L+R+L
Sbjct: 950 GGRTAHSRFGIPLNPNEFTTCNMKVGSDRANLVKEASLIIWDEAPMMSRYCFESLDRSLS 1009
Query: 114 DIMRQEDESNMDKPFGGKVVVLGGDFRQILPVIPKGGRQDIVSATVNSSDLWKYCKVLKL 173
DI D +KPFGGKVVV GG L +
Sbjct: 1010 DICGNGD----NKPFGGKVVVFGG---------------------------------LSV 1032
Query: 174 TKNMRLCTTGSPELATEIKEFAD*ILKIGDGDFESNERGESDIEIPKDLLIHDSENPL 231
++ A +IKEF++ IL +GDG GE+ I IP + LI +++P+
Sbjct: 1033 SE------------AKDIKEFSEWILAVGDGRIVEPNDGEALIVIPSEFLITKAKDPI 1078
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.342 0.152 0.494
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,546,593
Number of Sequences: 26719
Number of extensions: 317181
Number of successful extensions: 1066
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 28
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1000
Number of HSP's gapped (non-prelim): 43
length of query: 369
length of database: 11,318,596
effective HSP length: 101
effective length of query: 268
effective length of database: 8,619,977
effective search space: 2310153836
effective search space used: 2310153836
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0026.17


